Abstract
The aim of the present study was to investigate the presence and biological function of microRNA-92a (miR-92a) in chondrogenesis and cartilage degeneration. Human adipose-derived mesenchymal stem cells (hADSCs) in micromass and chondrocyte-like ATDC5 cells were induced to chondrogenesis, and primary human/mouse chondrocytes (PHCs/PMCs) and chondrogenic ATDC5 cells were stimulated with interleukin-1β (IL-1β). An miR-92a mimic/inhibitor was transfected into the ATDC5 cells using lipofectamine 2000. Gene expression was analyzed using reverse transcription-quantitative polymerase chain reaction. Alcian blue was used to stain the cartilage nodules and chondrogenic micromass. The potential target genes, signaling pathways and functions of miR-92a were examined using miRanda, miRDB, CLIP-Seq, TargetScan and Kyoto Encyclopedia of Genes and Genomes. The expression of miR-92a was elevated in the chondrogenic ATDC5 cells and hADSCs, and also in the IL-1β-induced ATDC5 cells, PMCs and PHCs. Forced expression of miR-92a enhanced the expression levels of col9a2 and aggrecan. A total of 279 genes were predicted as potential target genes of miR-92a. The phosphoinositide 3-kinase/PI3K)-Akt, ErbB and focal adhesion kinase pathways, extracellular matrix (ECM)-receptor interaction and the mammalian target of rapamycin (mTOR) signaling pathway were suggested to mediate the effects of miR-92a on chondrogenesis and cartilage degeneration. These results demonstrated that miR-92a was involved in chondrogenesis and the chondrocyte response induced by IL-1β. miR-92a positively contributed to the expression of col9a2 and of aggrecan.
Keywords: chondrogenesis, cartilage, osteoarthritis, microRNA-92a, col9a2
Introduction
Cartilage tissues are degenerated and are destroyed in osteoarthritic joints, which are more prevalent in elderly individuals (1). Although arthroplasty can efficiently relieve the symptoms of osteoarthritis, implant loosening is inevitable in the years following arthroplasty (1). Tissue engineered cartilage has been suggested as an improved substitution for conventional arthroplasty. Therefore, it is necessary to understand the molecular mechanisms underlying cartilage generation and degeneration.
Subsequent to mesenchymal condensation, mesenchymal stem cells sense cell-cell and cell-extracellular matrix (ECM) contact, which is termed focal adhesion (2), followed by differentiation into chondrocytes and expression of ECM. There are other exogenous stimuli and intracellular signaling pathways regulating chondrogenesis and cartilage degeneration, including the phosphoinositide 3-kinase(PI3K)-Akt, mammalian target of rapamycin (mTOR) and epidermal growth factor pathways (3–7).
MicroRNAs (miRNAs) are short, non-coding single-stranded RNAs, which have been identified as important post-transcriptional regulators. miRNAs specifically bind to the 3′-untranslational region (UTR) of target gene mRNAs by complementary base pairing in the RNA-induced silencing complex, and they degrade mRNA or repress the translation of target genes (8). miRNAs are essential for multiple biological processes, including cartilage formation and degeneration (9).
A previous study profiled the miRNA expression levels in chondrogenic human adipose-derived mesenchymal stem cells (hADSCs), and observed that the expression levels of miRNA-92a (miR-92a) were significantly altered (10). The present study hypothesized that miR-92a is involved in chondrogenesis and cartilage degeneration, and investigated the presence and biological function of miR-92a in chondrogenesis and cartilage degeneration.
Materials and methods
The Ethics Committee of Sun Yat-Sen University (Guangzhou, China) approved the experiments performed in the present study. Procedures involving human subjects were performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Informed consent was obtained from the patients prior to inclusion in the study. The experiments involving mice were performed in accordance with the Laboratory Animal Center of Sun Yat-Sen University and the Guide for the Care and Use of Laboratory Animals.
Primary chondrocyte isolation
Subsequent to obtaining informed consent, primary human chondrocytes (PHCs) were isolated from the cartilage of patients undergoing hip surgery. The patients included two females, aged 31 and 24 years, who were undergoing surgery for a femoral neck fracture at the First Affiliated Hospital of Sun Yat-Sen University. Patients with degraded cartilages, local or systemic immunological disorders or tumors were excluded from the investigation. The cartilage was carefully cut into sections and digested sequentially in pronase (cat. no. 10165921001; Roche Diagnostics, Basel, Switzerland) for 90 min and collagenase P (cat. no. 11213865001; Roche Diagnostics) for ~7 h on a 37°C stirring-plate. The chondrocytes were then collected by centrifugation (1,000 × g for 3 min) of the digestion solution and then were rinsed with Ca/Mg-free phosphate-buffered saline (cat. no. 14190-094; Gibco Life Technologies, Paisley, UK) three times. The chondrocytes were seeded into flasks containing Dulbecco's modified Eagle's medium (DMEM/F12; cat. no. SH30023.01B; GE Healthcare Life Sciences, Logan, UT, USA) with 5% fetal bovine serum (FBS; cat. no. 12657; Gibco Life Technologies), 2% penicillin and streptomycin (cat. no. 15140-122; Gibco Life Technologies,) and ITS+ Premix (cat. no. 354352; BD Biosciences, Franklin Lakes, NJ, USA).
The isolation of primary mouse chondrocytes (PMCs) was performed, as previously reported (11). A total of 25 male mice were purchased from the Laboratory Animal Center of Sun Yat-Sen University (Guangzhou, China), and were maintained in five isolator cages in pathogen-free conditions. Mice were fed by the Laboratory Animal Center, and maintained at 18–22°C under 12 h light/dark cycles, until the end of the experiments. The mice were washed with 70% ethanol prior to sacrifice. The mice were sacrificed via cervical dislocation under anesthesia with a sponge of 50 ml anhydrous diethyl ether (Laboratory Animal Center of Sun Yat Sen University). Cartilage was carefully collected from the femoral head, femoral condyle and tibial plateau, and was digested in 3 mg/ml collagenase D (cat. no. 11088866001; Roche Diagnostics) at 37°C with agitation for 90 min, followed by further digestion in collagenase D overnight. The cells were collected, filtered through a 48 µm nylon mesh, and were then seeded into a culture flask at a density of 8,000 cells/cm2 in M199 (Gibco Life Technologies; 10% FBS, 1% penicillin and streptomycin).
