Abstract
microRNAs (miRNAs/miRs) are a class of single-stranded non-coding RNA molecules of 19–24 nucleotides (nt) in length. They are widely expressed in animals, plants, bacteria and viruses. Via specific mRNA complementary pairing of target genes, miRNAs are able to regulate the expression of mRNA levels or inhibit protein translation following transcription. miRNA expression has a time- and space specificity, and it is involved in cell proliferation and differentiation, apoptosis, development, tumor metastasis occurrence and other biological processes. miR-26b is an miRNA of 22 nt and is important in the regulation of cellular processes. With the advancement of molecular biology techniques in recent years, there have been extensive investigations into miR-26b. Numerous studies have observed that miR-26b is involved in early embryonic development, cell proliferation regulation, pituitary hormone secretion and other physiological activities. miRNAs are associated with the function of propagation. The present study used reverse transcription quantitative polymerase chain reaction to detect the relative expression levels of miR-26b in the pituitary tissue of Yanbian cattle at different developmental stages. The 2−ΔΔCt method was used to calculate the relative gene expression levels. The miRNA target gene database TargetScan and RNA22 were used for prediction of the miR-26b target gene and selective recognition was also performed. The results demonstrated that miR-26b is expressed in the pituitary tissues of Yanbian cattle at 6 and 24 months of age. The relative expression levels of miR-26b in the pituitary tissues of 24-month-old Yanbian cattle were 2.41 times that of those in the six-month-old Yanbian cattle, demonstrating significant differences in the relative expression (P<0.01). The relative expression of the candidate target genes, EphA2 and miR-26b, exhibited the opposite expression pattern. The relative expression levels in the pituitary tissues of six-month-old Yanbian cattle were 3.34 times that of those in 24-month-old Yanbian cattle (P<0.01). There are miR-26b binding sites in the 3′-untranslated region (3′-UTR) of EphA2 in bovine, human, murine and other mammalian mRNAs, suggesting that the EphA2 gene may be a target gene of miR-26b. The results of a Luciferase reporter system assay revealed that miR-26b is able to suppress EphA2 expression at the transcription level. Following the site-directed mutagenesis of plasmid EphA2 3′-UTR pmirGLO-MUT- and miR-26b mimic-transfected HeLa cells, the dual-luciferase reporter gene assay revealed that there were three consecutive nucleotide mutations in the 3′-UTR, binding with the predicted seed region. This may have caused the miR-26b inhibition of luciferase activity to decrease from 60% in the wild-type to 26%, suggesting that miR-26b achieved its function via binding with the TACTTGAA sequence of the 3′-UTR in EphA2. In conclusion, the present study successfully assessed the expression pattern of miR-26b in the pituitary tissue of Yanbian cattle, and also confirmed that EphA2 was a target gene of miR-26b in Yanbian cattle in vitro. The present study provided the theoretical basis to further investigate the role of miR-26b in early embryonic development, pituitary hormone secretion and other reproductive functions.
Keywords: Yanbian cattle, microRNA-26b, quantitative polymerase chain reaction, ephrin type-A receptor 2, luciferase activity
Introduction
MicroRNAs (miRNAs/miRs) are a class of small RNAs found in a diverse range of eukaryotes, including animals and plants, as well as DNA viruses, which range in size between 19 and 24 nucleotides (nt) (1,2). miRNAs regulate mRNA or protein levels either by promoting mRNA degradation or by attenuating protein translation (3,4). As with mRNAs, certain miRNAs are differentially expressed among tissues or developmental stages (5). miRNAs are important in the regulation of numerous cellular processes, including cell proliferation and differentiation, apoptosis, development, tumor metastasis and other biological processes (6,7). miR-26b, at a length of 22 nt, is important in the regulation of cell biology processes. With the development of novel molecular biology techniques, the investigation of miR-26b has gained specificity. A number of studies have indicated that miR-26b is involved in early embryonic development, cell proliferation regulation, hormone expression regulation in the anterior pituitary and other physiological activities (8–11). Therefore, miR-26b is essential for reproductive function. The present study provides a basis for the further investigation into the function of miR-26b in embryonic development, pituitary hormone secretion and other reproductive processes.
Materials and methods
Sample collection
The present study enrolled Yanbian cattle reared under the same conditions and sacrificed by the Haoyue Company, three were six months old and three were 24 months old. The bovine pituitary gland was quickly collected in a liquid nitrogen tank and stored at −80°C prior to total RNA extraction. The animal slaughter experiments were conducted in accordance with the guidelines of Jilin University (Changchun, China) on the Review of Welfare and Ethics of Laboratory Animals approved by the Jilin Province Administration Office of Laboratory Animals. All animal procedures were approved by Institutional Animal Care and Use Committee of Jilin University (permit no. 20140204). All surgery was performed under 50 mg/kg sodium pentobarbital (Sigma-Aldrich, St. Louis, MO, USA) anesthesia, and all efforts were made to minimize suffering.
Reagents
TRIzol reagent, pMD18-T vector, LA Taq DNA polymerase, dNTP mixture, DNA marker, T4 DNA ligase, XhoI and XbaI were purchased from Takara Bio Inc. (Otsu, Japan). The ReverTra Ace qPCR RT kit and SYBR® Green Realtime PCR master mix (QPK-201) were purchased from Toyobo Co., Ltd. (Osaka, Japan) for reverse transcription quantitative polymerase chain reaction (RT-qPCR). The endotoxin-free plasmid extraction kit was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). The pmirGLO Dual-Luciferase miRNA Target Expression vector and Dual-Luciferase® Reporter Assay system were purchased from Promega Corporation (Madison, WI, USA). Dulbecco's modified Eagle's medium: Nutrient mixture F-12 (DMEM/F12) was purchased from Gibco-BRL (Invitrogen Life Technologies, Carlsbad, CA, USA). Fetal bovine serum (FBS) was purchased from Hyclone (GE Healthcare, Logan, UT, USA). miR-26b and the negative controls were synthesized by Shanghai Jima International Trading Co., Ltd. (Shanghai, China). The X-tremeGENE small interfering (si)RNA transfection reagent was purchased from Roche Diagnostics (Basel, Switzerland).
Main instruments
A myECL gel imaging system was purchased from Alpha Innotech (San Leandro, CA, USA). The Genesys 10 Nanodrop spectrophotometer, Arktik PCR instrument, 37°C CO2 incubator, −80°C freezer and high-speed desktop centrifuge were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). A PCR 5332 instrument, micropipettes and a Mastercycler Ep Realplex qPCR instrument were purchased from Eppendorf (Hamburg, Germany). The multifunctional microplate reader Mithras LB960 was purchased from Berthold Technologies GmbH & Co. KG (Bad Wildbad, Germany). A 37XB inverted microscope was purchased from Shanghai Yiguang Optical Instrument Co., Ltd. (Shanghai, China).
