Abstract
The present study aimed to investigate the association between xanthine dehydrogenase (XDH) gene polymorphism and essential hypertension in the rural Han Chinese population of Fuxin, Liaoning. Han Chinese individuals, who had lived in rural areas of Fuxin, were selected as subjects for the present study. A total of 521 unrelated patients with hypertension were selected, along with a further 533 unrelated individuals with normal blood pressure, in order to serve as controls. Five tag single nucleotide polymorphisms (SNP) of the XDH gene were selected. An estimation of SNP allele frequency was determined using DNA pooling and pyrosequencing methods. Prior to Bonferroni correction, T allele frequency for rs206811 was significantly higher in patients with hypertension, as compared with the controls (64.1 vs. 59.4%; P=0.031); C allele frequency for rs1042039 was significantly higher in patients with hypertension, as compared with the controls (66.1 vs. 60.6%; P=0.011), C allele frequency for rs1054889 was significantly lower in patients with hypertension, as compared with the controls (38.8 vs. 44.8%; P=0.007); and A allele frequency for rs2073316 was significantly lower in patients with hypertension, as compared with the controls (29.2 vs. 34.4%; P=0.013). However, once a Bonferroni correction for multiple testing was applied, the XDH gene polymorphisms rs1042039, rs1054889, and rs2073316 were shown to be associated with hypertension (P=0.044, 0.035, and 0.039, respectively). These results suggest that the XDH gene polymorphisms rs1042039, rs1054889, and rs2073316 may be associated with hypertension in the rural Han Chinese population.
Keywords: xanthine dehydrogenase gene, hypertension, Han Chinese, association
Introduction
Essential hypertension (EH) represents a serious public health burden. Furthermore, EH is one of the most common and significant risk factors for coronary heart disease, stroke, and kidney diseases (1). In China, the prevalence of hypertension in adults was 33.5% in 2010, and ~4.5×106 people succumb to hypertension annually (2). A previous study suggested that an important risk factor for hypertension is genetic predisposition (3). In addition, environmental factors, including diet and various lifestyles modify blood pressure levels. The long-term average heritability estimates of systolic blood pressure (SBP) and diastolic blood pressure (DBP) are 0.66 and 0.60, respectively (4). Within the last few decades, the analysis of candidate genes has allowed the prediction of modifiable risk factors, as well as the development of novel treatments for hypertension. However, the extent of the role that genetic predisposition has in EH remains to be elucidated.
EH is currently accepted to be the result of numerous genetic and environmental factors, gene interactions, and gene-environmental interactions. The majority of the mutations responsible for EH directly affect the signal conducting system, or the function of the corticosteroid receptors. Increased renal salt reabsorption is accompanied by increased water reabsorption, resulting in increased blood pressure. It is well-known that hyperuricemia and oxidative stress participate in the pathophysiology of hypertension, and xanthine dehydrogenase (XDH) produces urate in its oxidase isoform, which is a type of reactive oxygen species (5). XDH is associated with the oxidative metabolism of purines, and belongs to the group of molybdenum-containing hydroxylases. XDH converts to xanthine oxidase by reversible sulfhydryl oxidation, or by irreversible proteolytic modification. Previous epidemiological studies have identified an association between increased levels of serum urate and cardiovascular risk among patients with hypertension (6–8), thus suggesting that the XDH gene may be associated with constitutional susceptibility to EH. The human XDH gene is located on chromosome 2p23.1 (9), and consists of 36 exons that encode a 1,333 amino acid protein. The present study carried out the genotyping of tag single nucleotide polymorphisms (SNPs) in both a large hypertensive population and a general population sample, in order to further investigate the association between the XDH gene and EH. The participants were recruited from the rural areas of Fuxin County, Liaoning, China.
Materials and methods
Population selection
The rural areas of Fuxin county, located in the northeast of China, are regions with high mortality due to EH. The prevalence of hypertension in Fuxin is 38.6% (10). During a mean follow-up period of 28 months, ~29.6% of men and 23.4% of women developed EH in the rural areas of Fuxin (11). The salt intake of the population was >15 g/day, and the genetic background was normal. Subjects were selected from the residents living in 49 villages, in order to carry out the present study.
