Abstract
Considering the limitations of liver biopsy, reliable non-invasive serum biomarkers of liver fibrosis are required for early diagnosis. The present study analyzed the expression profile of circulating micro (mi)RNAs during the development and progression of hepatic fibrosis in patients with chronic hepatitis B virus (HBV) infection, aiming to identify novel earlier diagnostic biomarkers. Fresh plasma samples were collected from 50 patients diagnosed with chronic HBV infection and hepatic fibrosis. These patients were classified into five groups (S0, S1, S2, S3 and S4; n=10 per group) based on Scheuer's staging criteria. The differential expression of the circulating miRNAs was determined by performing miRNA microarray hybridization. Finally, the target genes of the miRNAs were predicted and classified using gene ontology analysis. A total of 140 miRNAs were detected in the S1–S4 patient groups, and their expression levels were >2-fold higher compared with those in the S0 group. The numbers of miRNAs differentially expressed in the S1–S4 patient groups were 48, 97, 84 and 56, respectively, with 12 miRNAs differentially expressed at all stages, 10 of which were upregulated and two of which were downregulated. The target genes of the miRNAs identified were found to be involved in 100 signal transduction pathways, the majority of which affected hepatic fibrosis via the TGF-/Smad, Wnt, MAPK, Jak/STAT and VEGF pathways. The differential expression levels of miRNAs were closely associated with the staging of hepatic fibrosis. The results of the present study provide evidence to facilitate the development and application of non-invasive biomarkers for earlier diagnosis of hepatic fibrosis.
Keywords: hepatic fibrosis, chronic hepatitis B virus infection, microRNAs, expression profile
Introduction
Chronic hepatitis B virus (HBV) infection is one of the most life threatening diseases affecting humans, which may lead to hepatic cirrhosis, hepatocellular carcinoma (HCC) or even mortality. It is estimated that of the 6,000,000,000 individuals worldwide, 2,000,000,000 are infected with HBV and 350,000,000 individuals suffer from chronic HBV infection. The mortality rate of HBV resulting from hepatic failure, cirrhosis or primary HCC is 1,000,000/year (1). The mortality rate of hepatic cirrhosis in China is estimated to be 9.5/100,000. Therefore, earlier diagnosis and sustained follow up of the progression of hepatic fibrosis is key for the effective improvement of the prognosis of those with chronic HBV infection.
MicroRNA (miRNA) is endogenous small single-stranded non-coding RNA of between 18 and 24 nucleotides in length. miRNA controls the expression levels of target genes at the post-transcriptional level by degrading target mRNA or suppressing mRNA translation following its specific binding to target mRNA (2). It has been demonstrated that miRNA is also involved in the development of fibrosis of the lung and kidney (3,4). It has been reported that variable expression levels of miRNA are observed in patients with chronic hepatic injury, with changes in the relevant expression levels of miRNA accelerating the development and progression of fibrosis (4,5). The present study aimed to examine the plasma miRNA biomarkers associated with fibrosis in patients with chronic HBV infection using Agilent miRNA microarray scanning.
Patients and methods
Clinical cohort establishment
A total of 50 patients were recruited in the clinical cohort between October 2012 and October 2013 through multi-center cooperation (Shanghai First People's Hospital and Shanghai Ruijin Hospital). The cohort comprised male and female (17:8) patients with a mean age of 41 years. The patient inclusion criteria was a diagnosis of chronic HBV infection or a sustained positive result in hepatitis Be antigen (HBsAg) and/or HBV DNA tests subsequent to a six month medical history of positive HBsAg. Diagnosis was made according to the Guideline of Prevention and Treatment of Chronic HBV Infection (6). The exclusion criteria included: HIV or hepatitis C infection, alcohol assumption of >30 g/day, hepatic injury due to metabolism or other causes, history of treatment for hepatic diseases or a liver biopsy not suitable for analysis. Fasting venous blood was collected from each patient in the morning 1 week prior to liver biopsy. Whole blood and biochemical assessment, HBV quantification and HBV DNA quantification were also performed, and the plasma samples were stored at −80°C. All patients with chronic HBV infection underwent a liver biopsy followed by pathological analysis, and the degree of hepatic fibrosis was determined according to Scheuer's Criteria (S0, no liver fibrosis; S1, expansion of portal fibrotic area; S2, fibrosis observed in and around portal area, may have formed fibrous septum; S3, clear fibrosis and hepatic lobule structure disorder but no cirrhosis; S4, early cirrhosis) (7). The present study was approved by the ethics committee of The First People's Hospital Affiliated to Shanghai Jiao-Tong University (Shanghai, China). Written informed consent was provided by the patients prior to the collection of samples.