Total RNA extraction of pituitary tissue from Yanbian cattle
The pituitary tissue was removed from the liquid nitrogen and placed in a mortar with 5 ml diethylpyrocarbonate-treated water. TRIzol was added, grinding was performed and total RNA was extracted. The total RNA was then dissolved in 20.0 μl RNase-free water and stored at −80°C. The RNA was separated by 1.0% agarose gel electrophoresis and the blots were visualized under ultra violet light, following ethidium bromide staining.
Specific identification of pituitary tissues
Based on the mRNA sequence of the follicle stimulating hormone (FSH) β gene in cattle available on GenBank (http://www.ncbi.nlm.nih.gov/genbank/), the upstream and the downstream primer sequences, with an annealing temperature of 60°C, are presented in Table I. The primers were synthesized by Huada Limited Co. (Shanghai, China)
Table I.
Reverse transcription-quantitative polymerase chain reaction primer sequences.
| Name | Primer sequence | Product length (bp) |
|---|---|---|
| miR-26b | RT: 5′-GTCGTATCCAGTGCGTGTCGTGGAGCGCAATTGCACTGGATACGACAACCTAT-3′ | 67 |
| F: 5′-GGGGTTCAAGTAATTCAGG-3′ | ||
| R: 5′-CAGTGCGTGTCGTGGAGT-3′ | ||
| U6 | RT: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | 89 |
| F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ | ||
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | ||
| EphA2 qPCR | F: 5′-CAACGACGACATCAAGAGGAT-3′ | 176 |
| R: 5′-CAGAAGGAAGCCAGTCCATTT-3′ | ||
| GADPH | F: 5′-ATTCTGGCAAAGTGGACATCG-3′ | 214 |
| R: 5′-ACATACTCAGCACCAGCATCAC-3′ | ||
| FSHβ | F: 5′-CAGCAAGGCCCAATATCCA-3′ | 817 |
| R: 5′-CCAGAGGAGGAGAGGCGAAACA-3′ | ||
| EphA2 PCR | F: 5′-TTTCTCGAGGACAGCCCTCGCA-3′ | 725 |
| R: 5′-GCTCTAGAGGAATGCCCGAAGC-3′ | ||
| Epha2 3′-UTR plasmid site-detected primers | F: 5′-ATCTGCTGAGAATACTTTCCAAATGGACTGGCTTC-3′ | 713 |
| R: 5′-GAAGCCAGTCCATTTGGAAAGTATTCTCAGCAGAT-3′ |
miR, microRNA; RT, reverse transcription primers; F, forward primers; R, reverse primers; qPCR, quantitative polymerase chain reaction.
The ReverTra Ace qPCR RT kit was used according to the manufacturer's instructions and a 10.0-μl RT system was developed and optimized. cDNA was synthesized and stored at −20°C. PCR was used for the amplification of FSHβ fragments. The cycling conditions were as follows: Initial denaturation for 1 min at 95°C, followed by 35 cycles of denaturation at 95°C for 35 sec, annealing at 60°C for 35 sec and extension at 72°C for 35 sec. Final extension was performed at 72°C for 5 min.
miR-26b-specific stem-loop RT primers and qPCR primer design and cDNA synthesis via RT
Based on the mature sequence of miR-26b provided by miRBase (http://www.mirbase.org/), miR-26b and U6-specific stem-loop RT primers were designed and are shown in Table I.
The ReverTra Ace qPCR RT kit was used according to the manufacturer's instructions and the miR-26b and U6-specific RT primers were used. cDNA was synthesized and stored at −20°C.
Based on the stem-loop primer and miR-26b sequences, the quantitative primers were designed and the primer sequences are shown in Table I.
miR-26b RT-qPCR detection in pituitary tissues of cattle at different developmental stages
The Realplex RT quantitative PCR instrument was used and a three-step method was adopted to perform the miR-26b expression analysis. The qPCR reaction system was as follows: Diethylpyrocarbonate-treated water (10.5 μl), SYBR® Green Realtime PCR master mix (12.5 μl), upstream primer (0.5 μl), downstream primer (0.5 μl) and 10 μmol/l cDNA (1 μl) in a total volume of 25.0 μl. A total of three biological replicates were included for each time period and three replicates were included in each sample. The reaction conditions were as follows: 95°C denaturation for 1 min; 95°C denaturation for 15 sec, 68°C annealing for 15 sec and 72°C extension for 45 sec for 50 cycles. Subsequently, melting curve analysis of the PCR products was performed to assess the specificity of the PCR products.
miR-26b target gene prediction
According to the miRNA sequence provided by miRBase, the two miRNA target gene databases TargetScan (http://www.targetscan.org/) and RNA22 (https://cm.jefferson.edu/rna22/) were used for the prediction of the target genes of miR-26b (12). The binding model (complementary region) was predicted using RNA mfold software (http://mfold.rna.albany.edu/?q=mfold).
Relative expression levels of candidate target gene mRNA
Based on the mRNA sequence of EphA2 in cattle, qPCR primers were designed. GAPDH was used as internal reference and the specific sequences are shown in Table I. The primers were synthesized by BGI (Shenzhen, China).
The EphA2 qPCR system was the same as that for miR-26b. Subsequently, the results of the qPCR were analyzed.
Construction of a dual-luciferase reporter gene vector of miR-26b of the target gene
In order to perform PCR amplification of the target gene, a target gene 3′-untranslated region (3′-UTR) was designed and synthesized, which covered a sequence of miRNA loci. XhoI endonuclease was added in the 5′ region and XbaI endonuclease was added in the 3′ region. The primers were synthesized by BGI. The specific sequences are presented in Table I. Following purification, the PCR product was ligated into the vector pMD18-T and DH5α competent cells were added and agitated for vector amplification for 12 h. The bacterial liquid was then subjected to PCR amplification. In addition, plasmid DNA was extracted for XhoI and XbaI double digestion and purified gene fragments were recovered. XhoI and XbaI double digestion of pmirGLO Dual-Luciferase miRNA target expression vector was then performed. The recovered target gene fragments were ligated into digested vectors, which were transformed into Escherichia (E.) coli, and monoclonal E. coli was selected. The positive clones were extracted and sent to BGI for sequencing. The correct plasmid was termed EphA2 3′-UTR pmirGLO.
Site-directed mutagenesis and construction of miR-26b target gene dual-luciferase reporter plasmid
The recombinant EphA2 3′-UTR pmirGLO plasmid was used and three consecutive bases on the binding sites of the miR-26b and EphA2 3′-UTR were mutated with a site-directed mutagenesis kit. The primer sequences used in site-directed mutagenesis are presented in Table I.
The PCR amplification reaction system consisted of 34.0 μl nuclease-free water, 5.0 μl 10X reaction buffer, 5.0 μl mutated template plasmid, 4.0 μl desoxyribonucleotide triphosphate mix (2.5 mM/l) and 1.0 μl up- and downstream primer (10 μM) each (total volume, 49.0 μl). 1.0 μl Pyrococcus furiosus DNA polymerase (10 μmol/l) was added to achieve a total volume of 50.0 μl. The reaction conditions were as follows: Initial denaturation for 1 min at 95°C, followed by 18 cycles of denaturation at 95°C for 40 sec, annealing at 60°C for 1 min and extension at 68°C for 8 min. Final extension was performed at 72°C for 10 min. 1.0 μl Dpn I was added directly to the PCR products, and following mixing, they were incubated at 37°C for 1 h.
