Abstract
Pokeweed antiviral protein (PAP) belongs to the family of type I ribosome-inactivating proteins (RIPs): Ribotoxins, which function by depurinating the sarcin-ricin loop of ribosomal RNA. In addition to its antibacterial and antifungal properties, PAP has shown promise in antiviral and targeted tumor therapy owing to its ability to depurinate viral RNA and eukaryotic rRNA. Several PAP genes are differentially expressed across pokeweed tissues, with natively isolated seed forms of PAP exhibiting the greatest cytotoxicity. To help elucidate the molecular basis of increased cytotoxicity of PAP isoenzymes from seeds, the present study used protein sequencing, mass spectroscopy and X-ray crystallography to determine the complete covalent structure and 1.7 Å X-ray crystal structure of PAP-S1aci isolated from seeds of Asian pokeweed (Phytolacca acinosa). PAP-S1aci shares ~95% sequence identity with PAP-S1 from P. americana and contains the signature catalytic residues of the RIP superfamily, corresponding to Tyr72, Tyr122, Glu175 and Arg178 in PAP-S1aci. A rare proline substitution (Pro174) was identified in the active site of PAP-S1aci, which has no effect on catalytic Glu175 positioning or overall active-site topology, yet appears to come at the expense of strained main-chain geometry at the pre-proline residue Val173. Notably, a rare type of N-glycosylation was detected consisting of N-acetyl-D-glucosamine monosaccharide residues linked to Asn10, Asn44 and Asn255 of PAP-S1aci. Of note, our modeling studies suggested that the ribosome depurination activity of seed PAPs would be adversely affected by the N-glycosylation of Asn44 and Asn255 with larger and more typical oligosaccharide chains, as they would shield the rRNA-binding sites on the protein. These results, coupled with evidence gathered from the literature, suggest that this type of minimal N-glycosylation in seed PAPs and other type I seed RIPs may serve to enhance cytotoxicity by exploiting receptor-mediated uptake pathways of seed predators while preserving ribosome affinity and rRNA recognition.
Keywords: pokeweed antiviral protein, N-glycosylation, glycoproteins, N-acetyl-D-glucosamine, ribosome-inactivating proteins, X-ray crystal structure
Introduction
Pokeweed antiviral protein (PAP) is an N-glycosidase of the ribosome-inactivating protein (RIP) superfamily, a widely-distributed family of toxins found in plants, fungi and bacteria (1,2). RIPs function by irreversibly depurinating the sarcin-ricin loop (SRL), a highly conserved sequence found in the RNA of all large ribosomal subunits (3). Depurination prevents ribosomes from completing the translocation step of the elongation cycle due to the inability to bind elongation factor 2 (4), leading to a ribotoxic stress response and RIP-induced apoptosis (5). In addition to targeting the SRL, PAP can depurinate a wide variety of RNA substrates, including viral RNA (6) and eukaryotic mRNA (7,8). The strong antiviral activity of PAP exists against a wide range of viruses, including human immunodeficiency virus-1, influenza, herpes, polio and human T-cell leukemia virus I (9,10). Investigations into the phytotherapeutic and prophylactic usage of PAP in this context have shown promise (11,12).
Based on their structure, RIPs are traditionally classified as either type I or type II. Type I RIPs, including PAP, are enzymatically active ~30 kDa monomeric toxins. By contrast, type II RIPs, including ricin and Shiga toxin, possess a dimeric quaternary structure with an active A-subunit (homologous to type I RIPs) and a larger lectin B-subunit that attaches to galactose-containing surface receptors of target cells. The mechanisms of cell entry and intracellular trafficking of type II RIPs have been investigated in detail, although less is known about these processes in type I RIPs, including PAP (5). Due to the absence of the cell-binding B subunit, type I RIPs are less toxic compared with type II RIPs; however, cellular uptake and cytotoxicity can be markedly enhanced through conjugation with monoclonal antibodies, lectins, hormones or growth factors (2,13).
Several studies on American pokeweed (Phytolacca americana) have revealed differential PAP gene expression at various developmental stages, with PAP-I, PAP-II, PAP-S1/PAP-S2 and PAP-R representing the different isozymes that appear in spring leaves, summer leaves, seeds and roots, respectively. In vivo toxicity studies in a preclinical mouse model revealed that the natively-isolated seed isoform, PAP-S1, has far greater toxicity than PAP-I and the A-subunit of ricin (14). The crystal structures of PAP-I and PAP-S1 from Phytolacca americana have been determined (15–17), although significantly less is known about the tissue distribution, biochemical properties and structural features of potentially therapeutic PAPs from other species. To address this deficiency, the present study combined protein sequencing, mass spectrometry and X-ray crystallography to elucidate the complete covalent and 1.7 Å X-ray crystal structure of a PAP homolog isolated from seeds of Asian pokeweed (Phytolacca acinosa). The sequence and structure of this RIP, termed PAP-S1aci, was compared to other known RIPs including the American pokeweed seed homolog (Phytolacca americana). An unusual type of post-translational carbohydrate modification identified in PAP-S1aci is discussed within the context of type-I RIP cytotoxicity, and more generally, is compared to well-established eukaryotic N-glycosylation patterns and pathways. Finally, the potential application of these findings in the development of specific RIP-based therapies is presented herein.