Criteria of subject inclusion and collection of data
Han individuals with a long-standing ancestry in the rural areas of Fuxin were selected as participants for the present study. A total of 521 unrelated hypertensive patients >18 years-old were selected with the following criteria for hypertension: Resting (sitting) systolic blood pressure (SBP) of ≥140 mm Hg, resting (sitting) diastolic blood pressure (DBP) of ≥90 mm Hg, self-report of current antihypertensive treatment, or any combination of the above, and no clinical or biochemical signs of secondary hypertension. A total of 533 unrelated control subjects >40 years-old were also selected with the following criteria: Resting (sitting) SBP of <140 mm Hg, and DBP of <90 mm Hg. Each subject gave a detailed interview regarding their medical history, smoking status, and alcohol use. All subjects were self-identified as having four Han Chinese grandparents. Pregnant women and those with a history of cancer or congestive heart failure were excluded from the present study. The study was approved by the ethics committee of Shengjing Hospital of China Medical University (Shenyang, China), and all subjects provided written informed consent.
A standard questionnaire was used to collect demographic data (age, ethnicity), information on alcohol consumption, smoking, diabetes, and hypertension history including medication usage. All participants underwent complete physical examinations prior to blood sampling. Body weight, height, and waist circumference (WC) were measured with the participants wearing light clothing, without shoes. The body mass index (BMI) of the subjects was calculated using the following equation: BMI = weight (kg)/height (m2). The blood pressure of subjects was measured when they were in a seated position, with three blood pressure values being measured in a single visit, and the average of the last two measurements was recorded. The subjects were required to fast for ≥12 h prior to blood sampling. The levels of serum total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglyceride (TG), serum creatinine, uric acid (UA), and fasting plasma glucose (FPG) were analyzed using an Olympus AU640 autoanalyzer (Olympus Corporation, Tokyo, Japan). Genomic DNA was extracted using a TIANamp Blood DNA kit (Tiangen Biotech, Co., Ltd., Beijing, China).
Hypertension is defined as an average SBP (≥140 mm Hg) and/or an average DBP (≥90 mm Hg), and/or the self-report of current treatment for hypertension with antihypertensive medication (12).
Construction of DNA pooling
Large-scale genotyping was performed on pooled DNA samples. DNA quality and quantity were determined using a NanoDrop® spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). DNA samples with a 260/280 ratio of 1.65–2.0, and a 260/230 ratio of 1.0–2.2 were considered acceptable for pooling. A total of 21 DNA samples from hypertensive subjects and 33 DNA samples from control subjects were excluded from the present study, due to the fact that the DNA was unsuccessfully extracted or unqualified. Therefore, the DNA samples from 1,000 subjects were statistically analyzed. The DNA samples were serially diluted to a concentration of 25 ng/µl. Following dilution, each DNA sample was quantified twice using the NanoDrop® spectrophotometer, and the mean was used to determine the concentration. A total of 100 ng individual DNA was assigned to individual DNA pools, yielding a total of ten independent DNA pools. The ten independent DNA pools were composed of five hypertensive pools and five controls pools, and each DNA pool included the DNA samples from 100 individuals.
Selection of SNPs and estimation of allele frequency
All the candidate loci of the XDH gene were assessed for tag SNPs using the HapMap database (http://www.hapmap.org/). SNPs which tagged common variants (minor allele frequency [MAF] ≥0.1) were selected in the HapMap CHB reference panel with r2≥0.9. The following tag SNPs of XDH were selected for further study: rs206811, rs1042039, rs1054889, rs2073316 and rs7574920. The allele frequency estimation of the tag SNPs in each DNA pool was determined using a pyrosequencing (PSQ) assay (Qiagen China, Co, Ltd., Shanghai, China) of polymerase chain reaction (PCR)-amplified genes according to the manufacturer's instructions (13). Allele frequency was measured three times for each pooled sample. PSQ assay software (PSQ96™ System; Pyrosequencing AB, Uppsala, Sweden) was used to design the PCR amplification and PSQ primers (Table I). All primer sequences were provided in the 5′-3′ direction and were purchased from Takara (Shiga, Japan). Each biotinylated primer was indicated with 'Bio' preceding the oligonucleotide sequence. The PCR reaction mix (final volume, 40 µl) consisted of 1 µl cDNA sample, 4 µl 10X buffer (Takara), 3.2 µl dNTP (2.5 mmol/l; Takara), 0.4 µl forward primer (10 mmol/l), 0.4 µl reverse primer (10 mmol/l), 0.4 µl Takara hotstart Taq, and 30.6 µl ddH2O. The PCR was initially performed for 3 min at 95°C, followed by 45 cycles of 10 sec at 95°C, 20 sec at 60°C, 30 sec at 72°C, and a final 5 min elongation step at 72°C.