A total of 10 μl of the above digested products were added to 100 μl competent cells (E. coli; DH5α, 100 μl, 5×107/ml), and transformation and cloning were performed for plasmid extraction. XhoI and XbaI restriction enzymes were used to perform double digestion site-directed mutagenesis of the recombinant plasmid. Following electrophoresis, positive test plasmids were sent to BGI for sequencing. The correct plasmid was termed EphA2 3′-UTR pmirGLO-MUT.
Cell culture and luciferase activity detection
HeLa cells were cultured in DMEM/F12 medium containing 5% FBS at 37°C and 5% CO2. HeLa cells (Saiqi Cellbio Co., Ltd., Shanghai, China) in the logarithmic phase were seeded into a 96-well cell culture plate. After 24 h, when adherent cells had reached 70% confluence, X-tremeGENE siRNA transfection reagents were used for transfection. The miR-26b mimics and EphA2 3′-UTR pmirGLO plasmid containing the 3′-UTR or the EphA2 3′-UTR pmirGLO-MUT plasmid were co-transfected into HeLa cells. The negative control group was set to co-transfect the miR-26b control and the EphA2 3′-UTR pmirGLO plasmid or EphA2 3′-UTR pmirGLO-MUT plasmid. A total of three replicates were performed for each combination. After 24 h, the HeLa cell culture DMEM/F12 medium containing fresh 5% FBS was replaced. After 48 h, the Dual-Luciferase® reporter assay system was used to perform the reporter gene assay, according to the manufacturer's instructions. After 48 h, the cells were lysed and the multifunctional microplate reader Mithras LB960 was used to detect the fluorescence intensity. All experiments were repeated three times.
Statistical analysis
The data were expressed as the mean ± standard deviation. Differences were assessed by Student's two-tailed t-test using the SPSS software, version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Total RNA extraction
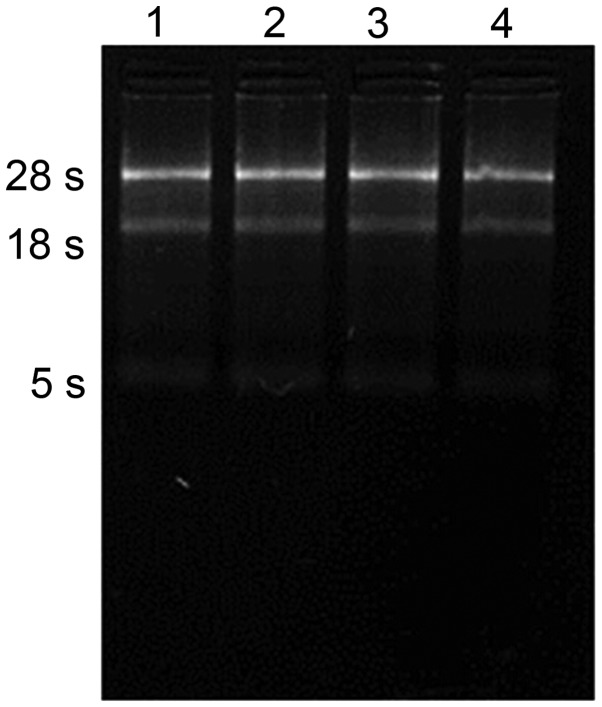
Total RNA in pituitary tissue of Yanbian cattle was extracted and the electrophoretic analysis is shown in Fig. 1. The optical density at 260 nm (OD260)/OD280 ratio of the total RNA concentration was 1.8–2.0, indicating that the integrity of the RNA was good and had not been contaminated with genetic material, proteins or other impurities. It was therefore deemed suitable for use in subsequent experiments.
Figure 1.
Electrophoretic analysis of total RNA indicating 28S and 18S units. Lanes 1 and 2 show samples from six-month-old cattle and lanes 3 and 4 show samples from 24 month-old cattle.
Identification of pituitary tissue
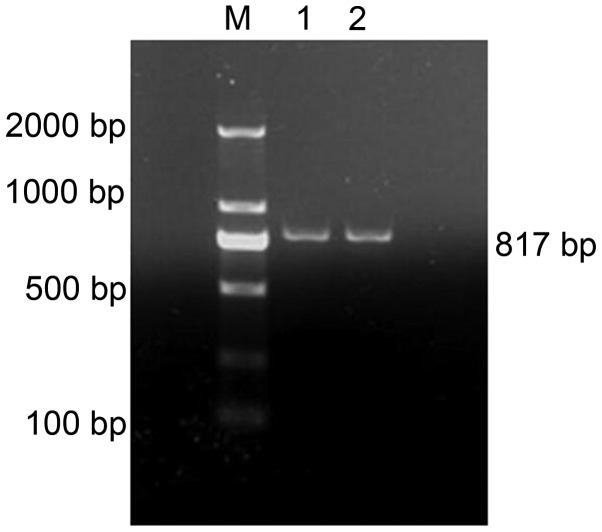
After the total RNA was reverse transcribed into cDNA, FSHβ gene primers were used for PCR amplification and the PCR electrophoresis results are shown in Fig. 2. They were amplified in order to confirm that specific bands were present, which were consistent in size with the designed amplified fragments. No apparent dimers or non-specific bands were observed, suggesting that the pituitary tissue samples were accurately identified, and were therefore deemed suitable for use in subsequent experiments.
Figure 2.
PCR analysis of pituitary follicle-stimulating hormone β gene. M is a molecular weight marker of 2,000 bp, 1 and 2 are PCR products from 6 and 24 months, respectively. PCR, polymerase chain reaction.
miR-26b RT-qPCR
RT-qPCR was used to analyze the relative expression of mature miR-26b. A total of three samples were assessed at each developmental stage and three replicates were performed for each sample. U6 was selected as a reference gene for normalization of RT-qPCR reactions and samples. The 2−ΔΔCt method (13) was used to calculate the relative expression levels of miR-26b. As shown in Figs. 3 and 4, the U6 and miR-26b samples exhibited a good amplification curve and the melting curve suggested that the designed primers had a high specificity.
Figure 3.