Materials and methods
Purification of PAP-S1aci for primary structure determination
The two isoenzymes of PAP-Saci were isolated from mature seeds of P. acinosa as previously described (18). Unless otherwise indicated, all chemicals were purchased from Carl Roth GmbH & Co. KG (Karlsruhe, Germany). The individual isoenzymes were subsequently separated by reversed phase high performance liquid chromatography (HPLC) on a Vydac C4 column (4.6×250 mm, 5 μm; Vydac; Grace Davison Discovery Sciences, Lokeren, Belgium) equilibrated with 0.1% trifluoroacetic acid (TFA) and eluted with an acetonitrile gradient (0–60% acetonitrile; 1 ml/min; 60 min; A280 nm). The higher molecular weight isoenzyme (PAP-S1aci) eluted first at ~35% acetonitrile and the two major peaks were individually collected and dried under a vacuum. The PAP-S1aci and PAP-S2aci fractions, which were ~95% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were transferred onto polyvinylidene fluoride (PVDF) membranes, and the area corresponding to PAP-S1aci was extracted for N-terminal sequencing. The PAP-S1aci elution fraction produced an N-terminal sequence identical to the blotted sample and was therefore used for subsequent sequencing experiments.
Preparation of PAP-S1aci cleavage peptides
For cyanogen bromide (CNBr) cleavage, the dried PAP-S1aci sample (5 mg) was dissolved in 400 μl of 70% formic acid containing 400 μg of CNBr, sealed under nitrogen and incubated for 24 h at room temperature in the dark. The mixture was desiccated under a vacuum and the residue was dissolved in 500 μl of 0.1% TFA and immediately separated by reversed phase HPLC on a Vydac C4 column as described above except that elution was monitored at 230 nm. A total of eight fractions were collected and desiccated. For further enzymatic digestion, the CNBr fragments were dissolved in 50 mM ethylmorpholine-acetate (pH 8.0) and digested with Arg-C sequencing grade Arg-C proteinase (Boehringer Ingelheim Deutschland GmbH, Ingelheim am Rhein, Germany; substrate:enzyme ratio 100:1) for 18 h at 37°C. Digestion was terminated by desiccation and the peptides (totaling 16 individual peaks) were separated as described above.
Automated Edman degradation
N-terminal sequencing of proteins and peptides was performed on an automatic LF3600D Protein Sequencer (Beckman Coulter, Miami, FL, USA). Cysteine residues were modified by acrylamide and detected as cys-S-β-propionamide (19). For sequencing of the C-terminal peptide, the arginine peptide R12 prepared from the CNBr fragment M6 was immobilized to a Sequelon AA membrane (Millipore Corporation, Billerica, MA, USA) using carbodiimide condensation.
In-gel protein digestion
Selected spots were excised from Coomassie gels and the gel plugs were minced, washed with water and soaked in 100 mM ethylmorpholine acetate buffer (pH 8.5) in 50% acetonitrile. Following complete gel destaining in a sonication bath, the gel sections were repeatedly washed with water and dehydrated in acetonitrile. The supernatant was removed and the gel was partially dried under a vacuum. Gel sections were swollen in a digestion buffer containing 50 mM ethylmorpholine acetate (Sigma-Aldrich, Munich, Germany), pH 8.0, 1 mM CaCl2, 10% acetonitrile and sequencing grade trypsin (Roche Diagnostics, Mannheim, Germany; trypsin:protein ratio=1:75). Following overnight digestion (agitation at 37°C) the resulting peptides were extracted from the gel by increasing the acetonitrile concentration to 50% and addition of TFA to a final concentration of 1%. Following sonication for 15 min, the liquid phase containing the extracted peptides was dried and the peptides were redissolved in aqueous 1% TFA and desalted on ZipTip microcolumns (Millipore Corporation, Hesse, Germany).
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)
A saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) in aqueous 50% acetonitrile/0.2% TFA was used as a MALDI matrix. A total of 0.5 μl of a 1:1 sample:matrix mixture was placed on the sample target and dried at ambient temperature. Positive ion MALDI mass spectra were measured on a Bruker BIFLEX reflectron time-of-flight mass spectrometer (Bruker-Franzen, Bremen, Germany) equipped with a SCOUT 26 sample inlet, a gridless delayed extraction ion source and a nitrogen laser (337 nm; Laser Science, Cambridge, MA, USA). The ion acceleration voltage and reflectron voltage were set to 19 and 20 kV, respectively. Spectra were calibrated externally using the monoisotopic [M+H]+ ion of α-cyano-4-hydroxycinnamic acid and a peptide standard (angiotensin II; Bachem, Bubendorf, Switzerland).
Tandem mass spectrometry
Peptides were loaded onto a capillary column (0.18 mm inner diameter) packed with 10 cm of reversed-phase resin (MAGIC C-18, 200 Å, 5 μm; Michrom Bioresources, Auburn, CA, USA) and separated using a 5–40% gradient of acetonitrile in 0.5% acetic acid (50 min; 2 μl/min). The column was interfaced with an LCQ DECA ion trap mass spectrometer (ThermoQuest Corp., San Jose, CA, USA) set to acquire a full MS spectrum over the mass range 300–1,800 a.m.u. followed by full MS/MS scans of the two most intense ions from the preceding MS scan. Dynamic exclusion was set to two repeat counts with a repeat duration of 0.5 min and a 3 min exclusion duration window. The spray voltage was held at 1.5 kV and the tube lens potential was −2 V. The heated capillary was kept at 175°C with a voltage of 13 V. Spectra were interpreted manually and/or with the Sequest™ v. B22 software (Thermo Finnigan, San Jose, CA, USA) and MS/MS MASCOT error tolerant search engine (http://www.matrixscience.com) (20).