Table I.
Polymerase chain reaction amplification primers, and pyrosequencing reaction extended primers (5′-3′).
| SNP | Primer sequence |
|---|---|
| rs206811 | |
| Forward | ATACTGACACCTTGTGGCAAACCA |
| Bio extended | AAGTAGGTAACAGATTTGCAGCTTACAT |
| Reverse | TTCATAAAGGGCATCTC |
| rs1042039 | |
| Forward | AGTAATCTTGCTTTATGCAGCTTCACA |
| Bio extended | AAATTTTGCAATGAGGCAGTGG |
| Reverse | CTTTCCAATGATTCAAAG |
| rs1054889 | |
| Forward | CCCTTTTATTGCTCCAAATGGTGAT |
| Bio extended | TCTCTGGCAGAAGGTTGGATTTATACA |
| Reverse | TCACTGTCACATGCCT |
| rs2073316 | |
| Forward | TGCCATCACCGACTCACC |
| Bio extended | CCTGGGGGTAATCTGAGACTTAGAG |
| Reverse | CCAGGGACATGGAGC |
| rs7574920 | |
| Forward | TTGTTTCCCATTCACCACTTGAT |
| Bio extended | CCCATGAAAATGCGATCATTTCTT |
| Reverse | GCTGAATCTGTGGTGAG |
Statistical analysis
All data were expressed as the mean ± standard deviation. All statistical analyses were performed using SPSS 13.0 (SPSS, Inc. Chicago, IL, USA). A Fisher's exact test was applied to examine the association between the XDH gene SNPs and hypertension. Bonferroni correction was used for multiple testing as five polymorphisms were associated with hypertension. P<0.05 was considered to indicate a statistically significant difference.
Results
No significant difference was observed in the age, gender, FPG, HDL-C, and TG of subjects with EH, as compared with the controls (P>0.05). Conversely, WC, BMI, TC, LDL-C, Cr, and UA were significantly higher in the hypertensive patients, as compared with the controls (P<0.05). The basic characteristics of the subjects are summarized in Table II.
Table II.
Basic characteristics of the selected subjects.
| Parameter | Case (n=500) | Control (n=500) |
|---|---|---|
| Age (years) | 60±11 | 59±9 |
| Gender (male/female) | 249/251 | 238/262 |
| SBP (mmHg) | 162±21 | 120±10a |
| DBP (mmHg) | 98±23 | 76±8a |
| WC (cm) | 89±16 | 81±13a |
| BMI (kg/m2) | 23.92±3.55 | 22.81±2.90a |
| FPG (mmol/l) | 5.75±2.02 | 5.72±2.01 |
| TC (mmol/l) | 5.38±1.08 | 5.16±1.02a |
| LDL-C (mmol/l) | 3.05±0.85 | 2.81±0.71a |
| HDL-C (mmol/l) | 1.41±0.31 | 1.47±0.35 |
| TG (mmol/l) | 1.70±1.17 | 1.68±1.05 |
| Cr (µmol/l) | 90.11±12.35 | 78.55±9.78a |
| UA (µmol/l) | 435.6±102.3 | 387.3±83.7a |
SBP, systolic blood pressure; DBP, diastolic blood pressure; WC, waist circumference; BMI, body mass index; FPG, fasting plasma glucose; TC, total cholesterol; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TG, triglyceride; Cr, serum creatinine; UA, uric acid.
P<0.05, vs. hypertensive subjects.