Cell culture and chondrogenic differentiation
The hADSCs were purchased from Cyagen Bioinformatics (Suzhou), Inc. (cat. no. HUXMD-01001; Taicang, China). The hADSCs were maintained to permit expansion in DMEM (Cyagen Bioinformatics (Suzhou), Inc. 11965–092; Gibco Life Technologies) with 10% FBS, 100 µ/ml penicillin and 100 mg/ml streptomycin at 37°C in 5% CO2 at saturated humidity. The cells were subcultured at 80% confluence and third generation cells were used for the subsequent characterization and chondrogenesis experiments.
The third generation hADSCs were harvested and resuspended in incomplete mesenchymal stem cell chondrogenic differentiation medium [cat. no. GUXMX-90041; Cyagen Bioinformatics (Suzhou), Inc.; 194 ml basal medium, 20 µl dexamethasone, 600 µl ascorbic acid, 2 ml ITS, 200 µl sodium pyruvate, 200 µl proline, 2 ml transforming growth factor-β3] at 2×107 cells/ml. Droplets (12.5 µl) were then carefully added to each well of a 24-well plate. The hADSCs were allowed to adhere at 37°C for 90 min, followed by the addition of 500 µl chondrogenic differentiation medium (10,12–14), which was replaced every 3 days.
ATDC5 mouse cells (Riken Cell Bank, Ibaraki, Japan) were cultured with DMEM/F12, 5% FBS and 1% penicillin and streptomycin in a 37°C, 5% CO2 humidified atmosphere. The culture medium was replaced every 2 days and the cells were subcultured when cells reached 90–100% confluence during the expansion culture. All the experiments were completed within 20 passages. The chondrogenic differentiation was induced using ITS+ Premix (15–17). The chondrogenic culture medium was then replaced daily.
PHCs were cultured in DMEM/F12, 5% FBS, 1% penicillin and streptomycin and ITS+ at 37°C in a 5% CO2 humidified atmosphere.
The PMCs were cultured for expansion in M199 (cat. no. 11150-059; Gibco Life Technologies), 10% FBS, 1% penicillin and streptomycin, basic fibroblast growth factor (cat. no. 450-33; PeproTech, Oak Park, CA, USA) and epidermal growth factor (cat. no. 315-09; PeproTech), at 37°C in a 5% CO2 humidified atmosphere.
Interleukin-1β (IL-1β)-treated chondrocytes
ATDC5 cells were maintained in chondrogenic medium with 1% ITS+ for 14 days at 37°C to form chondrogenic ATDC5 cells. Chondrogenic ATDC5 cells, PHCs and PMCs, at the fourth passage, were treated with recombinant IL-1β (cat. no. 200-01B; PeproTech) at 1 ng/ml for 4 h (18–20).
Morphological analysis
The stained ATDC5 cells were fixed in formalin for 4 h at room temperature, and were stained with 1 mg/ml alcian blue 8GX for 20 min at room temperature, followed by examination using microscopy (Axio Imager Z1; Carl Zeiss AG, Oberkochen, Germany). The micromass was harvested at 0, 7 and 14 days. The macromorphology was examined by imaging with the M205 FA microscope [Leica Microsystems AG, Heerbrugg, Switzerland]. The micromass was fixed in formalin, embedded in paraffin, and stained with alcian blue (14). Images were then captured under microscopy.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays
Total RNA was extracted from the cells using an miRNeasy Mini kit (cat. no. 217004; Qiagen, Hilden, Germany), according to the manufacturer's instructions. The concentration and purity of the extracted RNA was analyzed using an Epoch Multi-Volume Spectrophotometer System (BioTek Instruments, Inc., Winooski, VT, USA). The cDNA of was obtained from mRNA and miRNAs using a PrimeScript® miRNA cDNA Synthesis kit (cat. no. DRR350; Takara Bio, Inc., Otsu, Japan), according to the manufacturer's instructions.
Semi-qPCR was performed using SYBR® Premix Ex Taq™ II (cat. no. DRR081; Takara Bio, Inc.) and a Bio-Rad IQ5 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The concentration of reagents and cycling conditions were according to the manufacturer's instructions. The cycles began at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Ten nanograms of cDNA was added into the 25 µl reaction volume. The primer sequences are presented in Table I. The reverse primer for the miRNAs was Uni-miR qPCR Primer (cat. no. D352; Takara Bio, Inc.). Quality control was performed by monitoring the melting curve. Fold differences in mRNA expression were calculated using the ΔΔCt method (21). All samples were measured in triplicate.
Table I.