Crystallization and X-ray structure determination
Protein preparation, crystallization and preliminary phasing have been described previously (18). In brief, multiple rounds of ion exchange and gel filtration chromatography on P. acinosa seed homogenate yielded an approximate equimolar mixture of PAP-S1aci and PAP-S2aci. Crystallization of PAP-S1aci was achieved at 289 K by hanging-drop vapor diffusion of a solution containing 10 mg/ml of the isoenzyme mixture, 21–22% polyethylene glycol (PEG) 4 K, 0.2 M sodium citrate and 100 mM phosphate buffer (pH 7.2) over reservoirs containing 42–44% PEG 4K and 100 mM phosphate buffer (pH 7.2). An X-ray diffraction dataset to 1.7 Å was collected at 100 K at the joint University of Hamburg-IMB Jena-EMBL Beamline X13 at DESY (Hamburg, Germany). The crystals belonged to space group I222 with unit cell constants a=78.63, b=84.19 and c=90.88 Å and one monomer per asymmetric unit. The structure was solved by molecular replacement with the atomic coordinates of P. americana PAP-I (15; Protein Data Bank code 1PAF) as a search model. Preliminary X-ray sequencing and model rebuilding were conducted prior to determination of the full protein sequence. The preliminary X-ray sequenced model had correct residue assignments at 246 of 261 positions (with five errors due to Asn/Asp and Gln/Glu side-chain ambiguities). Final model building into σA-weighted 2Fo-Fc and Fo-Fc maps (21) was conducted with the molecular graphics program Crystallographic Object-Oriented Toolkit (Coot) (22). Crystallographic refinement was performed in CNS 1.1 (23) using anisotropic bulk solvent correction, positional and isotropic atomic B-factor protocols. The final PAP-S1aci model, which consists of 261 amino-acid residues, 1 N-linked GlcNAc and 464 water molecules, exhibits good geometry as determined by MolProbity (http://molprobity.biochem.duke.edu/) (24). An exception is the pre-Pro residue Val173, which violates the pre-Pro Ramachandran plot despite clear electron density supporting its refined main-chain conformation (see Results and Discussion). Final R and Rfree values are 16.9 and 21.1% for data in the 29.7–1.7 Å range (with 5% of the observed data reserved for calculation of Rfree). Final model statistics are presented in Table I. Secondary structure assignments were performed with the program DSSP (25). Structural superpositions were performed with LSQKAB (26), as implemented in the CCP4 suite software (http://www.ccp4.ac.uk) (27). Crystal contact analyses and surface area calculations were conducted with the CCP4 programs CONTACT and AREAIMOL, respectively. Molecular figures were generated with PyMOL (28). The refined coordinates of PAP-S1aci have been deposited in the RCSB Protein Data Bank under the ID code 2Q8W.
Table I.
Data collection and refinement statistics.
| X-ray data | Value |
|---|---|
| Wavelength (Å) | 0.8033 |
| Resolution (Å) | 29.73–1.70 (1.74–1.70) |
| Space group | I222 |
| Unit-cell dimensions | |
| a (Å) | 78.63 |
| b (Å) | 84.19 |
| c (Å) | 90.88 |
| Overall reflections | 417,357 |
| Unique reflections | 33,596 |
| Multiplicity | 12.4 |
| Completeness (%) | 99.9 (100.0) |
| Rmergea (%) | 4.3 (38.9) |
| I/σ (I) | 55.7 (7.1) |
|
| |
| Model refinement statistics | Value |
|
| |
| Resolution (Å) | 29.73–1.70 (1.78–1.70) |
| No. of reflections | 32,752 (1,625) |
| Overall completeness | 97.7 (93.4) |
| Rcrystb | 0.169 (0.230) |
| Rfreeb | 0.211 (0.277) |
| R.m.s.d. from ideal geometry | |
| Bonds (Å) | 0.012 |
| Angles (Å) | 1.7 |
| Protein atoms | 2,103 |
| GlcNAc atoms | 14 |
| Solvent atoms | 464 |
| Ramachandran plotc | |
| Favored (%) | 96.2 |
| Allowed (%) | 99.6 |
| Disallowed (%) | 0.4 |
Values in parentheses are for the highest resolution shell.
Rmerge = ∑hkl ∑i | I(hkl)i - 〈I(hkl)〉|/∑hkl ∑i I(hkl)i, where I(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over all equivalent reflections.
Rcryst = ∑hkl | Fo(hkl) - Fc(hkl)ǀ/∑hkl Fo(hkl). Rfree was calculated for a test set of reflections (5%) omitted from the refinement.
As defined by MolProbity (24). GlcNAc, N-acetylglucosamine; R.m.s.d., root-mean square deviation.