Collection and storage of plasma samples
A total of 2 ml peripheral blood was collected from each patient and was transferred into EDTA anticoagulation tubes. Following collection, the following procedure was performed either within 1 h at room temperature or 2 h in a 4°C environment: The samples were centrifuged at 820 × g for 10 min, following which 1 ml supernatant was collected and transferred into 1.5 ml centrifuge tubes. The samples were centrifuged again at 16,000 × g for 10 min and the supernatant was carefully collected and transferred into centrifuge tubes, which were then stored at −80°C.
RNA extraction of plasma
RNA extraction was performed using an mirVana PARIS kit (Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's instructions. The quality of the extracted RNA was subsequently confirmed using electrophoresis and was deemed suitable for use if 2100 RNA integrity number 6.0 and 28S/18S≥0.7. An Agilent miRNA microarray kit (Agilent Technologies, Inc.) was then used for fluorescent labeling of the miRNA from the suitable RNA samples.
Microarray hybridization and scanning
The miRNA conjugated with the fluorescent label were applied to the microarray following probe denaturation with dimethyl sulfoxide. The microarray was incubated in a hybridization oven at 55°C with 20 rpm agitation for 20 h. Following hybridization, the microarray was washed and scanned using an Agilent Microarray Scanner (Agilent Technologies, Inc.).
Analysis of miRNA microarray data
Screening of the different miRNAs was performed by applying the digital gene expression differential method, described by Audic et al (8) to the analysis. The genes defined as differentially expressed were those with a false discovery rate (FDR) ≤0.05 and fold change ≥2. Based on the expression status in the different groups, those miRNAs with expression levels exhibiting linear correlation with grouping were screened.
Cluster analysis
MeV software, version 4.7.4 (http://www.tm4.org/mev.html) was applied to the cluster analysis of the different miRNAs. Strata cluster, a method of non-monitoring cluster analysis, was used to achieve the clustering of the different miRNAs and groups of samples, enabling the detection of the differential effects of miRNA in the different groups.
Trend analysis
Log standardization of different miRNAs was performed to facilitate trend analysis. The mean value of miRNA expression was calculated in the S0–S4 groups. The base 1 logarithm of the mean value from S1–S4 - the logarithm of the mean value of S0 was applied to a plot graph, which demonstrated the trend of miRNA change in the different groups.
Gene ontology (GO) analysis
In order to detect the target genes of those miRNAs with different expression levels and to evaluate their function, GO annotation was applied to the target gene, according to the GO database (http://www.geneontology.org/). The GO values involved with these genes were determined and the Fisher exact test and χ2 test were used to determine the significance level and error rate of each GO value to enable screening of the significant GO terms reflected by the target gene. P<0.01 was considered to indicate a statistically significant difference.
Pathway analysis
In order to determine the cell pathways affected by the target gene with differential miRNA expression, a Fisher exact test and χ2 square test were used to determine the significance of the pathways, which the target gene is involved in, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. P<0.05 was considered to indicate a statistically significant difference.
Statistical analysis
Feature Extraction software 10.7 (Agilent Technologies, Inc.) was used to determine the expression data from the microarray, based on the plasma sample. Data normalization was performed using Quantile (9). The data are expressed as the mean ± standard deviation. Statistical analysis was performed using SPSS 18.0 (IBM SPSS, Chicago, IL, USA). Group comparison was performed using an independent samples t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of the clinical cohort
The clinical cohort of miRNA was established using male and female (17:8) patients with the mean age of 41 years. All subjects received a percutaneous liver biopsy followed by pathological diagnosis to determine the staging of fibrosis, according to Scheuer's criteria. The subjects were then divided into five groups, depending on the staging of fibrosis (S0–S4). Detailed clinical data of the patients are presented in Table I.
Table I.
Clinical data of 50 patients with HBV infection and hepatic fibrosis.