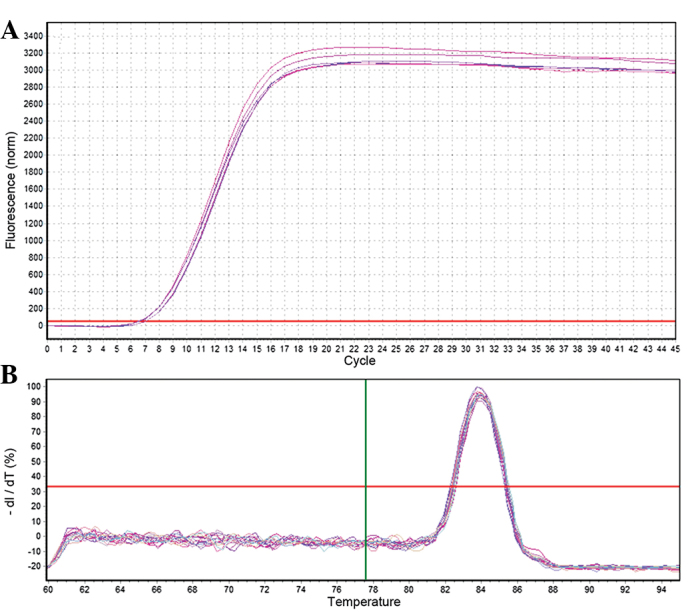
Quantification curve and melting curve for U6. (A) U6 amplification curve. (B) U6 melting curve.
Figure 4.
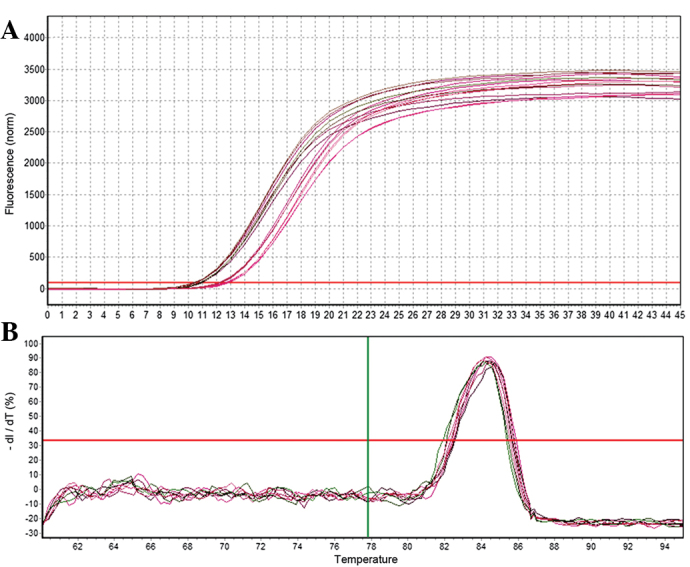
Quantification curve and melting curve for miR-26b. (A) Quantification curve for miR-26b. (B) Melting curve for miR-26b. miR, microRNA.
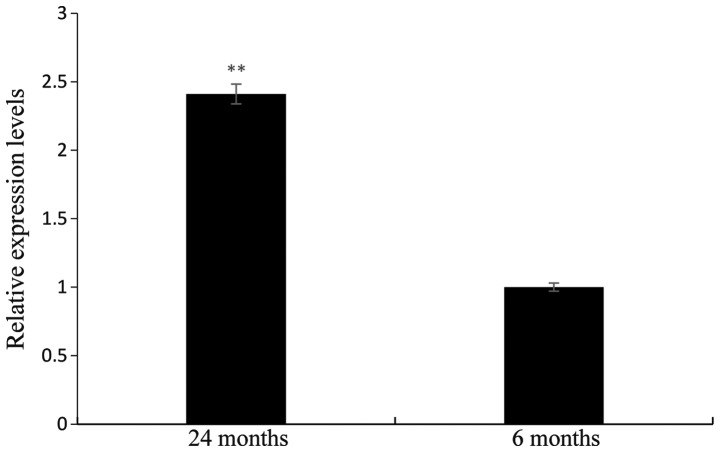
Relative expression levels of miR-26b in pituitary tissues of Yanbian cattle at different developmental stages
RT-qPCR analysis was used to detect the relative expression of miR-26b in the pituitary tissue of Yanbian cattle at six and 24 months of age. The results revealed that miR-26b was expressed in the pituitary tissue of Yanbian cattle at different developmental stages, revealing significant differences (P<0.01). The relative expression levels in 24-month-old cattle were 2.41 times greater than those in the six-month-old cattle, as shown in Fig. 5.
Figure 5.
Relative expression levels of microRNA-26b in the pituitary tissue of Yanbian cattle at different developmental stages. The expression at 6 months was set as 1. Values are expressed as the mean ± standard error of triplicate measurements from three independent experiments. **P<0.01.
Predicted target genes of miR-26b
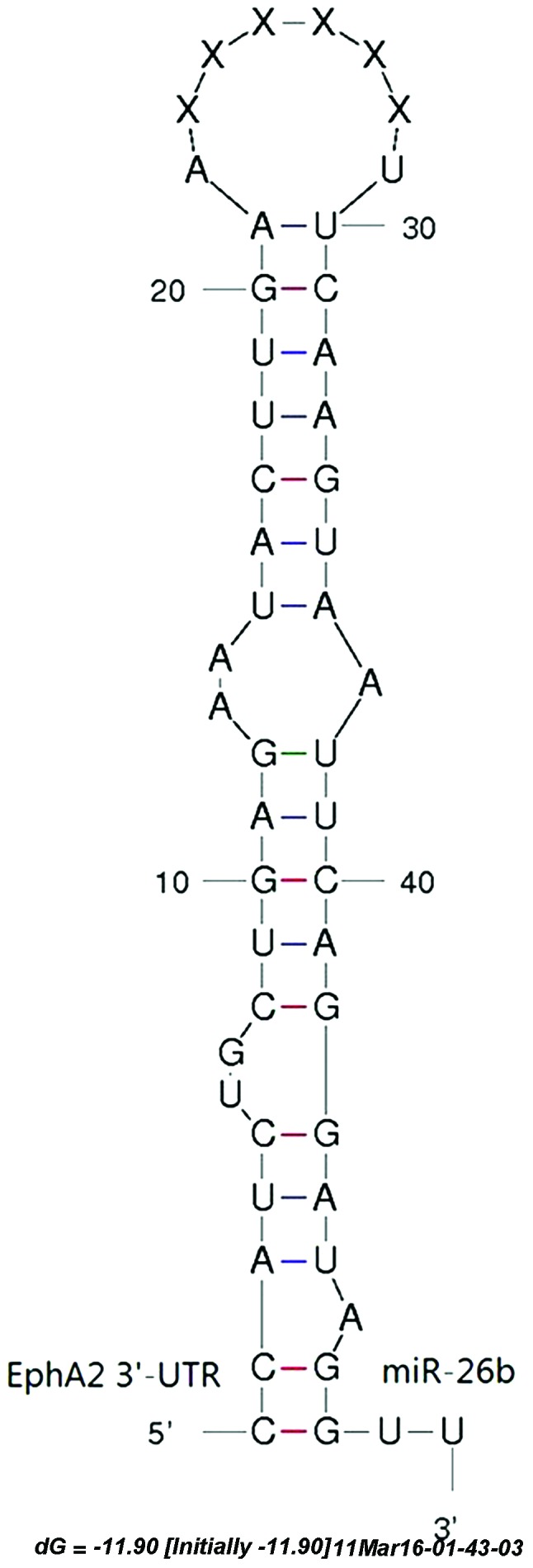
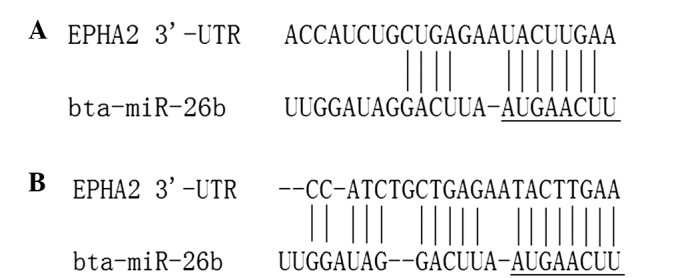
Using TargetScan, 561 predicted target genes were identified in the prediction database of the miRNA target gene. The target genes associated with reproductive traits were selected as the alternative target gene and the RNA22 software was used for further analysis. The genes predicted by the two software programs were identified. Following the gene function analysis, EphA2 was selected as the candidate target gene. The 3′-UTR of the EphA2 gene had a conserved seed sequence at the binding site of miR-26b. The predicted results of TargetScan and RNA22 are shown in Fig. 6. The mfold program was used to predict the seed region complementary of EphA2 mRNA and miR-26b, and predicted the secondary structure, as shown in Fig. 7.