Results
Preliminary protein sequencing of PAP-S1aci
Initial purification of an approximate equimolar mixture of seed PAP isoenzymes, PAP-S1aci and PAP-S2aci, was reported (18). Since the two isoenzymes were of similar size as determined by SDS-PAGE, they were separated by reversed-phase HPLC. Different gradients were used in an attempt to optimize separation, however, it was difficult to identify optimal conditions; hence the PAP-S1aci sample used for primary structure analysis had an estimated final purity of ~95%. In order to define the N-terminus of PAP-S1aci, samples from reversed-phase HPLC were separated by SDS-PAGE, electroblotted onto PVDF and used for PVDF sequencing, producing the initial sequence INTITFDAGXATIN, where X was an empty cycle (see below). Since the sequence length obtainable from a PVDF blot is limited, Protein Support sequencing was performed on the bulk PAP-S1aci sample obtained from HPLC. This yielded the more extensive N-terminal sequence INTITFDAGXATINKYATEMESLR NEAKDPSLKXYGXPXXP (X=unknown amino acid). A protein-protein BLAST search (29) on the N-terminal sequence revealed 85% sequence identity to the seed isoenzyme PAP-S1 from P. americana (30). The full sequence of this protein was used for subsequent planning of the sequencing strategy.
As the PAP-S1 homolog from P. americana has 261 amino-acid residues and five internal Met residues, CNBr cleavage was performed followed by fragment separation and analysis. Of the eight peaks that appeared during HPLC separation following CNBr cleavage, M1, M2 and M8 represented nonpeptide contaminants. Fragment M3 covered the sequence Glu21-Gly36 (residue numbering reflects the final sequence of PAP-S1aci), fragment M4 (Mr ~10 kDa by MALDI-MS) encompassed the entire C-terminal region of the protein (Val173-Thr261) and fragment M5 (Mr ~10 kDa by MALDI-MS) had the extended internal sequence Gly75-Gln171. The fragment MU6 (Ile1-Met20) was already determined by N-terminal sequencing and fragment MU7 comprised Leu40-Met74. Thus, the five CNBr fragments covered the entire PAP-S1aci protein sequence. The sixth Met peptide anticipated from the homology to PAP-S1 does not exist in PAP-S1aci due to the presence of an Ile residue (Ile65) in this position. Arg-C-generated peptides were used to complete the amino acid sequence in areas not available for MS or MS/MS analysis. Partial sequencing of two of these peptides covered the regions Asn25-Asn44 and Asn135-Lys164. Peptide resequencing following Cys modification confirmed the positions of four Cys residues within the sequence. Finally, the C-terminal sequence of the protein was derived from peptide sequencing following Cys modification and immobilization on arylamino-PVDF. Altogether, 241 of 261 amino-acid residues were covered by Edman degradation, yielding a sequence coverage of ~92%.
Protein sequence verification and completion by mass spectrometry
Certain sequences within PAP-S1aci would be difficult to cover by Edman methods due to the requirement for multiple rounds of cleavage and purification. To circumvent this complication, peptide MS was used to identify the small percentage of missing sequences and to verify sequences derived from automated Edman degradation. By employing μLC-ESI-MS/MS tandem mass spectrometry on different protease digests (trypsin, Glu-C and Asp-N) additional sequence data were generated for Asn70-Met74, Lys128-Arg134, Phe165-Met172 and Asp232-Arg240. In all instances, an overlap with the Edman data was evident. Altogether the MS/MS data covered ~90% of the PAP-S1aci sequence. Subsequent MALDI-MS was used to verify the pre-determined amino acid sequence by measuring the mass of peptide fragments. This analysis covered the PAP-S1aci sequence by another 93%.
Analysis of N-glycosylation sites
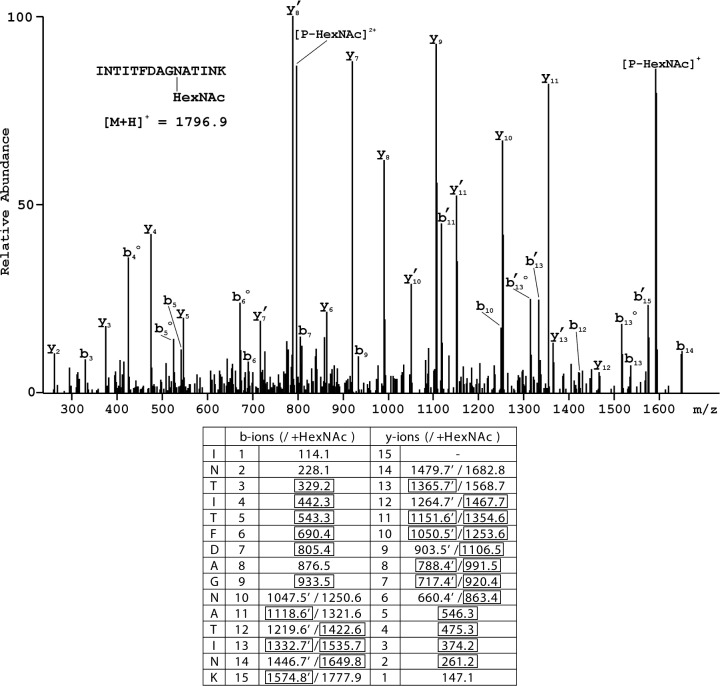
It was difficult to identify amino-acid residues at three positions within the PAP-S1aci sequence (residues 10, 44 and 255). These positions were characterized by low yields of phenylthiohydantoin (PTH)-amino acids with a similar elution profile to PTH-Asp. Although this may be indicative of any post-translational modification, they could be identified as Asn-GlcNAc linkages - an extremely rare type of N-glycosylation. Large carbohydrates attached to Asn residues may prevent extraction of the corresponding PTH-Asn-oligosaccharide from the cartridge (genuine 'empty' cycle), however, PTH-Asn-GlcNAc is extractable (albeit at a lower yield), and would possibly elute earlier than PTH-Asn in reversed-phase HPLC. Definitive evidence for this assignment at each position in the sequence was provided by tandem MS. An example of the MS/MS spectrum for the N-terminal glycosylated peptide having the structure INTITFDAG [N-HexNAc (L-asparagine-N-acetyl-hexosamine)]ATINK is shown in Fig. 1, where extensive sequence coverage by fragments generated following the initial loss of HexNAc as well as those containing the modification is evident.