| Index | S0 (n=10) | S1 (n=10) | S2 (n=10) | S3 (n=10) | S4 (n=10) |
|---|---|---|---|---|---|
| Male (%) | 80 | 70 | 80 | 70 | 60 |
| Age (years) | 38.6±9.1 | 40.9±12.3 | 40.1±9.6 | 37.7±5.7 | 49.2±8.6 |
| ALT (U/l) | 35.7±17.4 | 38.5±16.3 | 60.8±19.5 | 52.4±21.8 | 48.2±24.1 |
| AST (U/l) | 30.9±10.6 | 31±7.3 | 60.3±62.3 | 37±9.2 | 50.1±32.7 |
| GGT (U/l) | 35.3±36.1 | 18.7±7.0 | 52.3±47.1 | 38.7±19.4 | 29.4±24.1 |
| AKP (U/l) | 74.6±32.7 | 63.3±19.3 | 70±29.9 | 67.9±15.1 | 74±22.5 |
| TBil (µmol/l) | 14.8±3.2 | 19.89±8.2 | 21.44±9.9 | 14.02±3.0 | 17.42±9.2 |
| DBil (µmol/l) | 2.5±0.5 | 3.3±1.3 | 6.6±9.2 | 1.9±1.0 | 2.92±2.9 |
| TP (g/l) | 71±4.9 | 69±6.1 | 74±4.1 | 72±4.4 | 71±5.7 |
| Alb (g/l) | 43.4±1.9 | 43.7±3.0 | 42.5±3.7 | 42±2.3 | 42.7±4.5 |
| HBV-DNA (IU/ml) | 1.71×107±3.19×107 | 1.22×108±2.37×108 | 3.99×106±5.74×106 | 7.51×106±1.57×106 | 4.05×106±8.42×106 |
| HBeAg + (%) | 40 | 70 | 90 | 70 | 50 |
Data are expressed as the mean ± standard deviation. ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, γ glutamyl transpeptidase; AKP, alkline phosphatase; TBil, total bilirubine; DBil, direct bilirubin; TP, total protein; Alb, albumin; HBeAg, hepatitis Be antigen; HBV, hepatitis B virus.
miRNA microarray analysis
Agilent miRNA bio-microarray scanner and extraction software were used for analysis of the miRNA microarray. An inter-group t-test was performed to screen the different miRNAs. The results revealed that there werere 140 miRNAs with >2-fold change in the S1–S4 groups, compared with the S0 group. Compared with the SO group, the number of miRNAs differentially expressed in the the S1 group was 48 (28 upregulated; 20 downregulated); In the S2 group, the number of miRNAs differentially expressed, compared with the S0 group was 97 (49 upregulated; 48 downregulated), with five upregulated >5 fold and four downregulated to ≤20%. In the S3 group, the number of miRNAs differentially expressed, compared with the S0 group, was 84 (57 upregulated and 27 downregulated), with six upregulated >10 fold. In the S4 group, the number of miRNAs differentially expressed, compared with the S0 group, was 56 (40 upregulated and 16 downregulated). In total, 12 miRNAs were differentially expressed in all stages (10 upregulated; 2 downregulated), as shown in Table II. The miRNAs, which were upregulated >5 fold, compared with the S0 group, were hsa-miR-4651 and hsa-miR-4695-5p, and those downregulated >5 fold, compared with the S0 group, were hsa-miR-486-3p and hsa-miR-497-5p, (P<0.05).
Table II.
MicroRNAs exhibiting differential expression levels in the S1–S4 groups compared with the S0 group (fold change and expression trend).
| MicroRNA | Fold change
|
P-value | Trend of expression | |||
|---|---|---|---|---|---|---|
| S1/S0 | S2/S0 | S3/S0 | S4/S0 | |||
| hsa-miR-2861 | 2.37 | 2.60 | 2.48 | 3.12 | 0.0004 | Upregulation |