For each pooled sample, the peak height of the SNP alleles was determined using the PSQ96 AQ software, and the ratio of one allele peak height to the sum of the heights of both allele peaks was calculated. Peak values of five SNPs of controls and hypertensives were shown in Figs. 1–10. Using the standard curve, the SNP allele peak height ratios were converted to allele frequencies in the DNA pools. Allele frequency for each DNA pool was calculated as the mean value of the three replicates of each pooled sample. Using the previously determined allele frequencies of each DNA pool, the mean value of five DNA pools was then calculated in order to represent the allele frequency in hypertensives and controls. Prior to the application of Bonferroni correction, T allele frequency for rs206811 was significantly higher in hypertensives, as compared with controls (64.1 vs. 59.4%, P=0.031); C allele frequency for rs1042039 was significantly higher in hypertensives, as compared with controls (66.1 vs. 60.6%, P=0.011), C allele frequency for rs1054889 was significantly lower in hypertensives, as compared with controls (38.8 vs. 44.8%, P=0.007); and A allele frequency for rs2073316 was significantly lower in hypertensives, as compared with the controls (29.2 vs. 34.4%, P=0.013). Following Bonferroni correction for multiple testing, the rs1042039, rs1054889, and rs2073316 polymorphisms of the XDH gene were associated with hypertension (P=0.044, 0.035, and 0.039, respectively). All results are shown in Table III.
Figure 1.
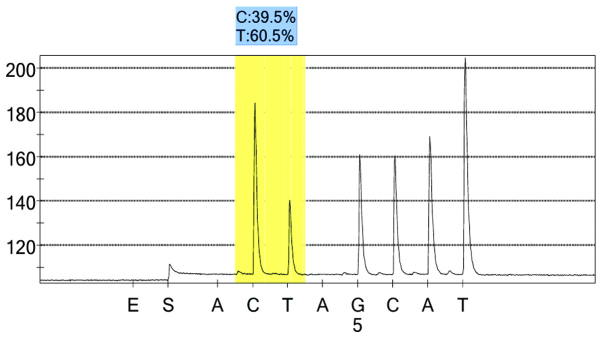
rs206811 control group 1 peak value.
Figure 2.

rs206811case group 1 peak value.
Figure 3.
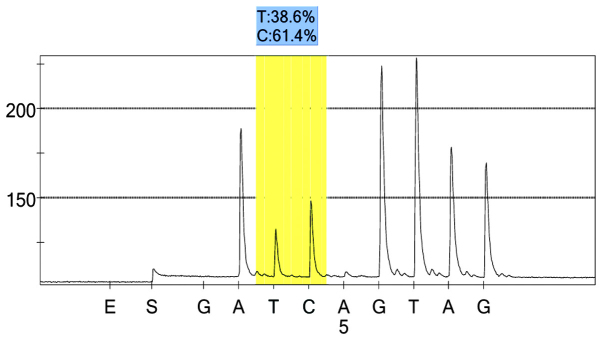
rs1042039 control group 1 peak value.
Figure 4.
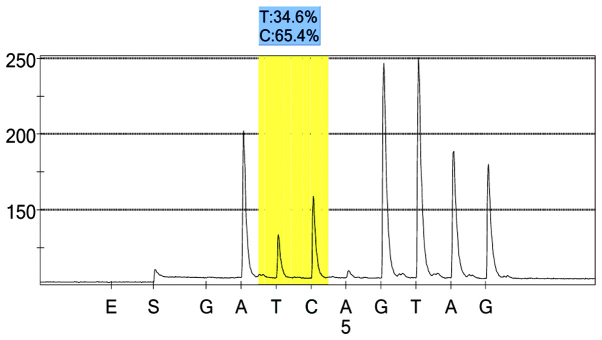
rs1042039 case group 1 peak value.
Figure 5.
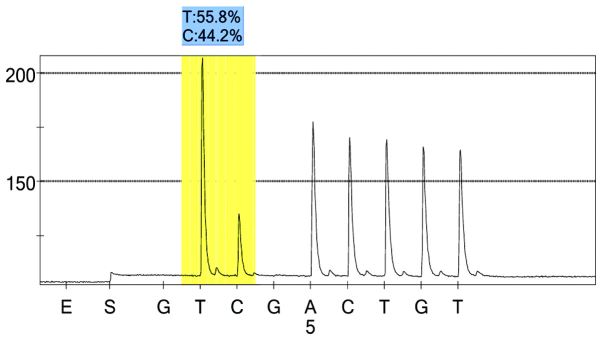
rs1054889 control group 1 peak value.
Figure 6.