Figure 1.
Example of the analysis of glycosylation at Asn10 by tandem MS. MS/MS spectrum of peptide INTITFDAGNATINK with HexNAc attached to Asn10. The two ion series result either from fragmentation of peptide-HexNAc or by initial loss of HexNAc. Accordingly, there are two series of fragment ions in the corresponding parts of the spectrum; those assigned to peptides with the initial loss of HexNAc are marked by apostrophes. Open circles indicate peaks corresponding to loss of water from the indicated fragment ion. P, parent ion. Numerical values for the two series of fragment ions are provided in the accompanying table, with those identified in the spectrum highlighted in boxes. HexNAc, N-acetylhexosamine; MS, mass spectrometry.
Correction of Edman/MS sequencing errors by X-ray sequencing
The high-quality X-ray data allowed us to identify three positions in the sequence where the Edman/MS data were evidently erroneous (residues 65, 161 and 260). On the basis of clear electron density and/or H-bonding features, the original assignments of Leu65 (derived by Edman and MS/MS), Ile161 (Edman) and Ala260 (Edman) were corrected to Ile65, Thr161 and Thr260. Electron density at these positions is presented in Fig. 2, illustrating the utility of high-quality X-ray data as a complementary sequencing method, particularly for direct visualization of isomeric (Ile/Leu) or isobaric (Gln/Lys) residues, which are frequently misannotated by other protein sequencing methods.
Figure 2.
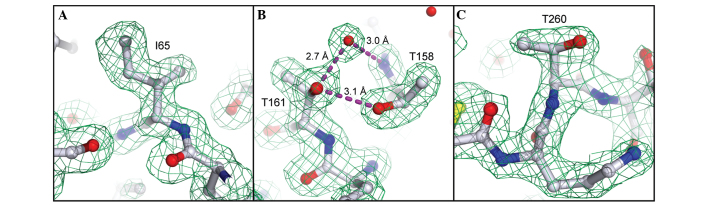
X-ray sequencing of PAP-S1aci at positions 65, 161 and 260. The final model is presented as ball-and-stick and shown in CPK coloring. Water molecules are depicted as red spheres and H-bonds are presented as dashed lines. Simulated-annealing omit maps (Fo-Fc; green; 3σ) are superimposed over the final model. (A) A Leu→Ile correction was applied to residue 65 on the basis of clear electron density features. (B) Electron density and a clear hydrogen-bond network prompted an Ile→Thr correction at position 161. (C) Electron density at position 260 indicated the presence of Thr rather than Ala.
Sequence and structure of PAP-S1aci
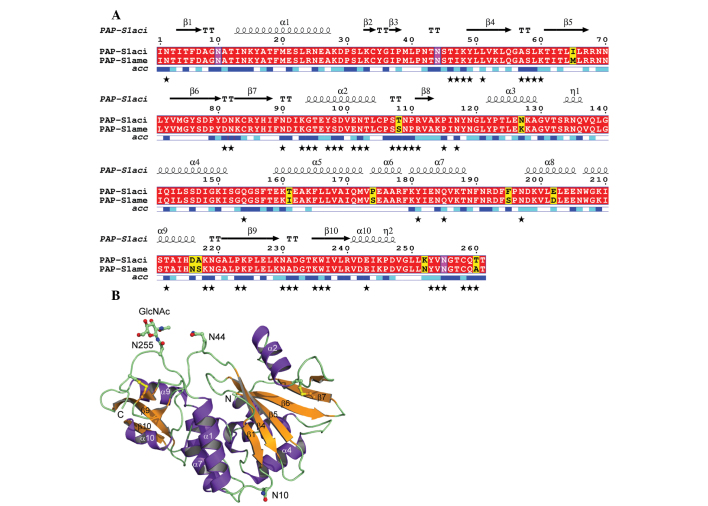
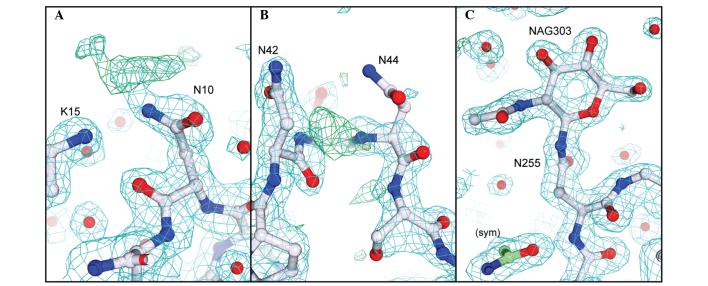
The sequence and X-ray structure of PAP-S1aci are presented in Fig. 3. The figure was generated with DSSP (25), Multalin (31) and ESPript 2.2 (32). PAP-S1aci contains invariant active-site residues of the RIP superfamily (corresponding to Tyr72, Tyr122, Glu175 and Arg178 in PAP-S1aci), which are critical for catalysis (15,33). Four conserved Cys residues forming two intramolecular disulfide bonds (Cys34-Cys258 and Cys84-Cys105) are also present. A protein-protein BLAST search (34) was performed to determine the closest known sequence relative, identifying P. americana PAP-S1 (30) with 96% sequence identity (250 of 261 residues) to PAP-S1aci (Fig. 3A). The 1.8 Å crystal structure of PAP-S1 from P. americana has been elucidated and includes a fully resolved carbohydrate structure consisting of three GlcNAc monosaccharide linkages at Asn10, Asn44 and Asn255 (17). The current results demonstrated that the same three N-glycosylation sequons are also found in PAP-S1aci and that each site is similarly modified with a single GlcNAc residue. In our crystal form there is clear electron density for the sugar at Asn255 due to its involvement in crystal packing (18,35), however, Asn10 and Asn45 are freely exposed to solvent and the corresponding electron density for the linked GlcNAc residues is weak (Fig. 4).