Primer sequences for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence (5′-3′) |
|---|---|
| Mmu/hsa-U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| Mmu-GAPDH | Forward: TGTGTCCGTCGTGGATCTGA |
| Reverse: TTGCTGTTGAAGTCGCAGGAG | |
| Mmu/hsa-mir-92a | TATTGCACTTGTCCCGGCCTG |
| Mmu-col2a1 | Forward: CCCGCCTTCCCATTATTGAC |
| Reverse: GGGAGGACGGTTGGGTATCA | |
| Mmu-Sox9 | Forward: GGGGGTGAGCTTTGATTAATTC |
| Reverse: GGGATTTAAGGCTCAAGGTGTTT | |
| Mmu-Col10a1 | Forward: TTCTGCTGCTAATGTTCTTGACC |
| Reverse: GGGATGAAGTATTGTGTCTTGGG | |
| Mmu-Runx2 | Forward: ATGCTTCATTCGCCTCACAAA |
| Reverse: GCACTCACTGACTCGGTTGG | |
| Mmu-mmp13 | Forward: ATGCATTCAGCTATCCTGGCCA |
| Reverse: AAGATTGCATTTCTCGGAGCCTG | |
| Mmu-TNF-α | Forward: GACGTGGAACTGGCAGAAGAG |
| Reverse: TTGGTGGTTTGTGAGTGTGAG | |
| Hsa-GAPDH | Forward: GGAGCGAGATCCCTCCAAAAT |
| Reverse: GGCTGTTGTCATACTTCTCATGG | |
| Hsa-mmp13 | Forward: TCCTGATGTGGGTGAATACAATG |
| Reverse: GCCATCGTGAAGTCTGGTAAAAT | |
| Hsa-col2a1 | Forward: GAGGGCAATAGCAGGTTCACGTA |
| Reverse: TGGGTGCAATGTCAATGATGG | |
| Hsa-col10a1 | Forward: CACCAGGCATTCCAGGATTCC |
| Reverse: AGGTTTGTTGGTCTGATAGCTC |
Transfection assays
The condition and efficiency of transfection assays were verified using a CY3-labelled siR-Ribo™ Transfection Control (cat. no. siN05815122149-1-1; Guangzhou RiboBio Co., Ltd., Guangzhou, China). The ATDC5 cells (4×104) were seeded into a 6-well plate with DMEM/F12 with 10% FBS, and were allowed to grow at 37°C until they had reached 50–70% confluence. Lipofectamine® 2000 transfection reagent (cat. no. 11668; Invitrogen Life Technologies, Carlsbad, CA, USA) was then used to transfect the micrON™ mmu-miR-92a-3p mimic/inhibitor (cat. nos. miR10000539-1-2 and miR20000539-1-2; Guangzhou RiboBio Co., Ltd.) and micrON™ mimic/inhibitor negative control (cat. no. miR01101-1-2 and miR02101-1-2; Guangzhou RiboBio Co., Ltd.) into the cells, according to the manufacturer's instructions. Subsequent to 6 h transfection, chondrogenic differentiation was induced by replacing the medium with chondrogenic medium containing 1% ITS+ Premix.
Target prediction
The potential target genes of miRNAs were predicted using the following online algorithms: miRanda (August 2010 release; http://www.microrna.org/), miRDB (MirTarget2_v4.0; http://www.mirdb.org/miRDB/), CLIP-Seq (2012-03-28; http://mirtarclip.mbc.nctu.edu.tw/) and TargetScan (version 6.2; http://targetscan.org/). Genes predicted by three or four separate algorithms were considered as potential target genes.
Based on these predicted target genes, the signaling pathways potentially regulated by miR-92a were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, kobas2.0-20120208; http://www.genome.jp/kegg/) database and the possible function of miR-92a was predicted.
Statistical analysis
All experiments were performed in triplicate. The quantitative data was expressed as the mean ± confidence interval (mean ± 1/2 CI). Differences between the groups were analyzed using Student's t-text or analysis of variance with SPSS, version 13.0 (SPSS, Inc., Chicago, IL, USA). The least significant difference test and Tamhane's T2 test were used in conditions with, and without, equal variances, respectively. The qualitative data was analyzed using Fisher's Exact test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-92a is elevated in chondrogenic differentiation
The micromass produced from the hADSCs in chondrogenic medium was embedded in paraffin, cut into sections and stained with alcian blue (Fig. 1A). The expression levels of miR-92a, col2a1 and col10a1 increased in the chondrogenic hADSCs cells (Fig. 1B). The expression levels of mir-92a and the chondrogenic marker of col2a1 peaked at day 7 of chondrogenic induction, and the hypertrophic marker of col10a1 peaked at day 14.
Figure 1.
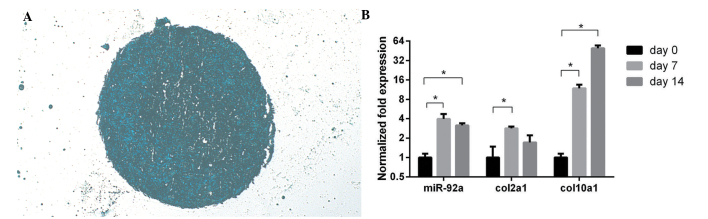
Chondrogenic adipose-derived mesenchymal stem cell micromass and the expression of miR-92a. (A) Subsequent to chondrogenesis for 7 days, the micromass was embedded in paraffin and cut into sections, followed by alcian blue staining (magnification, ×100). (B) RNA was isolated from the micromass and the expression levels of miR-92a, col2a1, and col10a1 were examined using reverse transcription-quantitative polymerase chain reaction. Error bars indicate the mean ± standard deviation. *P<0.05. miR-92a, microRNA-92a.
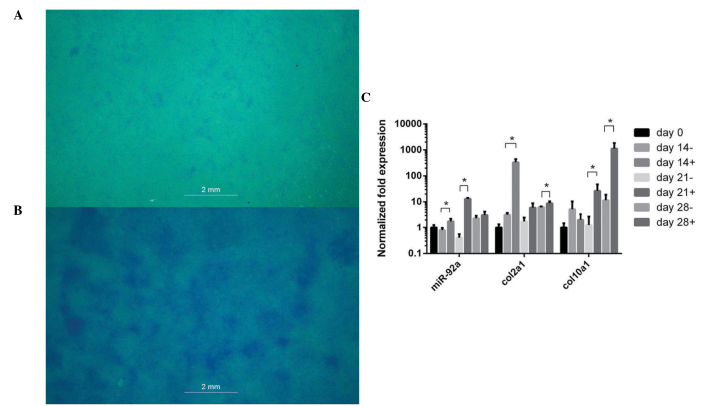
Following 14 days of chondrogenic differentiation with ITS+ Premix, ATDC5 cells exhibited marked staining with alcian blue, compared with the cells without ITS+ Premix (Fig. 2A and B). The expression of col2a1 peaked at day 14, col10a1 peaked at day 28 and miR-92a peaked at day 21 (Fig. 2C).
Figure 2.