| hsa-miR-345-3p | 2.62 | 2.62 | 2.47 | 2.19 | 0.0053 | Upregulation |
| hsa-miR-3620-3p | 4.60 | 8.09 | 8.09 | 3.76 | 0.0013 | Upregulation |
| hsa-miR-3656 | 2.22 | 2.19 | 2.23 | 2.22 | 0.0018 | Upregulation |
| hsa-miR-371a-5p | 3.55 | 4.10 | 4.02 | 2.41 | 0.0057 | Upregulation |
| hsa-miR-4646-5p | 3.15 | 2.96 | 4.47 | 2.21 | 0.0052 | Upregulation |
| hsa-miR-4651 | 5.13 | 7.78 | 7.26 | 4.39 | 0.0025 | Upregulation |
| hsa-miR-4695-5p | 5.20 | 9.01 | 9.81 | 6.77 | 0.0016 | Upregulation |
| hsa-miR-4800-5p | 2.63 | 2.58 | 3.69 | 2.34 | 0.0044 | Upregulation |
| hsa-miR-638 | 2.42 | 2.59 | 2.65 | 2.88 | 0.0016 | Upregulation |
| hsa-miR-497-5p | 0.23 | 0.23 | 0.21 | 0.35 | 0.0058 | Upregulation |
| hsa-miR-486-3p | 0.23 | 0.14 | 0.37 | 0.45 | 0.0043 | Upregulation |
Cluster analysis
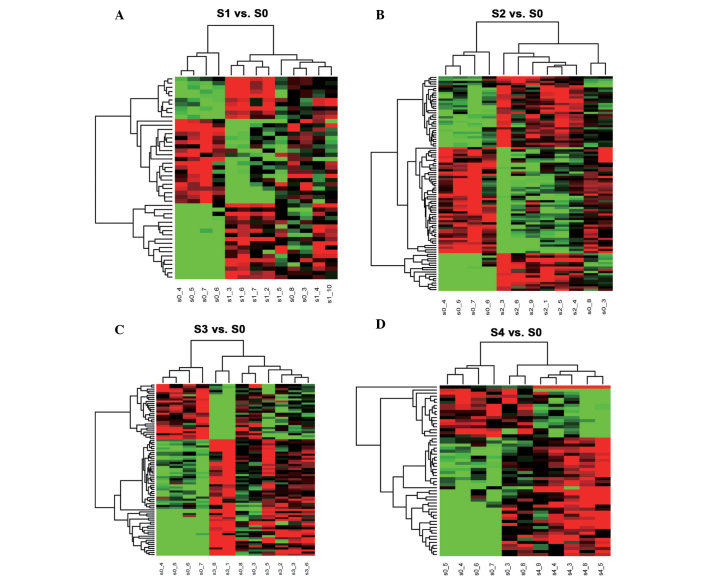
Surveillance-based cluster analysis was performed using the method of Euclidean Distance to analyze the miRNAs with different expression levels in terms of hepatic fibrosis. A hierarchical cluster graph of the miRNAs with different expression levels and fibrosis grouping revealed the characteristics of the expression levels of miRNA in different Scheuer stages. The plasma samples with lower levels of fibrosis, according to Scheuer's criteria, were concentrated to the left of the graph. The plasma samples with higher levels of fibrosis, according to Scheuer's criteria, were concentrated to the right of the graph. As shown in Figs. 1 and 2, the intensity of miRNA expression is highlighted in green (lower level of expression) and red (higher level of expression).
Figure 1.
Cluster analysis of miRNAs in patients with hepatitis B virus infection and hepatic fibrosis. The horizontal axis on the graph represents the Scheuer stages, the vertical axis indicates plasma miRNAs with altered levels of expression. The intensity of miRNA expression is indicated in green (reduced level of expression) and red (higher level of expression). (A) S1, vs. S0, (B) S2, vs. S0, (C) S3, vs. S0, (D) S4 vs. S0. miRNA, microRNA.
Figure 2.
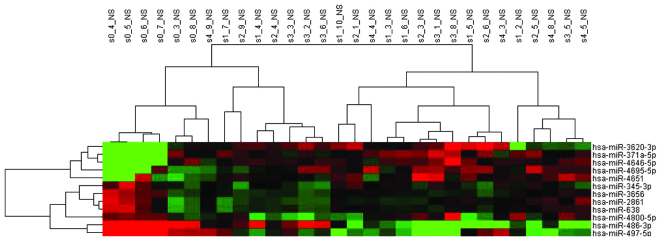
Cluster analysis of differentially expressed miRNAs in different Scheuer's stages. The horizontal axis represents different Scheuer stages (S0–S4), the vertical axis indicates the differentially expressed miRNAs. The intensity of the miRNA expression is indicated in green (lower level of expression) and red (higher level of expression).
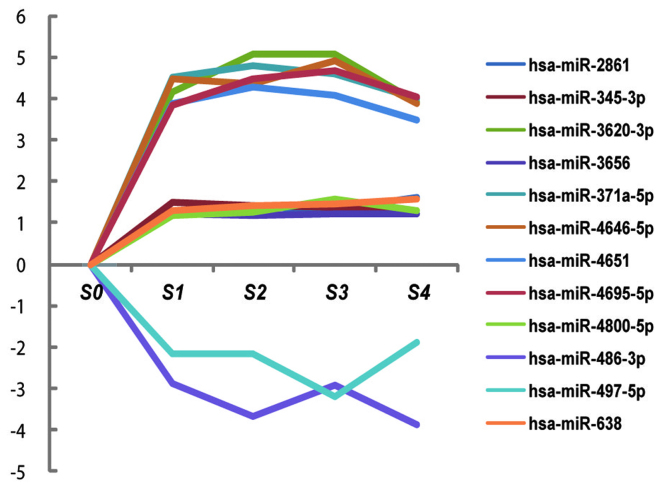
Trend analysis
Log standardization was applied to the different miRNAs to eliminate the effect from varied baseline values and to facilitate trend display in the different groups (Fig. 3). This demonstrated that hsa-miR-4695-5p, hsa-miR-486-3p and hsa-miR-497-5p were suitable for use as markers to differentiate between the different stages of hepatic fibrosis due to HBV infection.