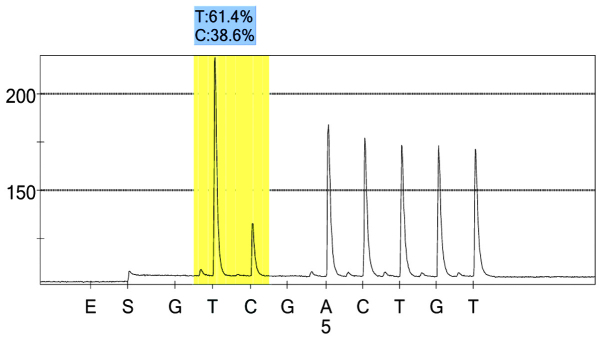
rs1054889 case group 1 peak value.
Figure 7.
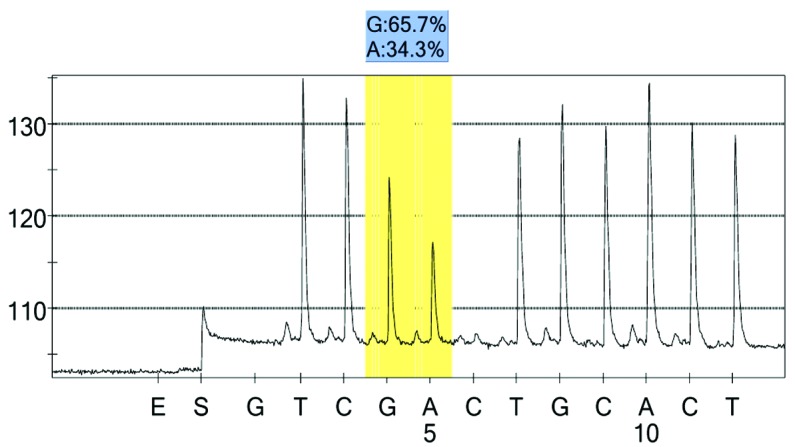
rs2073316 control group 1 peak value.
Figure 8.
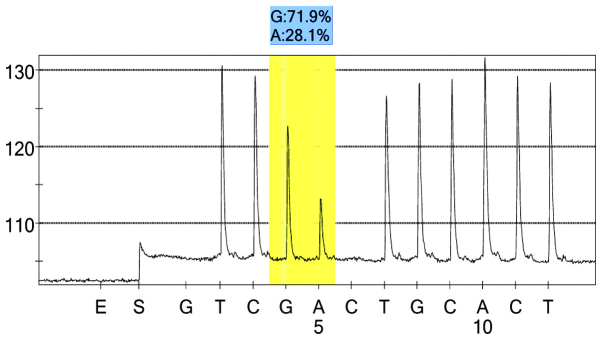
rs2073316 case group 1 peak value.
Figure 9.
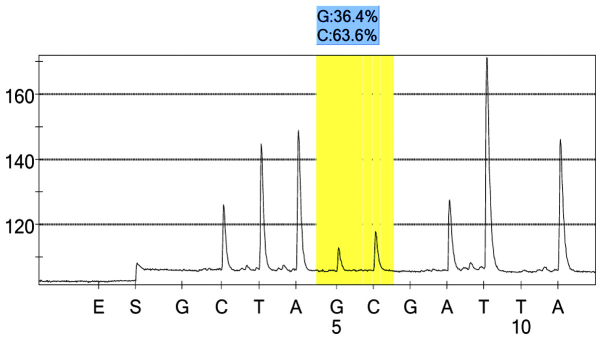
rs7574920 control group 1 peak value.
Figure 10.
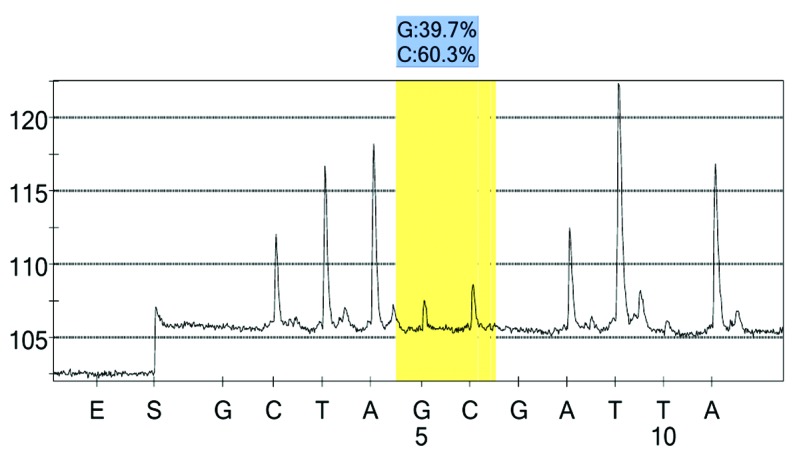
rs7574920 case group 1 peak value.