Chondrogenic ATDC5 and miR-92a expression. Chondrogenic ATDC5 cells were induced with ITS+ Premix, fixed with formalin and stained with alcian blue. (A) ATDC5 without ITS+ Premix exhibited low levels of alcian blue staining. (B) Chondrogenic ATDC5 cells exhibited more marked staining. (C) Expression levels of miR-92a, col2a1 and col10a1 in chondrogenic ATDC5 cells, examined using reverse transcription-quantitative polymerase chain reaction, compared with the negative control of ATDC5 without ITS+ Premix. Error bars indicate the mean ± standard deviation. *P<0.05. miR-92a, microRNA-92a.
Expression of miR-92a is increased in IL-1β-treated chondrocytes
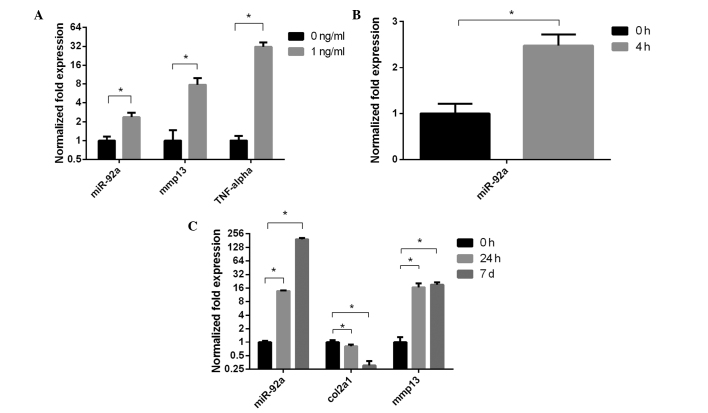
Expression levels of miR-92a and mmp13 were upregulated in the PHCs and PMCs treated with 1 ng/ml IL-1β for 4 h, compared with the control (Fig. 3A and B). The expression levels of miR-92a and mmp13 were elevated, and that of col2a1 was suppressed in a time-dependent manner in the chondrogenic ATDC5 cells treated with 1 ng/ml IL-1β (Fig. 3C).
Figure 3.
Experssion of miR-92a in chondrocytes with IL-1β. Error bars indicate the mean ± standard deviation. (A) Expression of miR-92a was elevated in primary mouse chondrocytes treated with 1 ng/ml IL-1β for 4 h. (B) Expression of miR-92a was elevated in primary human chondrocytes treated with 1 ng/ml IL-1β for 4 h. No mmp13 was detected in the control group. (C) Expression of miR-92a was elevated in a time-dependent manner in the chondrogenic ATDC5 cells treated with 1 ng/ml IL-1β. miR-92a, microRNA-92a; IL-1β, interleukin-1β.
Col9a2 and aggrecan may be regulated by miR-92a
In order to investigate the effects of miR-92a on chondrogenesis, the expression levels of miR-92a were manipulated via transfection with a mimic or inhibitor. The altered expression of miR-92a affected chondrogenic markers, increasing the expression levels of col9a2 and aggrecan with the miR-92a mimic in a dose-dependent manner, compared with the untransfected control. In addition, the miR-193b-3p inhibitor reduced the expression levels of col9a2 and aggrecan in a dose-dependent manner, compared with the control (Table II).
Table II.
Relative mRNA expression levels in ATDC5 cells transfected with miR-92a mimic or inhibitor.
| Gene | Mimic (50 nM)
|
Mimic (100 nM)
|
Inhibitor (50 nM)
|
Inhibitor (100 nM)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change | P-value | SD | Fold change | P-value | SD | Fold change | P-value | SD | Fold change | P-value | SD | |
| Col2a1 | 0.89 | 0.24 | 0.14 | 0.74 | 0.02 | 0.10 | 1.11 | 0.14 | 0.10 | 0.40 | <0.001 | 0.10 |
| Col10a1 | 1.10 | 0.17 | 0.06 | 1.11 | 0.12 | 0.01 | 1.36 | <0.001 | 0.12 | 0.75 | 0.001 | 0.05 |
| Comp | 0.61 | <0.001 | 0.07 | 1.24 | 0.001 | 0.07 | 0.91 | 0.014 | 0.06 | 0.24 | <0.001 | 0.04 |
| Agc | 3.52 | 0.001 | 0.62 | 6.89 | <0.001 | 0.76 | 0.56 | <0.001 | 0.01 | 0.26 | <0.001 | 0.01 |
| Mmp-13 | 1.94 | 0.004 | 0.47 | 3.03 | <0.001 | 0.14 | 1.00 | 0.994 | 0.03 | 0.67 | <0.001 | 0.02 |
| Col9a2 | 3.80 | 0.01 | 0.60 | 14.97 | <0.001 | 1.95 | 0.37 | <0.001 | 0.09 | 0.10 | <0.001 | 0.02 |
| Sox9 | 2.45 | <0.001 | 0.26 | 4.15 | <0.001 | 0.50 | 1.18 | 0.15 | 0.16 | 1.10 | 0.40 | 0.23 |
| Runx2 | 1.59 | <0.001 | 0.16 | 2.16 | <0.001 | 0.12 | 0.91 | 0.19 | 0.02 | 1.00 | 0.98 | 0.13 |
Expression of chondrogenic markers in ATDC5 cells transfected with miR-92a mimic or inhibitor at the indicated doses. Subsequent to transfection, ATDC5 cells were cultured in chondrogenic medium with ITS+ Premix for 4 days. Chondrogenic markers were measured using reverse transcription-quantitative polymerase chain reaction. Col9a2 and aggrecan were markedly upregulated in the miR-92a mimic groups, but were downregulated in the inhibitor groups, in a dose-dependent manner. No clear trends were observed for the other markers. miR-92a, microRNA-92a; SD, standard deviation.
Predicted target genes, signaling pathways and functions of miR-92a
The four algorithms, miRanda, miRDB, CLIP-Seq and TargetScan, were used for prediction of the miR-92a target genes. The general distribution of the predicted potential target genes is shown in Fig. 4. A total of 279 genes were predicted by three or four algorithms and were considered as potential target genes of miR-92a.