Figure 3.
Sequence and structure of PAP-S1aci. (A) Pairwise sequence alignment between PAP-S1aci and PAP-S1. The secondary structure of PAP-S1aci is given above the alignment [α-helices and 310 (η) helices are presented as squiggles, β-strands as arrows and β-turns as 'TT']. Identical residues are presented on a red background; differences are presented on a yellow background. The three glycosylated Asn residues are framed in magenta. Relative solvent accessibility (acc) for residues of PAP-S1aci are given below the sequence alignment as a color-coded bar [full accessibility (blue), intermediate (cyan) and buried (white)]. Black stars indicate residues involved in crystal contacts (<4.2 Å from symmetry-related protein atoms) in the PAP-S1aci crystal form. (B) The 1.7 Å crystal structure of PAP-S1aci is depicted in the ribbon diagram (purple, helices; orange, β-strands; green, loops). Secondary structure elements are numbered according to the corresponding sequence alignment. The N- and C-termini are labeled and disulfide bonds are shown in yellow (ball-and-stick). Also presented as a ball-and-stick model are the three glycosylated Asn residues (Asn10, Asn45 and Asn255-GlcNAc). GlcNAc, N-acetylglucosamine.
Figure 4.
Electron density features of the three N-linked GlcNAc residues in PAP-S1aci. σA-weighted 2Fo-Fc (blue; 1 σ) and Fo-Fc (green; 3 σ) electron density maps are shown, with the final PAP-S1aci model presented as ball-and-stick. (A) Weak density was observed for an N-linked GlcNAc at Asn10. (B) The Asn44 side-chain and linked sugar are disordered. (C) The Asn255-GlcNAc303 linkage is clearly defined. The GlcNAc moiety is labeled 'NAG'; symmetry-related atoms are labeled 'sym' and distinguished in green. GlcNAc, N-acetylglucosamine.
As expected by the high sequence identity between PAP-S1aci and PAP-S1, the overall folds of the two proteins are identical within the limits of experimental detection. An overall Cα least-squares superposition between the two provided a Cα root-mean-square deviation (r.m.s.d.) of 0.48 Å, comparable with the summed cross-validated Luzatti estimates of the coordinate errors in the two structures (0.52 Å). A few small regional Cα displacements on the order of 0.8–1.3 Å can be attributed to localized areas of poor density or differences in crystal packing between the two crystal forms. A Cα geometrical validation of our refined model with MolProbity (24) indicated a significant outlier, Val173 (φ,Ψ= −114°, −83°), which lies outside the small α region of the pre-proline Ramachandran plot. Given that the nearby Glu175 is a conserved active-site residue believed to stabilize the developing oxocarbonium ion during substrate cleavage (15), the present study aimed to determine whether the intervening Pro174 was commonly found in RIP sequences. An extensive protein-protein BLAST search and alignment of non-redundant RIP sequences indicated the residue preceding the catalytic Glu175 was almost invariably Ser or Ala (corresponding to Ser174 in PAP-S1; Fig. 3A). Only three sequences out of all known bacterial and plant RIPs contain a Pro residue in this position: Ribosome inactivating protein 2 from Phytolacca insularis (Korean pokeweed), curcin from Jatropha curcas (spurge family) and ebulin from Sambucus ebulus (Dwarf Elder). Since the geometrically strained Val173 is situated in a break between helices α5 and α6 (Fig. 3A), the present study aimed to determine whether the Pro174 substitution in PAP-S1aci led to any notable structural deviations in the α5/α6 region or if the refined geometry of Val173 is fully supported by the X-ray data. A superposition analysis between the structures of PAP-S1aci and PAP-S1 revealed the Ser→Pro substitution in PAP-S1aci is accommodated without significant alterations in the positioning of Glu175 or in the overall topology of the active site (Fig. 5A). In addition, the lower than average temperature factors in this region (B 2 Val173=15.4 Å ; BProtein=22.3 Å2), coupled with excellent electron density, strongly supports the refined backbone conformation at Val173-Pro174 (Fig. 5B). Although the main-chain conformation does appear to be rather unusual, a more recent statistical analysis of protein pre-Pro geometry indicated that the φ,Ψ values observed for Val173 are within a broad sterically allowed area extending from the α region towards the lower left corner of the Ramachandran plot (36).