Table III.
Correlation analysis between xanthine dehydrogenase tag single nucleotide polymorphisms and hypertension.
| SNP | Minor allele frequency (%)
|
P-value | Pc-value | OR | 95% CI | |
|---|---|---|---|---|---|---|
| Case group | Control group | |||||
| rs206811 | T (64.1) | T (59.4) | 0.031 | 0.062 | 1.220 | 1.019–1.462 |
| rs1042039 | C (66.1) | C (60.6) | 0.011 | 0.044 | 1.268 | 1.056–1.521 |
| rs1054889 | C (38.8) | C (44.8) | 0.007 | 0.035 | 0.781 | 0.654–0.933 |
| rs2073316 | A (29.2) | A (34.4) | 0.013 | 0.039 | 0.786 | 0.651–0.950 |
| rs7574920 | C (60.9) | C (63.9) | 0.166 | 0.166 | 0.880 | 0.734–1.055 |
SNP, single nucleotide polymorphism; CI, confidence interval; OR, odds ratio; Pc, Pearson coefficient.
Discussion
PSQ provides an accurate and valid approach to allele frequency estimation in pooled DNA samples. Allele frequencies in pooled DNA samples are strongly correlated with those of individual DNA samples across a wide range of allele frequencies (2–50%). PSQ is able to detect an allele frequency difference of <2% between pools, indicating that this method may be sensitive enough for use in association studies involving complex diseases with a small difference in allele frequency between the subjects of interest, and the controls. Therefore, PSQ may provide an efficient method for reducing the labor and expense involved in similar experiments (14,15).
The sample size of the present study was small; however recruiting experimental subjects in rural China had benefits, insofar as the Han population exhibits relative genetic homogeneity. The selection of rural study sites was based on the homogeneity of the study population regarding their ethnicity and environmental exposures, including lifestyle, nutritional factors and habitual diet. The residents of these regions were of Han nationality, the ethnic majority in China.
Inherited XDH deficiency was first reported by Dent et al (16), following which the importance of the XDH gene became apparent (5,17–20). One of the mechanisms that underlies high-salt diet-induced high blood pressure is oxidative stress (21,22). In normotensive Sprague-Dawley rats with high-salt diets, a moderate blood pressure increase was observed (23). In the present study, the levels of UA in hypertensives was higher, as compared with the controls, which suggests that hyperuricemia may be associated with hypertension. Xanthine oxidoreductase is able to convert hypoxanthine to xanthine and xanthine to uric acid via its two inter-convertible isoforms, XDH and xanthine oxidase (XO). Reactive oxygen species (ROS) have important roles in the pathogenesis of numerous cardiovascular diseases, such as hypertension, atherosclerosis, diabetes mellitus, and heart failure (24). The conversion of XDH to XO, and the increase of XO activity have been reported in hypertension (25–27). Furthermore, treatment with XO inhibitors has been shown to normalize ROS levels in the microvessels of rats fed a high-salt diet (28). These results suggested that XO may be an important source of ROS in patients with hypertension.
In the present study, tag SNPs were selected to perform a population-based association analysis. Among the five SNPs, rs1042039 and rs1054889 were located on the 3′-untranslated region, and the other three SNPs were located in the introns. Preliminary results demonstrated that rs206811, rs1042039, rs1054889, and rs2073316 were significantly associated with hypertension. However, when a Bonferroni correction for multiple testing was applied, rs1042039, rs1054889, and rs2073316 were associated with hypertension. These results suggested that polymorphisms of rs1042039, rs1054889, and rs2073316 may serve as regulatory regions for the XDH gene, which may regulate XDH expression, and further participate in blood pressure regulation.
A previous study reported that the XDH gene was also associated with hypertension in the Japanese population (5). However, in the Japanese study, only common SNPs located on exons were selected. Many reasons may explain this, such as ethnicity, geography, and SNP selection differences.