Figure 4.
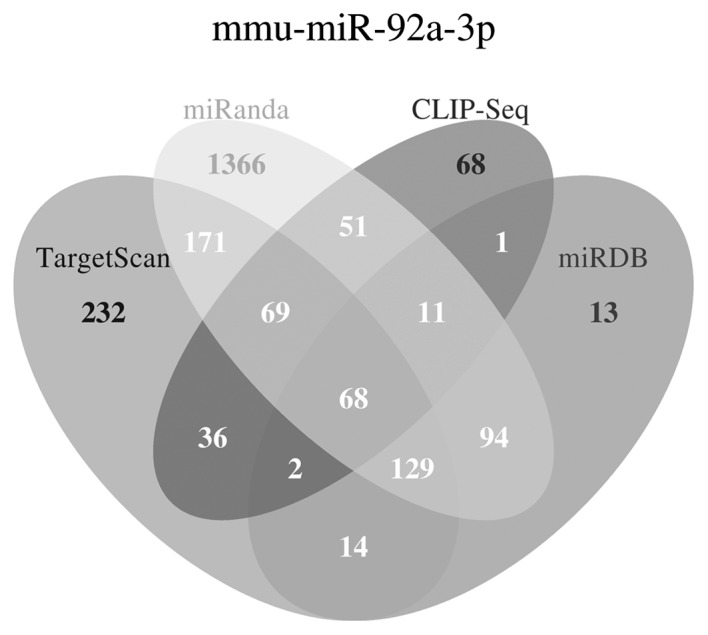
Potential target genes of miR-92a, predicted using four algorithms. In order to minimize possible false positive prediction, only genes predicted by three or four algorithms were identified as potential target genes of miR-92a. miR-92a, microRNA-92a. Numbers refer to genes predicted by one of the four software program.
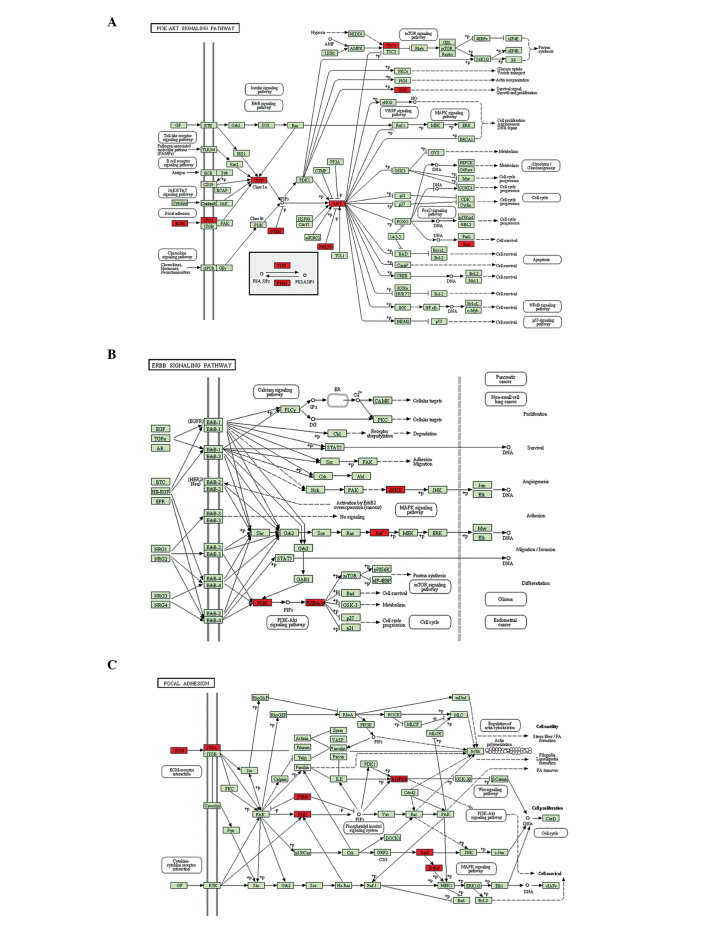
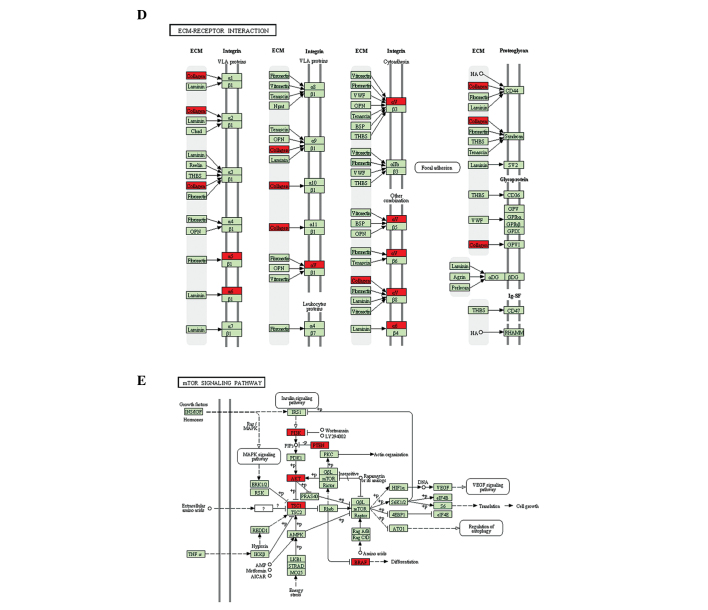
In addition to RT-qPCR, KEGG analysis was used to investigate the signaling pathways by which miR-92a may regulate chondrogenesis, based on the predicted potential target genes. The potential target genes were clustered based on their involvement in signaling pathways, and signaling pathways, which have been previously identified to be involved in chondrogenesis or cartilage degeneration were selected (5,22–25). The P-value was determined based on the number of potential target genes and the number of total genes in each signaling pathway. Of all of the predicted signaling pathways, the PI3K-Akt (P=0.064), ErbB (P=0.076) and focal adhesion kinase pathways (P=0.014), ECM-receptor interaction (P=0.024) and the mTOR signaling pathway (P=0.007) were identified as significant (Table III, Fig. 5).