Figure 3.
Trend analysis of different miRNAs. The expression trends of different miRNAs associated with the progression of hepatic fibrosis is shown. The expression levels of hsa-miR-2861, hsa-miR-345-3p, hsa-miR-3620-3p, hsa-miR-3656, hsa-miR-371a-5p, hsa-miR-4646-5p, hsa-miR-4651, hsa-miR-4695-5p, hsa-miR-4800-5p and hsa-miR-638 were upregulated and the expression levels of hsa-miR-486-3p and hsa-miR-497-5p were downregulated. miR/miRNA, microRNA.
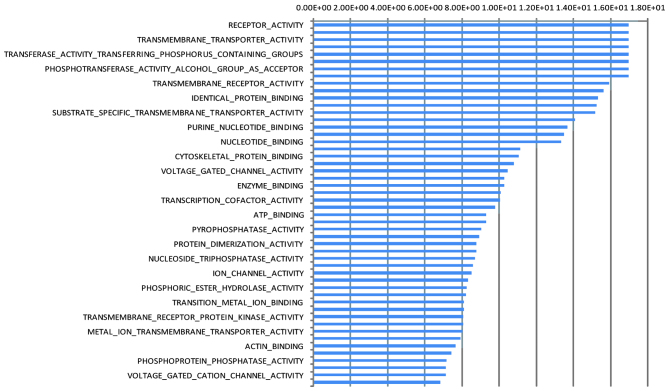
GO functional analysis revealed that the biological processes, with which the differentially expressed target gene was markedly involved included biopolymers, signal transduction, protein metabolism and lipid metabolism. The biological processes identified to have significant involvement of the differentially expressed target gene included molecule functional activation, transmembrane transport, phosphotransferase activity upgrade, binding with protein, purine nucleotide metabolism upgrade, peptidase activation, oxidoreductase activation, binding with L-amino acid peptidase and cytokine activation (Fig. 4).
Figure 4.
Determination of the GO molecular functions modulated by differentially expressed miRNAs in patients with hepatitis B virus infection and hepatic fibrosis. The target genes were selected based on the GO database. Each GO significance level and misjudgment rate was calculated using Fisher's exact test and a χ2 test (P<0.01). GO, gene ontology.
Pathway significance concentration analysis revealed that the differentially expressed genes were involved in 100 significant signal transduction pathways, including TGF-β/Smad, Wnt, MAPK, Jak/STAT and VEGF, which have a significant impact on hepatic fibrosis (Table III) (10–13).
Table III.
Top 15 channels identified as involved with target genes exhibiting differentially expressed mRNA patients with hepatitis B virus infection and hepatic fibrosis.
| Category | Signaling pathway | Target genes involved (n) | P-value | False discovery rate |
|---|---|---|---|---|
| PDGF | MAPK | 56 | 1.00−18 | 0.00 |
| PI/3K | 2.08−06 | 2.33−05 | ||
| PI3K/p85 | 6 | 1.40−04 | 6.26−04 | |
| Erk1/Erk2 Mapk | 14 | 4.59−11 | 1.15−09 | |
| Jak-STAT | 31 | 4.02−10 | 8.53−09 | |
| Ras | 7 | 1.74−04 | 7.16−04 | |
| Rho | Rho | 12 | 6.30−08 | 7.70−07 |
| TGF-β | TGF-β/Smad | 19 | 1.83−07 | 2.00−06 |
| mTOR | mTOR | 15 | 8.37−08 | 9.92−07 |
| AKT/mTOR | 8 | 5.94−06 | 4.20−05 | |
| Blood vessel regeneration | VEGF | 16 | 3.29−06 | 2.60−05 |
| VEGF, anoxia, angiogenesis | 9 | 1.72−05 | 1.05−04 | |
| Wnt | Wnt | 40 | 1.00−18 | 0.00 |
| Integrin | Integrin | 9 | 1.79−04 | 7.27−04 |
| Toll-like receptor | Toll-like receptor | 16 | 1.48−04 | 6.49−04 |
Discussion
Hepatic fibrosis is one of the common characteristics of chronic liver disease. Early diagnosis and sustained follow-up of the progression of hepatic fibrosis is essential in terms of preventing hepatic cirrhosis and end-stage hepatic disease, which result from chronic liver diseases. The follow up of the chronic progression of hepatic diseases by routine liver biopsy and histological assessment are associated with multiple limitations, including patient compliance, risks associated with the intervention, varied biopsy technique, inconsistency between 'inner-observer' and 'inter-observer', false negatives, and inconvenience in sustained assessment and follow up (14). Non-invasive diagnosis indices or predictive models for hepatic fibrosis, including the Forns index (15), Fibrotest (16) and APRI index, are all established based on chronic hepatitis C and alcoholic liver diseases. A non-invasive predictive model, which is specific for chronic HBV infection and hepatic fibrosis has not yet been developed.