In conclusion, rs1042039, rs1054889, and rs2073316 polymorphisms of the XDH gene may be associated with EH in the rural Han Chinese population in Fuxin, Liaoning. However, future association studies using larger sample sizes and more diverse populations are required in order to confirm these preliminary results.
Acknowledgments
The present study was supported by the Prophase of National Key Basic Research Development Plan (grant no. 2010CB535000) and the National Natural Science Fund of China (grant no. 81203002).
References
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: Analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)70151-3. [DOI] [PubMed] [Google Scholar]
- 2.Li YC, Wang LM, Jiang Y, Li XY, Zhang M, Hu N. Prevalence of hypertension among Chinese adults in 2010. Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46:409–413. In Chinese. [PubMed] [Google Scholar]
- 3.Rutherford PA. Genetic influences in human hypertension. J Hypertens. 2003;21:19–22. doi: 10.1097/00004872-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Kamide K, Kokubo Y, Takiuchi S, Horio T, Matayoshi T, Yasuda H, Miwa Y, Yoshii M, Yoshihara F, et al. Associations of hypertension and its complications with variations in the xanthine dehydrogenase gene. Hypertens Res. 2008;31:931–940. doi: 10.1291/hypres.31.931. [DOI] [PubMed] [Google Scholar]
- 6.Yü TF, Berger L, Dorph DJ, Smith H. Renal function in gout. V. Factors influencing the renal hemodynamics. Am J Med. 1979;67:766–771. doi: 10.1016/0002-9343(79)90732-0. [DOI] [PubMed] [Google Scholar]
- 7.Alderman MH, Cohen H, Madhavan S, Kivlighn S. Serum uric acid and cardiovascular events in successfully treated hypertensive patients. Hypertension. 1999;34:144–150. doi: 10.1161/01.HYP.34.1.144. [DOI] [PubMed] [Google Scholar]
- 8.Verdecchia P, Schillaci G, Reboldi G, Santeusanio F, Porcellati C, Brunetti P. Relation between serum uric acid and risk of cardiovascular disease in essential hypertension. The PIUMA study. Hypertension. 2000;36:1072–1078. doi: 10.1161/01.HYP.36.6.1072. [DOI] [PubMed] [Google Scholar]
- 9.Minoshima S, Wang Y, Ichida K, Nishino T, Shimizu N. Mapping of the gene for human xanthine dehydrogenase (oxidase) (XDH) to band p23 of chromosome 2. Cytogenet Cell Genet. 1995;68:52–53. doi: 10.1159/000133887. [DOI] [PubMed] [Google Scholar]
- 10.Sun Z, Zheng L, Detrano R, Zhang D, Zhang X, Xu C, Li J, Liu S, Li J, Hu D, Sun Y. The accelerating epidemic of hypertension among rural Chinese women: Results from Liaoning Province. Am J Hypertens. 2008;21:784–788. doi: 10.1038/ajh.2008.170. [DOI] [PubMed] [Google Scholar]
- 11.Sun Z, Zheng L, Detrano R, Zhang X, Xu C, Li J, Hu D, Sun Y. Incidence and predictors of hypertension among rural Chinese adults: Results from Liaoning province. Ann Fam Med. 2010;8:19–24. doi: 10.1370/afm.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu D, Reynolds K, Wu X, Chen J, Duan X, Muntner P, Huang G, Reynolds RF, Su S, Whelton PK, He J, InterASIA Collaborative Group The International Collaborative Study of Cardiovascular Disease in ASIA. Prevalence, awareness, treatment, and control of hypertension in china. Hypertension. 2002;40:920–927. doi: 10.1161/01.HYP.0000040263.94619.D5. [DOI] [PubMed] [Google Scholar]
- 13.Royo JL, Hidalgo M, Ruiz A. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc. 2007;2:1734–1739. doi: 10.1038/nprot.2007.244. [DOI] [PubMed] [Google Scholar]