Table III.
Predicted signaling pathways, based on the potential target genes of miR-92a.
| Signaling pathway | P-value | Predicted target gene | Function in chondrogenesis |
|---|---|---|---|
| PI3K-Akt | 0.064 | Sgk3, Phlpp2, Pten, Pik3r3, Tsc1, Itga5, Itga6, Col1a2, Akt1, Bcl2l11and Itgav | Synergizing with runx2 to enhance normal hypertrophic differentiation and endochondral bone growth; promoting matrix synthesis and chondrocyte survival in adult articular chondrocytes (22). |
| ErbB | 0.076 | Akt1, Pik3r3, Map2k4 and Braf | Contributing to expression of aggrecanases and matrix metalloproteinases, delayed chondrogenesis and inhibition of the PI3K-Akt signaling pathway via downstream MAPK activation (23). |
| Focal adhesion | 0.014 | Rap1b, Pten, Pik3r3, Itga5, Itga6, Braf, Col1a2, Akt1 and Itgav | Inhibiting chondrogenesis via expression of actin and activation of the RhoA/ROCK pathway (24). |
| ECM-receptor interaction | 0.024 | Col1a2, Sdc2, Itga5, Itga6 and Itgav | Inhibiting chondrogenesis via Itga5-mediated cellular-ECM interaction (25). |
| mTOR | 0.007 | Pten, Tsc1, Pik3r3, Braf and Akt1 | Reducing bone growth and hypertrophy; enhancing insulin-like growth factor I mediated proteoglycan synthesis in adult articular chondrocytes (5). |
PI3K, phosphoinositide 3-kinase; MAPK, mitogen-activated protein kinase; ROCK, Rho-associated protein kinase; ECM, extracellular matrix; mTOR, mammalian target of rapamycin.
Figure 5.
Predicted signaling pathways mediating the effects of miR-92a on chondrogenesis and cartilage degeneration. Predicted potential target genes of miR-92a are indicated by the red boxes. (A) PI3K-Akt, (B) ErbB and (C) focal adhesion signaling pathways were predicted based on the potential target genes of miR-92a. miR-92a, microRNA-92a; PI3K, phosphoinositide-3 kinase; mTOR, mammalian target of rapamycin. Predicted signaling pathways mediating the effects of miR-92a on chondrogenesis and cartilage degeneration. Predicted potential target genes of miR-92a are indicated by the red boxes. (D) ECM-receptor interaction and (E) mTOR signaling pathways were predicted based on the potential target genes of miR-92a. miR-92a, microRNA-92a; PI3K, phosphoinositide-3 kinase; mTOR, mammalian target of rapamycin.
Discussion
Previous studies have suggested a role for miR-92a in renal tumorigenesis via the gene expression of VHL (26), and in human acute promyelocytic leukemia via the expression of p63 (27). An additional study identified the positive effects of miR-92a on the proliferation, differentiation and survival of chondrogenic progenitors via the targeting of nog3, an inhibitor of the bone morphogenetic protein (BMP) signaling pathway (28). Although miR-92a was observed to contribute to chondrogenesis by enhancing the expression of col2a1 in the study by Ning et al (28), no significant trend in the expression of col2a1 was not observed in the present study following transfection of either the miR-92a mimic or inhibitor. This discrepancy may be due to differences in experimental subjects and signaling pathways. In the study by Ning et al (28), the BMP signaling pathway (smad1/5/8) and the inhibitor of BMP pathway (nog3) were observed to mediate the effects of miR-92a on in vivo pharyngeal chondrogenesis. In the present study, cultured ATDC5 cells were used for the investigation of miR-92a and chondrogenesis, which are associated with the autocrine transforming growth factor-β (smad2/3) signaling pathway (29).
In the study by Ning et al (28), the morphological defects resulting from the inhibition of nog3, one of the target genes of miR-92a's, were partially reversed by p53 co-inhibition, suggesting a contribution of miR-92a-nog3-apoptosis/proliferation to in vivo morphological regulation of pharyngeal cartilage formation. During chondrogenesis, high levels of type 9 collagen and aggrecan are expressed, along with additional matrix proteins to form the cartilage matrix, with col9a2 and aggrecan considered as chondrogenic markers (30). Col9a2 and aggrecan were previously demonstrated to be associated with a number of diseases, including osteoarthritis (31,32), degeneration of intervertebral discs (33,34) and multiple epiphyseal dysplasia, characterized as the deformed deposition of cartilage at the ends of the bones (32,35,36). The present study hypothesized that col9a2 may be another mediator of the degeneration of cartilage, followed by miR-92a knockdown. For the upstream regulation of col9a2 and aggrecan, Sox9 has been previously suggested as to be critical in initiating the expression of col9a2 and aggrecan (37), although multiple enhancers have been observed to initiate expression of aggrecan (38). However, more detailed information is required on the regulation of the expression levels of col9a2 and aggrecan in order to identify the cure for these diseases. In the present study, the results indicated that miR-92a may contribute to the upregulation of col9a2 and aggrecan, without enhancing the expression of sox9. These results provided novel insight into the upstream regulation of col9a2 and aggrecan, beyond what is already known about sox9 in relation to col9a2 and aggrecan. In addition, the results suggested another possible mechanism of a miR-92a-col9a2-cartilage deformity axis contributing to cartilage deformity following miR-92a knockdown. Further investigations are required in order to verify the effect of miR-92a on the in vivo expression levels of col9a2, aggrecan, and cartilage degeneration, and to determine the underlying mechanisms.