Figure 6.
Bioinformatics analysis of miR-26b acting on the EphA2 3′-UTR. UTR, untranslated region; miR, microRNA.
Figure 7.
Predicted duplex structure between miR-26b and the EphA2 mRNA target sequence. (A) Analysis using TargetScan software. (B) Analysis using RNA22 software. UTR, untranslated region; miR, microRNA. bta, bos taurus.
RT-qPCR assessment of candidate target genes of EphA2
RT-qPCR was used to analyze the relative expression of the candidate target gene of EphA2. A total of three samples were assessed at each developmental stage and three replicates were performed for each sample. GAPDH was selected as a reference gene to allow for differences between RT-qPCR reactions and samples. The 2−ΔΔCt method was used to calculate the relative expression of the candidate target gene of EphA2. As shown in Figs. 8 and 9, GAPDH and the candidate target gene EphA2 exhibited a good amplification curve and melting curve, suggesting that the designed primers had a high specificity.
Figure 8.
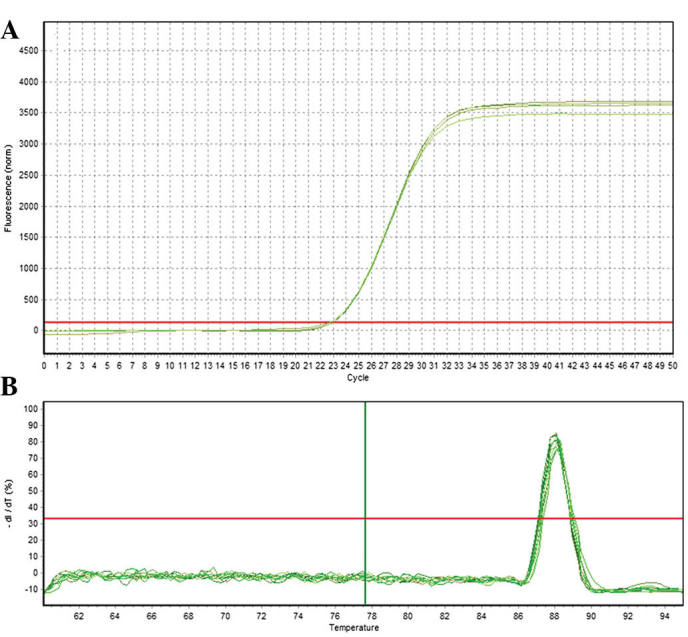
Quantification curve and melting curve for GAPDH. (A) GAPDH amplification curve. (B) GAPDH melting curve.
Figure 9.
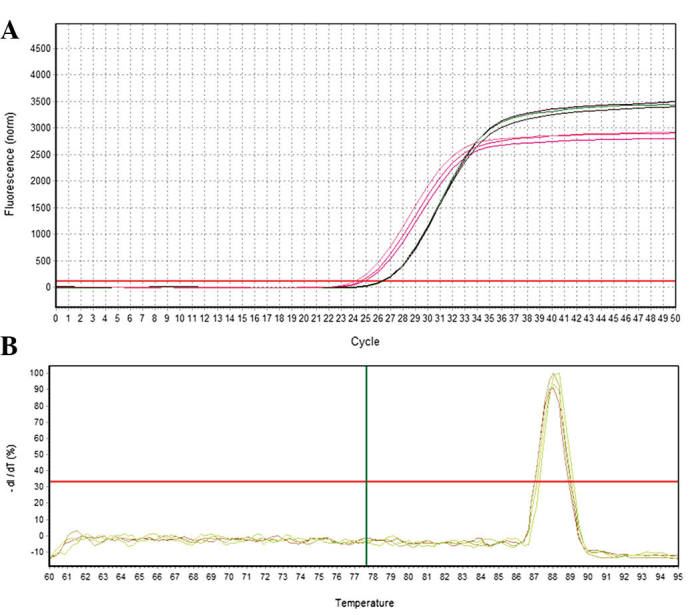
Quantification curve and melting curve for EphA2. (A) EphA2 amplification curve. (B) EphA2 melting curve.
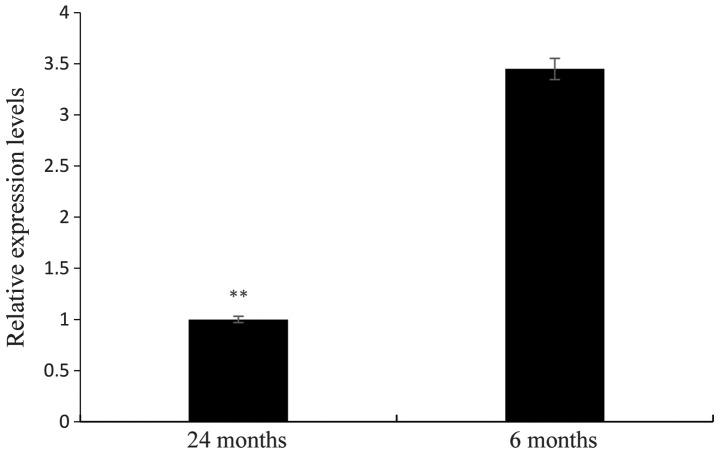
Relative expression levels of EphA2 in pituitary tissues of Yanbian cattle at different developmental stages
RT-qPCR was used to detect the relative expression of EphA2 in the pituitary tissue of Yanbian cattle at six or 24 months of age. The results revealed that EphA2 expression was significantly differentially in the pituitary tissue of Yanbian cattle at different developmental stages (P<0.01). The relative expression levels in 24-month-old cattle was 3.34 times greater than that in six-month-old cattle, as shown in Fig. 10.
Figure 10.
Relative expression levels of EphA2 in pituitary tissues of Yanbian cattle at different developmental stages. The expression at 24 months was set as 1. Values are expressed as the mean ± standard error of triplicate measurements from three independent experiments. **P<0.01.