miRNA is small non-coding RNA, comprising 21–25 nucleotides. miRNA controls the expression of target genes at the post-transcriptional level by binding to the non-coding region of the target gene (17). A previous study reported that one single miRNA can bind with hundreds of target mRNAs with varied functions, resulting in its modulation of mRNA (18). miRNA is involved in almost all pathological and physical activities in mammals, therefore, it is closely correlated with the development and progression of various diseases (19). It is now understood that there is an almost equal quantity of miRNA in human serum and plasma, which is termed circulative miRNA (20). This type of endogenous micromolecule non-coding RNA with modulation activity is characterized by high stability when combined with proteins in the serum and plasma, providing the possibility of evaluating circulative miRNA as biomarker. These miRNAs are not degraded by endogenous or exogenous ribonucleotidases in the blood and, are refractory to high or low temperatures, high pH, repetitive freeze thawing or extreme acid and base environments (21). In addition, the expression of the miRNA remains consistent among healthy individuals, without significant individual differences, while disease significantly affects the expression levels of circulative miRNA (20,22), leading to favorable repeatability and comparability. Different diseases are associated with different miRNA expression profiles, indicating that circulative miRNA can be used as an effective non-invasive diagnostic parameter and ideal marker in the blood for disease diagnosis.
It has been demonstrated that the expression levels of miR-141 in the serum can be used to diagnose advanced stage prostate cancer. In addition, differential expression levels of circulative miRNA have also been observed in patients with gastrointestinal cancer, including esophageal cancer, gastric cancer, hepatic cancer and large intestine cancer (20,23–25). A previous study on the miRNA expression levels in the serum of patients with chronic HBV infection by Zhang et al (26) demonstrated that miRNA-122 can be used as marker of hepatic tissue injury, with favorable sensitivity and specificity. Xu et al (27) demonstrated that the serum levels of miR-21, miR-122 and miR-223 are increased in patients with chronic HBV infection and in patients with hepatocarcinoma, therefore, they may be regarded as specific markers of hepatic injury, although they are not specific for hepatocarcinoma.
In the present study, a mathematical multiple analytical method was applied to variation analysis. miRNAs, which have a ratio between 0.5 and 2.0 are not considered indicative of significant differences in expression levels, while a ratio beyond 0.5–2.0 is regarded as a significant change in expression. Therefore, the present study screened for miRNAs with upregulated or downregulated expression levels with a fold change >2 and FDR <0.05 in patients with hepatic fibrosis (S1–4/S0), and for miRNAs exhibiting consistent expression in the S1–4 patients. The results demonstrated that those miRNAs exhibiting a trend of consistent expression were predominantly upregulated. Upregulated target genes are involved in signal transduction pathways, including JAK/STAT, MAPK and p53, while target genes with downregulated miRNAs are predominantly involved in fatty acid metabolism and intracellular protein transportation-associated pathways (28).
Among the 12 miRNAs identified with significant changes in expression levels, preliminary understanding of the effects of hsa-miR-345-3p, hsa-miR-371a-5p, hsa-miR-486-3p, hsa-miR-497-5p and hsa-miR-2861 have been previously obtained. Shiu et al (29) revealed that hsa-miR-345-3p facilitates the progression of hepatocarcinoma by negatively regulating p21waf1/Cip1 in cancer cells. Li et al (30) demonstrated that hsa-miR-371a-5p promotes the progression of nasopharyngeal carcinoma by its positive regulation of the TGF-β pathway. Other previous studies (31,32) have reported that hsa-miR-2861 exerts negative modulation on chordoma and basalioma through the mitogen-activated protein kinase pathway. The present study observed upregulated expression levels of hsa-miR-345-3p, hsa-miR-371a-5p and hsa-miR-2861 with the development of hepatic fibrosis, however, the detailed mechanism remains to be elucidated. Kanda et al (33) demonstrated that the expression of hsa-miR-486-3p in hepatic tissue is upregulated in mice with obstructive jaundice resulting from bile duct ligation, while its expression in hepatocarcinoma is downregulated (34). Another previous study (35) revealed that hsa-miR-486-3p exerts a positive regulatory effect on glioma via the nuclear factor-κB signaling pathway. The present study demonstrated a trend of downregulation of hsa-miR-486-3p with the progression of hepatic fibrosis, however, its clinical relevance remains to be elucidated. Furuta et al (36) reported that hsa-miR-497-5p exerts a negative regulatory effect on hepatocarcinoma by inhibiting the G1-S of cancer cells. Another previous study (37) demonstrated that hsa-miR-497-5p inhibits the proliferation, migration and invasion of colon cancer cells by negatively regulating IGF-1. Yadav et al (38) demonstrated that hsa-miR-497-5p is associated with apoptosis, resulting from the effect this has on the BCL family. In the present study, downregulation of hsa-miR-497-5p was observed in all stages of hepatic fibrosis, with a maximum fold change of 10. The detailed mechanism remains to be elucidated.