Figure 5.
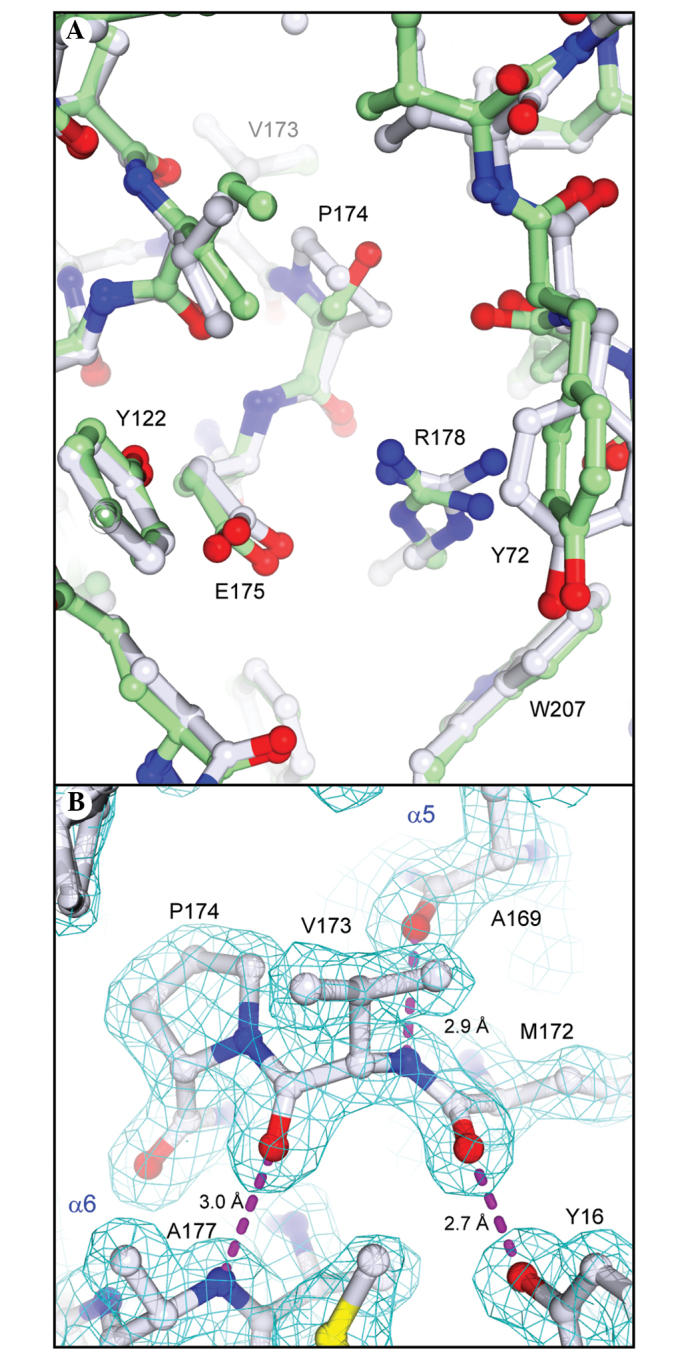
Active site of PAP-S1aci. (A) Superposition between PAP-S1aci (grey) and PAP-S1 (green; Protein Data Bank code 1gik). Active-site residues for PAP-S1aci are labeled. (B) Electron density features in the vicinity of Val173, a pre-proline Ramachandran outlier according to MolProbity analysis (24). The final model is shown in standard CPK coloring. The final σA-weighted 2Fo-Fc electron density map (cyan; 1.5σ) is overlaid. H-bonds are shown as dashed lines, with corresponding distances between non-hydrogen atoms given in Å.
Discussion
Using complementary methods, the present study detected the presence of N-GlcNAc residues in mature PAP-S1aci. This rare type of N-glycosylation has been previously identified in fungi (37), animals (38,39) and plants (40), including RIPs from the seeds of Phytolacca americana and Luffa cylindrica (sponge gourd) (41). The metabolic origin of this modification remains to be elucidated, although certain investigators have hypothesized that it originates from endogenous endo-β-N-acetylglucosaminidase (ENGase) activity on proteins bearing N-linked oligosaccharide chains (39,41). However, this theory has been recently challenged by a study demonstrating that proteins with N-GlcNAc modifications are produced in ENGase knockout strains of Arabidopsis, indicating the presence of a separate and as yet unresolved pathway (40). The homologous O-linked GlcNAc found in certain eukaryotic glycoproteins has gained recognition as a regulatory post-translational modification akin to reversible phosphorylation (42). O-GlcNAc linkages are produced by a single conserved nucleocytoplasmic enzyme, O-GlcNAc transferase, which catalyzes GlcNAc transfer from UDP-GlcNAc to the hydroxyl oxygen of Ser/Thr (42). Previous biochemical studies also described the isolation of a eukaryotic N-GlcNAc transferase (NGT) that transfers GlcNAc from UDP-GlcNAc to the carboxamide nitrogen of Asn in Asn-Xxx-Ser/Thr sequons (EC 2.4.1.94) (43–45). According to these studies, NGT activity was detected in various organisms and subcellular locations. Therefore, it remains plausible that the N-GlcNAc modifications identified in the present study, and in previous studies (37–41), may be associated with the activity of an uncharacterized NGT homolog.