Recombinant plasmid construction
PCR amplification of target fragments of EphA2 was performed and PCR products were obtained and detected using agarose gel electrophoresis. As no non-specific bands and dimers were identified, the fragments were deemed suitable for the plasmid construction of the dual-luciferase reporter gene. The PCR results are shown in Fig. 11.
Figure 11.
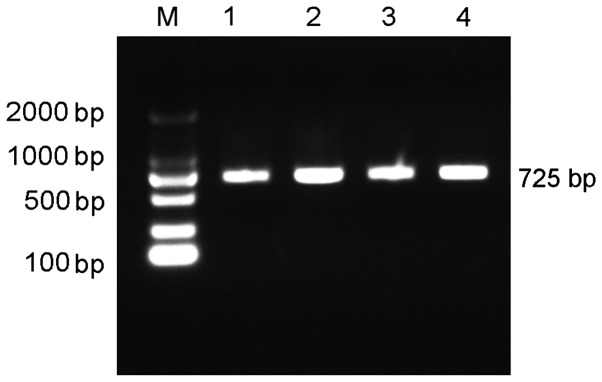
Agarose gel electrophoresis indicating the product size of polymerase chain reaction products. Lane M, DL 2000 DNA marker; lanes 1–4, polymerase chain reaction results of the dual-luciferase reporter gene.
For the construction and identification of miR-26b candidate target gene dual-luciferase reporter gene vectors and mutated vectors, the target gene was connected with the vector and E. coli was transformed to obtain the recombinant plasmid. PCR and restriction enzyme digestion were used to identify the positive clones and the bands with a consistent size with the target fragment are shown in Fig. 12. The recombinant plasmids were sequenced and confirmed by BGI to be identical with the 3′-UTR amplified sequences available in the GenBank database.
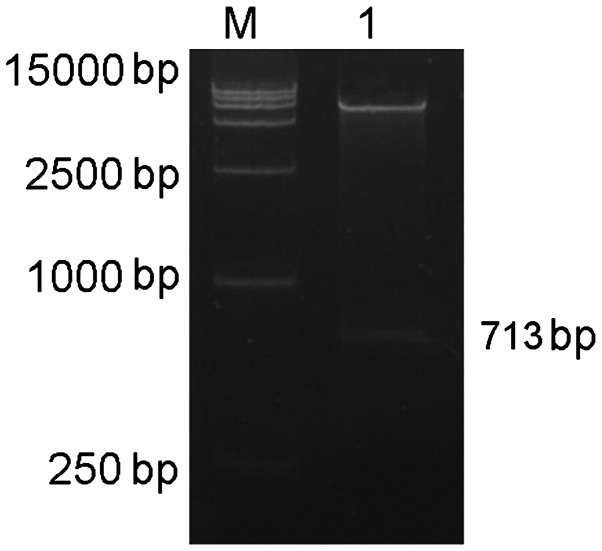
Figure 12.
Restriction enzyme digestion map of the EphA2 3′-UTR pmirGLO plasmid. M is a molecular weight marker of 15,000 bp and lane 1 shows the EphA2 3′-UTR pmirGLO plasmid. UTR, untranslated region; miR, microRNA.
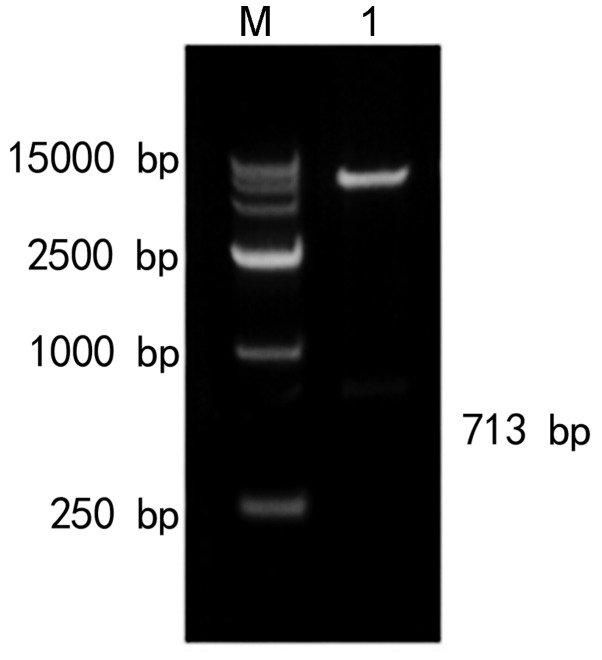
The XhoI and XbaI restriction enzymes were used to perform double digestion of the recombinant plasmid EphA2 3′-UTR pmirGLO-MUT. The recombinant plasmid EphA2 3′-UTR pmirGLO-MUT cleavage results revealed bands of the same size as the fragment, as shown in Fig. 13. The recombinant plasmid and sequencing results matched the expected results.
Figure 13.
Restriction enzyme digestion map of EphA2 3′-UTR pmirGLO-MUT plasmid. M is a molecular weight marker of 15,000 bp and lane 1 shows the EphA2 3′-UTR pmirGLO-MUT plasmid. UTR, untranslated region; miR, microRNA.
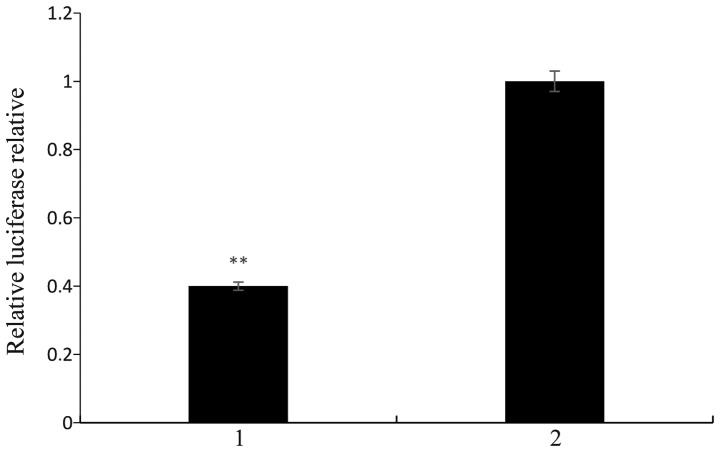
EphA2 identification of the miR-26b target gene
miR-26b mimics and negative control, as well as EphA2 3′-UTR pmirGLO were co-transfected into HeLa cells. Interaction of miR-26b with the candidate target gene EphA2 was indicated by a decrease in luciferase expression of the cells. The luciferase expression was significantly decreased in the miR-26b-transfected group compared with that in the control group (P<0.01), as shown in Fig. 14. This suggested that miR-26b may exert its functions through interacting with the 3′-UTR of EphA2.
Figure 14.