Several previous studies investigating miRNAs used one or two algorithms to predict the target genes, with subsequent mechanistic experiments, based on the predicted genes (39,40). However, each of these widely used algorithms has an intrinsic false positive rate. The false positive rate is 22–31% for TargetScan, 24–39% for miRanda and ~30% for PicTar (41). In the present study, four algorithms were used, and an intersection set of predicted genes from at least three algorithms was identified as a potential target gene. Based on the potential target genes, KEGG analysis was then used to predict several signaling pathways that possibly contribute to the effect of miR-92a on chondrogenesis. KEGG is a database, which is usually used for the prediction of function and signaling pathways from large scale molecular information of high-throughput experiments, including sequencing. This prediction method enables the minimization of false positive rates and assist in understanding the possible function of miR-92a in a wider context (42). Investigations of underlying mechanisms can be performed using these predictions, including luciferase reporter assays of miR-92a and 3′-UTR of Akt1.
In conclusion, the present study demonstrated the presence of miR-92a in chondrogenesis and the chondrocyte response induced by IL-1β. The positive contribution of miR-92a in the expression of col9a2 and aggrecan was observed and the PI3K-Akt, ErbB and focal adhesion kinase pathways, ECM-receptor interaction, and mTOR signaling pathway were indicated as potential mediators of the effects of miR-92a on chondrogenesis and cartilage degeneration.
Acknowledgments
The authors would like to thank to Dr Xuerong Li, Dr Shan Li and Dr Shang Mei at the Department of Parasitology, Zhongshan School of Medicine, Sun Yat-sen University (Guangzhou, China) for their technical assistance.
The present study was supported by the National Natural Science Foundation of China (grant nos. 81301558, 81371941 and 81171709), the Doctoral Scientific Fund Project of the Ministry of Education of China (grant no. 20130171120074) and the Natural Science Foundation of Guangdong Province, China (grant no. s2013040016269). The sponsors had no involvement in the study design; collection, analysis and interpretation of data; the writing of the manuscript; or in the decision to submit the manuscript for publication.
References
- 1.Li Y, Wei X, Zhou J, Wei L. The age-related changes in cartilage and osteoarthritis. Biomed Res Int. 2013;2013:916530. doi: 10.1155/2013/916530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathieu PS, Loboa EG. Cytoskeletal and focal adhesion influences on mesenchymal stem cell shape, mechanical properties and differentiation down osteogenic, adipogenic and chondrogenic pathways. Tissue Eng Part B Rev. 2012;18:436–444. doi: 10.1089/ten.teb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier F, Loeser RF. Biology and pathology of Rho GTPase, PI-3 kinase-Akt and MAP kinase signaling pathways in chondrocytes. J Cell Biochem. 2010;110:573–580. doi: 10.1002/jcb.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Crawford R, Xiao Y. Vertical inhibition of the PI3K/Akt/mTOR pathway for the treatment of osteoarthritis. J Cell Biochem. 2013;114:245–249. doi: 10.1002/jcb.24362. [DOI] [PubMed] [Google Scholar]
- 5.Rokutanda S, Fujita T, Kanatani N, Yoshida CA, Komori H, Liu W, Mizuno A, Komori T. Akt regulates skeletal development through GSK3, mTOR and FoxOs. Dev Biol. 2009;328:78–93. doi: 10.1016/j.ydbio.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Malemud CJ. Intracellular signaling pathways in rheumatoid arthritis. J Clin Cell Immunol. 2013;4:160. doi: 10.4172/2155-9899.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xian CJ. Roles of epidermal growth factor family in the regulation of postnatal somatic growth. Endocr Rev. 2007;28:284–296. doi: 10.1210/er.2006-0049. [DOI] [PubMed] [Google Scholar]
- 8.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage and osteoarthritis: Implications for tissue engineering. Tissue Eng Part B Rev. 2012;18:445–453. doi: 10.1089/ten.teb.2012.0116. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Kang Y, Zhang Z, Zhang H, Duan X, Liu J, Li X, Liao W. Expression of microRNAs during chondrogenesis of human adipose-derived stem cells. Osteoarthritis Cartilage. 2012;20:1638–1646. doi: 10.1016/j.joca.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Thirion S, Berenbaum F. Culture and phenotyping of chondrocytes in primary culture. Methods Mol Med. 2004;100:1–14. doi: 10.1385/1-59259-810-2:001. [DOI] [PubMed] [Google Scholar]
- 12.Zhang ZJ, Zhang H, Kang Y, Sheng PY, Ma YC, Yang ZB, Zhang ZQ, Fu M, He AS, Liao WM, et al. miRNA expression profile during osteogenic differentiation of human adipose-derived stem cells. J Cell Biochem. 2012;113:888–898. doi: 10.1002/jcb.23418. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Su P, Xu C, Yang J, Yu W, Huang D. Chondrogenic differentiation of human mesenchymal stem cells: A comparison between micromass and pellet culture systems. Biotechnol Lett. 2010;32:1339–1346. doi: 10.1007/s10529-010-0293-x. [DOI] [PubMed] [Google Scholar]
- 14.Estes BT, Diekman BO, Gimble JM, Guilak F. Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat Protoc. 2010;5:1294–1311. doi: 10.1038/nprot.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Y, Wang Y. ATDC5: An excellent in vitro model cell line for skeletal development. J Cell Biochem. 2013;114:1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 16.Newton PT, Staines KA, Spevak L, Boskey AL, Teixeira CC, Macrae VE, Canfield AE, Farquharson C. Chondrogenic ATDC5 cells: An optimised model for rapid and physiological matrix mineralisation. Int J Mol Med. 2012;30:1187–1193. doi: 10.3892/ijmm.2012.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atsumi T, Miwa Y, Kimata K, Ikawa Y. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ Dev. 1990;30:109–116. doi: 10.1016/0922-3371(90)90079-C. [DOI] [PubMed] [Google Scholar]