- 14.Lavebratt C, Sengul S. Single nucleotide polymorphism (SNP) allele frequency estimation in DNA pools using Pyrosequencing. Nat Protoc. 2006;1:2573–2582. doi: 10.1038/nprot.2006.442. [DOI] [PubMed] [Google Scholar]
- 15.Gruber JD, Colligan PB, Wolford JK. Estimation of single nucleotide polymorphism allele frequency in DNA pools by using Pyrosequencing. Hum Genet. 2002;110:395–401. doi: 10.1007/s00439-002-0722-6. [DOI] [PubMed] [Google Scholar]
- 16.Dent CE, Philpot GR. Xanthinuria, an inborn error (or deviation) of metabolism. Lancet. 1954;266:182–185. doi: 10.1016/S0140-6736(54)91257-X. [DOI] [PubMed] [Google Scholar]
- 17.Xu P, Huecksteadt TP, Hoidal JR. Molecular cloning and characterization of the human xanthine dehydrogenase gene (XDH) Genomics. 1996;34:173–180. doi: 10.1006/geno.1996.0262. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Xanthine dehydrogenase (XDH): Episodic evolution of a “neutral” protein. J Mol Evol. 2001;53:485–495. doi: 10.1007/s002390010239. [DOI] [PubMed] [Google Scholar]
- 19.Eggermann T, Spengler S, Denecke B, Zerres K, Mache CJ. Multi-exon deletion in the XDH gene as a cause of classical xanthinuria. Clin Nephrol. 2013;79:78–80. doi: 10.5414/CN106994. [DOI] [PubMed] [Google Scholar]
- 20.Boban M, Kocic G, Radenkovic S, Pavlovic R, Cvetkovic T, Deljanin-Ilic M, Ilic S, Bobana MD, Djindjic B, Stojanovic D, et al. Circulating purine compounds, uric acid, and xanthine oxidase/dehydrogenase relationship in essential hypertension and end stage renal disease. Ren Fail. 2014;36:613–618. doi: 10.3109/0886022X.2014.882240. [DOI] [PubMed] [Google Scholar]
- 21.Briones AM, Touyz RM. Oxidative stress and hypertension: Current concepts. Curr Hypertens Rep. 2010;12:135–142. doi: 10.1007/s11906-010-0100-z. [DOI] [PubMed] [Google Scholar]
- 22.Kawada N, Imai E, Karber A, Welch WJ, Wilcox CS. A mouse model of angiotensin II slow pressor response: Role of oxidative stress. J Am Soc Nephrol. 2002;13:2860–2868. doi: 10.1097/01.ASN.0000035087.11758.ED. [DOI] [PubMed] [Google Scholar]
- 23.Carlström M, Sällström J, Skøtt O, Larsson E, Persson AE. Uninephrectomy in young age or chronic salt loading causes salt-sensitive hypertension in adult rats. Hypertension. 2007;49:1342–1350. doi: 10.1161/HYPERTENSIONAHA.107.087213. [DOI] [PubMed] [Google Scholar]
- 24.Cai H, Harrison DG. Endothelial dysfunction in cardiovascular diseases: The role of oxidant stress. Circ Res. 2000;87:840–844. doi: 10.1161/01.RES.87.10.840. [DOI] [PubMed] [Google Scholar]
- 25.Laakso J, Mervaala E, Himberg JJ, Teräväinen TL, Karppanen H, Vapaatalo H, Lapatto R. Increased kidney xanthine oxidoreductase activity in salt-induced experimental hypertension. Hypertension. 1998;32:902–906. doi: 10.1161/01.HYP.32.5.902. [DOI] [PubMed] [Google Scholar]
- 26.Wallwork CJ, Parks DA, Schmid-Schönbein GW. Xanthine oxidase activity in the dexamethasone-induced hypertensive rat. Microvasc Res. 2003;66:30–37. doi: 10.1016/S0026-2862(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 27.Laakso JT, Teräväinen TL, Martelin E, Vaskonen T, Lapatto R. Renal xanthine oxidoreductase activity during development of hypertension in spontaneously hypertensive rats. J Hypertens. 2004;22:1333–1340. doi: 10.1097/01.hjh.0000125441.28861.9f. [DOI] [PubMed] [Google Scholar]
- 28.Lenda DM, Boegehold MA. Effect of a high-salt diet on oxidant enzyme activity in skeletal muscle microcirculation. Am J Physiol Heart Circ Physiol. 2002;282:H395–H402. doi: 10.1152/ajpheart.0354.2001. [DOI] [PubMed] [Google Scholar]