The present study had several limitations. Although the changes in the plasma miRNA expression levels was based on the screening of samples from patients with hepatic fibrosis due to HBV infection, whether it is specific to hepatic fibrosis due to HBV infection remains to be elucidated. Further investigations are required to exclude other confounding factors. In addition, hepatic fibrosis staging is the only categorization factor used in the present study, and the effects of inflammation and qualification of viral load on the results have not been excluded.
In the present study, a set of miRNAs, potentially correlating with the development of hepatic fibrosis due to HBV infection, were identified based on gene microarray screening, may be used as a potential diagnostic marker to differentiate patients with chronic HBV infection and hepatic fibrosis at different stages. Since miRNA is relatively stable and readily obtainable from serum/plasma, this method provides a novel non-invasive option for the diagnosis of chronic HBV infection with hepatic fibrosis and for the staging of this condition. Future investigations aim to screen miRNAs that may be correlated with the staging of hepatic fibrosis due to HBV infection and confirming the correlation between microarray results and hepatic fibrosis using methods, including quantitative polymerase chain reaction. Further investigations aim to expand on current knowledge regarding miRNA in the developmental mechanism of hepatic fibrosis due to HBV infection, and to identify novel serum molecular biological targets for the diagnosis of hepatic fibrosis caused by HBV infection.
Acknowledgments
This study was supported by the Development Program of China during the 11th and 12th Five-year Plan Period (grant. nos. 2008ZX10002-006, 2012ZX10002007-001-040 and 2013ZX10002004-002-003), the Science and Technology Commission of Shanghai Municipality (grant. nos. 10411955300 and 12DZ1941603) and the Shanghai Municipal Health Bureau (grant,. no. XBR2011012).
Abbreviations
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- GO
gene ontology
- EDTA
ethylene diamine tetraacetic acid
- ALT
alanine minotransferase
- AST
aspartate aminotransferase
- GGT
γ glutamyl transpeptidase
- AKP
alkline phosphatase
- TBil
total bilirubin
- DBil
direct bilirubin
- TP
total protein
- Alb
albumin
- HBeAg
hepatitis Be antigen
- FDR
error detection rate
References
- 1.Hou J, Liu Z, Gu F. Epidemiology and prevention of hepatitis B virus infection. Int J Med Sci. 2005;2:50. doi: 10.7150/ijms.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Vettori S, Gay S, Distler O. Role of MicroRNAs in Fibrosis. Open Rheumatol J. 2012;6:130–139. doi: 10.2174/1874312901206010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 5.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinese Society of Hepatology, Chinese Medical Association; Chinese Society of Infectious Diseases, Chinese Medical Association The guidelines of prevention and treatment for chronic hepatitis B. Chin J Hepatol. 2005;13:881–891. In Chinese. [PubMed] [Google Scholar]
- 7.Scheuer PJ. The nomenclature of chronic hepatitis: Time for a change. J Hepatol. 1995;22:112–114. doi: 10.1016/0168-8278(95)80269-X. [DOI] [PubMed] [Google Scholar]
- 8.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 9.Mar JC, Kimura Y, Schroder K, Irvine KM, Hayashizaki Y, Suzuki H, Hume D, Quackenbush J. Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics. 2009;10:110. doi: 10.1186/1471-2105-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JH, Yoon J, Lee KY, Park B. Effects of geniposide on hepatocytes undergoing epithelial-mesenchymal transition in hepatic fibrosis by targeting TGFβ/Smad and ERK-MAPK signaling pathways. Biochimie. 2015;113:26–34. doi: 10.1016/j.biochi.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Qu W, Huang H, Li K, Qin C. Danshensu-mediated protective effect against hepatic fibrosis induced by carbon tetrachloride in rats. Pathol Biol (Paris) 2014;62:348–353. doi: 10.1016/j.patbio.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura I, Zakharia K, Banini BA, Mikhail DS, Kim TH, Yang JD, Moser CD, Shaleh HM, Thornburgh SR, Walters I, Roberts LR. Brivanib attenuates hepatic fibrosis in vivo and stellate cell activation in vitro by inhibition of FGF, VEGF and PDGF signaling. PLoS One. 2014;9:e92273. doi: 10.1371/journal.pone.0092273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Li X, Xu W, Wang S, Hu Z, Zhang Q, Deng X, Wang J, Zhang J, Guo C. Antifibrotic effects of luteolin on hepatic stellate cells and liver fibrosis by targeting AKT/mTOR/p70S6K and TGFβ/Smad signalling pathways. Liver Int. 2015;35:1222–1233. doi: 10.1111/liv.12638. [DOI] [PubMed] [Google Scholar]