- 18.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simsa-Maziel S, Monsonego-Ornan E. Interleukin-1β promotes proliferation and inhibits differentiation of chondrocytes through a mechanism involving down-regulation of FGFR-3 and p21. Endocrinology. 2012;153:2296–2310. doi: 10.1210/en.2011-1756. [DOI] [PubMed] [Google Scholar]
- 20.MacRae VE, Farquharson C, Ahmed SF. The restricted potential for recovery of growth plate chondrogenesis and longitudinal bone growth following exposure to pro-inflammatory cytokines. J Endocrinol. 2006;189:319–328. doi: 10.1677/joe.1.06609. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Kita K, Kimura T, Nakamura N, Yoshikawa H, Nakano T. PI3K/Akt signaling as a key regulatory pathway for chondrocyte terminal differentiation. Genes Cells. 2008;13:839–850. doi: 10.1111/j.1365-2443.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- 23.Fisher MC, Clinton GM, Maihle NJ, Dealy CN. Requirement for ErbB2/ErbB signaling in developing cartilage and bone. Dev Growth Differ. 2007;49:503–513. doi: 10.1111/j.1440-169X.2007.00941.x. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi I, Onodera K, Sasano Y, et al. Effect of stretching on gene expression of beta1 integrin and focal adhesion kinase and on chondrogenesis through cell-extracellular matrix interactions. Eur J Cell Biol. 2003;82:182–192. doi: 10.1078/0171-9335-00307. [DOI] [PubMed] [Google Scholar]
- 25.Knudson CB. Hyaluronan and CD44: strategic players for cell-matrix interactions during chondrogenesis and matrix assembly. Birth Defects Res C Embryo Today. 2003;69:174–196. doi: 10.1002/bdrc.10013. [DOI] [PubMed] [Google Scholar]
- 26.Valera VA, Walter BA, Linehan WM, Merino MJ. Regulatory Effects of microRNA-92 (miR-92) on VHL Gene Expression and the Hypoxic Activation of miR-210 in clear cell renal cell carcinoma. J Cancer. 2011;2:515–526. doi: 10.7150/jca.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and inhibits cell proliferation in human acute promyelocytic leukemia through modulation of p63 expression. Mol Biol Rep. 2014;41:2799–2808. doi: 10.1007/s11033-014-3134-5. [DOI] [PubMed] [Google Scholar]
- 28.Ning G, Liu X, Dai M, Meng A, Wang Q. MicroRNA-92a upholds Bmp signaling by targeting noggin3 during pharyngeal cartilage formation. Dev Cell. 2013;24:283–295. doi: 10.1016/j.devcel.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Kawai J, Akiyama H, Shigeno C, Ito H, Konishi J, Nakamura T. Effects of transforming growth factor-beta signaling on chondrogenesis in mouse chondrogenic EC cells, ATDC5. Eur J Cell Biol. 1999;78:707–714. doi: 10.1016/S0171-9335(99)80039-9. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki K, Sandell LJ. Extracellular matrix gene regulation. Clin Orthop Relat Res. 2004;427(Suppl):S123–S128. doi: 10.1097/01.blo.0000144478.51284.f3. [DOI] [PubMed] [Google Scholar]
- 31.Nakki A, Videman T, Kujala UM, Suhonen M, Männikkö M, Peltonen L, Battié MC, Kaprio J, Saarela J. Candidate gene association study of magnetic resonance imaging-based hip osteoarthritis (OA): Evidence for COL9A2 gene as a common predisposing factor for hip OA and lumbar disc degeneration. J Rheumatol. 2011;38:747–752. doi: 10.3899/jrheum.100080. [DOI] [PubMed] [Google Scholar]
- 32.Gleghorn L, Ramesar R, Beighton P, Wallis G. A mutation in the variable repeat region of the aggrecan gene (AGC1) causes a form of spondyloepiphyseal dysplasia associated with severe, premature osteoarthritis. Am J Hum Genet. 2005;77:484–490. doi: 10.1086/444401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aladin DM, Cheung KM, Chan D, Yee AF, Jim JJ, Luk KD, Lu WW. Expression of the Trp2 allele of COL9A2 is associated with alterations in the mechanical properties of human intervertebral discs. Spine (Phila Pa 1976) 2007;32:2820–2826. doi: 10.1097/BRS.0b013e31815b75c5. [DOI] [PubMed] [Google Scholar]
- 34.Kim NK, Shin DA, Han IB, Yoo EH, Kim SH, Chung SS. The association of aggrecan gene polymorphism with the risk of intervertebral disc degeneration. Acta Neurochir (Wien) 2011;153:129–133. doi: 10.1007/s00701-010-0831-2. [DOI] [PubMed] [Google Scholar]
- 35.Fiedler J, Stöve J, Heber F, Brenner RE. Clinical phenotype and molecular diagnosis of multiple epiphyseal dysplasia with relative hip sparing during childhood (EDM2) Am J Med Genet. 2002;112:144–153. doi: 10.1002/ajmg.10554. [DOI] [PubMed] [Google Scholar]
- 36.Briggs MD, Choi H, Warman ML, Loughlin JA, Wordsworth P, Sykes BC, Irven CM, Smith M, Wynne-Davies R, Lipson MH, et al. Genetic mapping of a locus for multiple epiphyseal dysplasia (EDM2) to a region of chromosome 1 containing a type IX collagen gene. Am J Hum Genet. 1994;55:678–684. [PMC free article] [PubMed] [Google Scholar]
- 37.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 38.Hu G, Codina M, Fisher S. Multiple enhancers associated with ACAN suggest highly redundant transcriptional regulation in cartilage. Matrix Biol. 2012;31:328–337. doi: 10.1016/j.matbio.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge YZ, Xu LW, Xu Z, et al. Medicine. Vol. 94. (Baltimore); 2015. Expression Profiles and Clinical Significance of MicroRNAs in Papillary Renal Cell Carcinoma: A STROBE-Compliant Observational Study; p. e767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J, Tan ZH, Tang X, et al. MiR-374b-5p suppresses RECK expression and promotes gastric cancer cell invasion and metastasis. World J Gastroenterol. 2014;20:17439–17447. doi: 10.3748/wjg.v20.i46.17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bentwich I. Prediction and validation of microRNAs and their targets. Febs Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 42.ElHefnawi M, Soliman B, Abu-Shahba N, Amer M. An integrative meta-analysis of microRNAs in hepatocellular carcinoma. Genomics Proteomics Bioinformatics. 2013;11:354–367. doi: 10.1016/j.gpb.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]