EphA2 identification of the miR-26b target gene. 1 represents EphA2 and 3′-UTR pmirGLO and the miR-26b mimics co-transfected HeLa cells. 2 represents EphA2 3′-UTR pmirGLO and negative control co-transfected HeLa cells. Values are expressed as the mean ± standard error (n=3). **P<0.01. UTR, untranslated region; miR, microRNA.
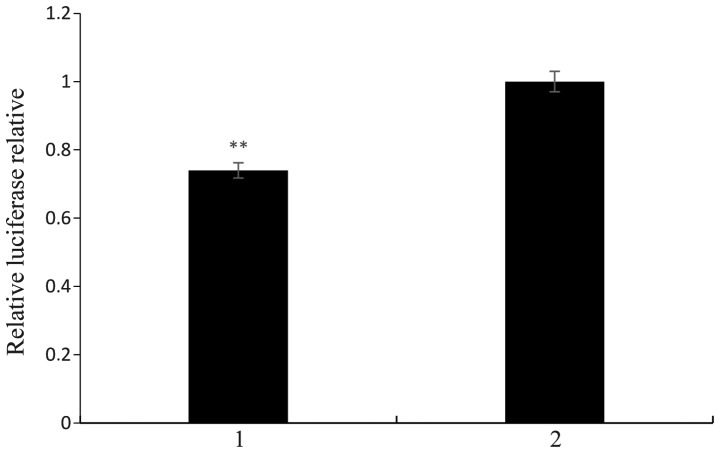
Interaction site recognition of the target gene EphA2 and miR-26b
The dual-luciferase gene reporter assay demonstrated that the seed region of miR-26b specifically binds with the mutation in the 3′-UTR of candidate target gene, causing miR-26b inhibition of the luciferase activity to decrease 60% (100–40%; Fig. 14) and 26% (100–74%; Fig. 15) in the wild-type. This suggested that the predicted binding sites were miR-26b activation sites. The cell luciferase relative expression results following transfection are shown in Fig. 15. The results suggested that the miR-26b seed sequence AUGAACUU exerted a function via binding with the TACTTGAA sequence of the 3′-UTR in EphA2. Therefore, it was hypothesized that the TACTTGAA sequence in the 3′-UTR of EphA2 was the target sequence of miR-26b.
Figure 15.
Identification of miR-26b target sites in EphA2. 1 represents EphA2 and 3′-UTR pmirGLO and the miR-26b mimics co-transfected HeLa cells. 2 represents EphA2 3′-UTR pmirGLO and negative control co-transfected Hela cells. Values are expressed as the mean ± standard error (n=3). **P<0.01. UTR, untranslated region; miR, microRNA.
Discussion
miR-26b is important in the regulation of a number of biological processes in cells (14). It is involved in the regulation of cell proliferation and the control of cell differentiation (15). With the advancement of molecular biology techniques in recent years, investigations into miR-26b have become increasingly in-depth and specific. A previous study using a miRNA chip combined with qPCR techniques found that the placental expression of miR-26b in women with severe pre-eclampsia was different from that in normal pregnant women (16). In additional studies, it was hypothesized that miR-26b was likely to be involved in the differentiation of embryonic stem cells and the regulation of nervous system development via inhibiting the expression of KLF4 and MYCBP (14–17). Subsequently, a study on pituitary and non-pituitary cell lines identified that miR-26b is able to target the 3′-UTR of lef-1, thereby inhibiting the expression of lef-1. In the GH3 cell line, the expression of lef-1 was inhibited while simultaneously promoting the expression of Pit-1 and GH (12). Numerous studies have revealed that miR-26b is involved in early embryonic development, cell proliferation regulation and pituitary hormone secretion, and among other physical activities, it is an miRNA associated with the function of propagation (18–20).
Therefore, the present study investigated the differences in the expression of miR-26b in the pituitary tissue of Yanbian cattle, predicted target genes of miR-26b and selected reproductive trait-associated candidate target genes for identification. In the present study, the RT-qPCR method was used to detect the relative expression levels of miR-26b in the pituitary tissues of Yanbian cattle at different developmental stages. The 2−ΔΔCt calculation method was used to perform the relative quantitative analysis. The results revealed that miR-26b was expressed in the pituitary tissue of six- and 24-month-old Yanbian cattle, and the relative expression levels of miR-26b exhibited significant differences between the pituitary tissues at the different developmental stages. The relative expression of miR-26b in the pituitary tissues of 24-month-old Yanbian cattle was 2.41 times that of six-month-old Yanbian cattle. As studies have revealed that miR-26b is capable of affecting the secretion of pituitary hormones in mice (18,21–23) and the relative expression of miR-26b revealed significant differences in the pituitary tissue of six- and 24-month-old Yanbian cattle in the present study, it was hypothesized that it may be associated with the development of the pituitary gland.
The TargetScan and RNA22 prediction databases were used to analyze and predict the miR-26b sequence provided on miRBase. It was identified that the TACTTGAA sequence in the 3′-UTR of EphA2 and miR-26b seed sequence AUGAACUU had conserved binding sites, suggesting that miR-26b was a target gene. EphA2 is a member of the receptor tyrosine kinase family. The Eph receptor and ligand-mediated signal transduction are involved in numerous physical processes, including nerve signal transduction, angiogenesis and regulation of the adhesion state among cells (24–26). The present study found that EphA2 was widely expressed in human epithelial cells, and after binding to its ligand, the signaling pathway may be activated, regulating cell differentiation and adhesion, inhibiting vascular endothelial growth factor and epithelial growth factor and inducing cell proliferation (27); therefore, it may be involved in embryo implantation (28). The RT-qPCR results revealed that in the pituitary tissue of six-and and 24-month-old Yanbian cattle, the relative expression of miR-26b exhibited the opposite expression pattern and the relative expression of miR-26b in the pituitary tissue of 24 month-old Yanbian cattle was 2.41 times that of the six-month-old Yanbian cattle. The relative expression of candidate target gene EphA2 in the pituitary tissue of six-month-old Yanbian cattle was 3.34 times that of 24-month-old Yanbian cattle. The results provided a theoretical basis for the EphA2 gene being a target gene of miR-26b. However, there are miR-26b binding sites in the EphA2 mRNA 3′-UTR of bovine, human, murine and other mammalian genomes, suggesting that the EphA2 gene may be a target gene of miR-26b.
The luciferase reporter system assay revealed that miR-26b is able to suppress EphA2 expression at the gene level. The 3′-UTR reporter plasmid of mutant candidate target gene EphA2 termed EphA2 3′-UTR pmirGLO-MUT and miR-26b mimics were co-transfected into HeLa cells. A dual-luciferase gene detection reporter assay revealed that there were three mutations in the binding sites of the miR-26b seed region and the 3′-UTR of the candidate target gene, EphA2. It may be the cause of the decrease in the miR-26b inhibition of luciferase activity from 60% in the wild-type to 26%, suggesting that miR-26b exerted its effects via binding with the 3′-UTR in EphA2. It also revealed that the TACTTGAA sequence in the 3′-UTR of EphA2 was the binding site of miR-26b. miR-26b is able to inhibit EphA2 mRNA and protein expression in human glioma cells, as it was previously demonstrated that EphA2 was the direct target gene of miR-26b in glioma cells (29); these results were consistent with those of the present study.