- 14.Zhou K, Lu LG. Assessment of fibrosis in chronic liver diseases. J Dig Dis. 2009;10:7–14. doi: 10.1111/j.1751-2980.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 15.Forns X, Ampurdanès S, Llovet JM, Aponte J, Quintó L, Martínez-Bauer E, Bruguera M, Sánchez-Tapias JM, Rodés J. Identification of chronic hepatitis C patients without hepatic fibrosis by a simple predictive model. Hepatology. 2002;36:986–992. doi: 10.1053/jhep.2002.36128. [DOI] [PubMed] [Google Scholar]
- 16.Imbert-Bismut F, Ratziu V, Pieroni L, Charlotte F, Benhamou Y, Poynard T, MULTIVIRC Group Biochemical markers of liver fibrosis in patients with hepatitis C virus infection: A prospective study. Lancet. 2001;357:1069–1075. doi: 10.1016/S0140-6736(00)04258-6. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Wang F, Huang Q, Cao Y, Zhao Y, Yang C. MicroRNAs contribute to hepatocellular carcinoma. Mini Rev Med Chem. 2015;15:459–466. doi: 10.2174/1389557515666150324125353. [DOI] [PubMed] [Google Scholar]
- 18.Park K, Kim KB. miRTar Hunter: A prediction system for identifying human microRNA target sites. Mol Cells. 2013;35:195–201. doi: 10.1007/s10059-013-2165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim T, Reitmair A. Non-Coding RNAs: Functional aspects and diagnostic utility in oncology. Int J Mol Sci. 2013;14:4934–4968. doi: 10.3390/ijms14034934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 25.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol- and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, et al. Circulating MicroRNAs, miR-21, miR-122 and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 28.Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci. 2014;21:95. doi: 10.1186/s12929-014-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down-regulates p21Waf1/Cip1 and inhibits curcumin-induced apoptosis through microRNA-345 targeting in human hepatoma cells. PloS One. 2013;8:e61089. doi: 10.1371/journal.pone.0061089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y, Liu Y. Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing. PLoS One. 2013;8:e84486. doi: 10.1371/journal.pone.0084486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long C, Jiang L, Wei F, Ma C, Zhou H, Yang S, Liu X, Liu Z. Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS One. 2013;8:e66676. doi: 10.1371/journal.pone.0066676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, Altmeyer P, Bechara FG. Expression of microRNAs in basal cell carcinoma. Brit J Dermatol. 2012;167:847–855. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 33.Kanda T, Ishibashi O, Kawahigashi Y, Mishima T, Kosuge T, Mizuguchi Y, Shimizu T, Arima Y, Yokomuro S, Yoshida H, et al. Identification of obstructive jaundice-related microRNAs in mouse liver. Hepatogastroenterology. 2010;57:1013–1023. [PubMed] [Google Scholar]
- 34.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song L, Lin C, Gong H, Wang C, Liu L, Wu J, Tao S, Hu B, Cheng SY, Li M, et al. miR-486 sustains NF-kappaB activity by disrupting multiple NF-kappaB-negative feedback loops. Cell Res. 2013;23:274–289. doi: 10.1038/cr.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo ST, Jiang CC, Wang GP, Li YP, Wang CY, Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32:1910–1920. doi: 10.1038/onc.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadav S, Pandey A, Shukla A, Talwelkar SS, Kumar A, Pant AB, Parmar D. miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem. 2011;286:37347–37357. doi: 10.1074/jbc.M111.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]