In conclusion, the results of the present study have confirmed in vitro that EphA2 is a target gene of miR-26b in Yanbian cattle. However, the function, regulatory mechanism and impact of miR-26b on reproductive traits, including early embryonic development and pituitary hormone secretion, remains to be further investigated.
Acknowledgments
The current study was supported by the National Key Technology R&D Program (grant nos. 2015BAI07B02 and 20150101102JC), the National Beef Cattle Industrial System of the China Agriculture Research System (grant no. 38) and the National High-tech R&D Program (grant no. 2013AA102505).
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beilharz TH, Humphreys DT, Preiss T. miRNA Effects on mRNA closed-loop formation during translation initiation. Prog Mol Subcell Biol. 2010;50:99–112. doi: 10.1007/978-3-642-03103-8_7. [DOI] [PubMed] [Google Scholar]
- 4.Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19:2009–2020. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trakooljul N, Hicks JA, Liu HC. Identification of target genes and pathways associated with chicken microRNA miR-143. Anim Genet. 2010;41:357–364. doi: 10.1111/j.1365-2052.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 6.Bizuayehu TT, Fernandes JM, Johansen SD, Babiak I. Characterization of novel precursor miRNAs using next generation sequencing and prediction of miRNA targets in Atlantic halibut. PLoS One. 2013;8:e61378. doi: 10.1371/journal.pone.0061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros-Carvalho GA, Paschoal AR, Marcelino-Guimarães FC, Hungria M. Prediction of potential novel microRNAs in soybean when in symbiosis. Genet Mol Res. 2014;13:8519–8529. doi: 10.4238/2014.October.20.28. [DOI] [PubMed] [Google Scholar]
- 8.Verghese ET, Drury R, Green CA, Holliday DL, Lu X, Nash C, Speirs V, Thorne JL, Thygesen HH, Zougman A, Hull MA, et al. MiR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol. 2013;231:388–399. doi: 10.1002/path.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Wernet P. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics. 2013;14:111. doi: 10.1186/1471-2164-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alijani S, Alizadeh S, Kazemi A, Khatib ZK, Soleimani M, Rezvani M, Minayi N, Karami F, Tayebi B. Evaluation of the effect of miR-26b up-regulation on HbF expression in erythroleukemic K-562 cell line. Avicenna J Med Biotechnol. 2014;6:53–56. [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Zhang L, Huang H, Huang Y, Huang L, Wang J, Huang S, He L, Zhou Y, Jia W. MiR-26b/KPNA2 axis inhibits epithelial ovarian carcinoma proliferation and metastasis through downregulating OCT4. Oncotarget. 2015 doi: 10.18632/oncotarget.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Schmittgen TD, Zakrajsek BA, Mills AG, et al. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of end-point and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, Li P, Hao S, et al. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47:923–929. doi: 10.1515/CCLM.2009.228. [DOI] [PubMed] [Google Scholar]
- 15.Ji Y, He Y, Liu L, Chong X. MiRNA-26b regulates the expression of cyclooxygenase-2 in desferrioxamine-treated CNE cells. FEBS Lett. 2010;584:961–967. doi: 10.1016/j.febslet.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Qian K, Tang ZP, et al. Bioinformatics and microarray analysis of microRNA expression profiles of murine embryonic stem cells, neural stem cells induced from ESCs and isolated from E8.5 mouse neural tube. Neurol Res. 2010;32:603–613. doi: 10.1179/174313209X455691. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Florez S, Gutierrez-Hartmann A, et al. MicroRNAs regulate pituitary development and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression. J Biol Chem. 2010;285:34718–34728. doi: 10.1074/jbc.M110.126441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Kim K, Li X, Moreno M, Sharp T, Goodheart MJ, Safe S, Dupuy AJ, Amendt BA. MicroRNA-26b represses colon cancer cell proliferation by inhibiting lymphoid enhancer factor 1 expression. Mol Cancer Ther. 2014;13:1942–1951. doi: 10.1158/1535-7163.MCT-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palagani A, Op De, Beeck K, Naulaerts S, Diddens J, Sekhar Chirumamilla C, Van Camp G, Laukens K, Heyninck K, Gerlo S, Mestdagh P, et al. Ectopic microRNA-150-5p transcription sensitizes glucocorticoid therapy response in MM1S multiple myeloma cells but fails to overcome hormone therapy resistance in MM1R cells. PLoS One. 2014;9:e113842. doi: 10.1371/journal.pone.0113842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G, Shi C, Ji C, Song G, Chen L, Yang L, Zhao Y, Guo X. Expression of microRNA-26b, an obesity-related microRNA, is regulated by free fatty acids, glucose, dexamethasone and growth hormone in human adipocytes. Mol Med Rep. 2014;10:223–228. doi: 10.3892/mmr.2014.2204. [DOI] [PubMed] [Google Scholar]
- 21.Palumbo T, Faucz FR, Azevedo M, Xekouki P, Iliopoulos D, Stratakis CA. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene. 2013;32:1651–1659. doi: 10.1038/onc.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song G, Xu G, Ji C, Shi C, Shen Y, Chen L, Zhu L, Yang L, Zhao Y, Guo X. The role of microRNA-26b in human adipocyte differentiation and prowliferation. Gene. 2014;533:481–487. doi: 10.1016/j.gene.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Xu G, Ji C, Song G, Shi C, Shen Y, Chen L, Yang L, Zhao Y, Guo X. Obesity associated microRNA26b regulates the proliferation of human preadipocytes via arrest of the G1/S transition. Mol Med Rep. 2015;12:3648–3654. doi: 10.3892/mmr.2015.3858. [DOI] [PubMed] [Google Scholar]
- 24.Kinch MS, Carles-Kinch K. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metastasis. 2003;20:59–68. doi: 10.1023/A:1022546620495. [DOI] [PubMed] [Google Scholar]
- 25.Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19:5614–5619. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbegr IM, Göke M, Kanai M, et al. Epithelial cell kinase-B61: an autocrine loop modulating intestinal epithelial migration and barrier function. Am J Physiol. 1997;273:G824–G832. doi: 10.1152/ajpgi.1997.273.4.G824. [DOI] [PubMed] [Google Scholar]
- 27.Miao H, Wei BR, Peehl DM, et al. Activation of EphA receptor tyrosine kinase inhibits the Ras/MAPK pathway. Nat Cell Biol. 2001;3:527–530. doi: 10.1038/35074604. [DOI] [PubMed] [Google Scholar]
- 28.Fujii H, Tatsumi K, Kosaka K, et al. EPh-ephrin A system regulates murine blastocyst attachment and sreading. Dev Dyn. 2006;235:3250–3258. doi: 10.1002/dvdy.20977. [DOI] [PubMed] [Google Scholar]
- 29.Wu N, Zhao X, Liu M, et al. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. [DOI] [PMC free article] [PubMed] [Google Scholar]