Further studies are required to determine whether N-GlcNAc modifications affect glycoprotein function by regulating enzyme activity, protein localization or protein-protein interactions. In the present study, a molecular modeling approach was undertaken to examine whether the N-GlcNAc modifications of PAP-S1aci or PAP-S1 could affect interactions with the ribosome. Previous studies have revealed the involvement of regions Asn42-Thr43 and Asn253-Val255 of PAP-I in the binding of the SRL of ribosomal RNA (33,46). The structural similarity between PAP-I and PAP-S1aci allowed us to map these rRNA-binding regions in PAP-I to the corresponding Asn44-GlcNAc and Asn255-GlcNAc residues in PAP-S1aci. Since PAP-I lacks glycosylation sequons and does not contain N-GlcNAc modifications, the present study aimed to determine whether larger oligosaccharide linkages at these positions in PAP-S1aci and PAP-S1 are able to impair ribosomal interactions. Our modeling studies suggested that the ribosome depurination activity of seed PAPs would be adversely affected by the N-glycosylation of Asn44 and Asn255 with larger and more typical oligosaccharide chains, as they would shield the rRNA-binding sites on the protein. Given our current structural data, the earlier in vitro evidence that PAP-I and PAP-S1aci exhibit similar ribosomal depurination activities (47,48) supports the hypothesis that smaller N-linked monosaccharides do not substantially affect ribosome binding. Taken together, this evidence suggests that N-GlcNAc modifications observed in certain type I seed RIPs, including PAP-S1aci, may serve a different biological function while also preserving ribotoxin activity.
Suggestions at a more plausible role for N-GlcNAc modifications stem from the observation that PAP-S1 exhibits far greater cytotoxicity in vivo than PAP-I or the A-chain of ricin (14). RIPs are hypothesized to be internalized by cells either through receptor-mediated or fluid-phase endocytosis. Double-chain RIPs, including ricin, are primarily taken up by hepatocytes via two main routes: i) The additional lectin B-chain interacts with galactosyl residues of the cell membrane and ii) mannose-containing oligosaccharides on the two chains exploit the mannose/GlcNAc-receptor-mediated uptake pathway (49). Non-glycosylated single-chain RIPs, including PAP-I, are hypothesized to enter cells by the less efficient route of fluid-phase endocytosis. The absence of carbohydrate chains on PAP-I, a cell-wall protein, may in fact reflect a specialized antiviral role involving a local suicide mechanism for compromised cells. Evidence demonstrating that PAP can efficiently inactivate pokeweed ribosomes supports this hypothesis (47). The superior cytotoxicity and biodistribution of PAP-S1 in animal models, however, favors an active receptor-mediated uptake mechanism characteristic of type-2 RIPs or fully glycosylated type-1 RIPs. Nevertheless, the question remains: Can N-linked GlcNAc residues effectively facilitate receptor-mediated uptake of seed RIPs, including PAP-S1, PAP-S1aci, α-luffin and β-luffin? A previous study on unrelated glycoproteins have demonstrated that ENGase-treated glycoproteins, which are trimmed to contain only Asn-GlcNAc monosaccharide linkages, are internalized via mannose/GlcNAc receptor-mediated pinocytosis with an even greater efficiency than their oligosaccharide-bearing counterparts (50). Collectively, this body of evidence supports our hypothesis that N-GlcNAc modifications may have evolved to enhance the cytotoxicity of type I seed RIPs by exploiting receptor-mediated uptake pathways of seed predators without compromising ribosome affinity. A deeper understanding of the different structural characteristics and uptake mechanisms that confer cytotoxic variability may prove beneficial in the development of modified RIPs tailored for specific therapeutic applications.
Acknowledgments
The authors would like to thank Dr. R. Hilgenfeld for support and early interest in the present study, Dr. K. Bezouska for protein sequencing and mass-spectrometry analysis and Dr. I.K. Smatanova for the preliminary crystallization experiments.
Abbreviations
- PAP
pokeweed antiviral protein
- GlcNAc
N-acetyl-D-glucosamine
- HexNAc
N-acetylhexosamine
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- r.m.s.d
root-mean square deviation
References
- 1.Puri M, Kaur I, Perugini MA, Gupta RC. Ribosome-inactivating proteins: current status and biomedical applications. Drug Discov Today. 2012;17:774–783. doi: 10.1016/j.drudis.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Stirpe F. Ribosome-inactivating proteins: from toxins to useful proteins. Toxicon. 2013;67:12–16. doi: 10.1016/j.toxicon.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Endo Y, Tsurugi K, Lambert JM. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: the RNA N-glycosidase activity of the proteins. Biochem Biophys Res Commun. 1988;150:1032–1036. doi: 10.1016/0006-291X(88)90733-4. [DOI] [PubMed] [Google Scholar]
- 4.Gessner SL, Irvin JD. Inhibition of elongation factor 2-dependent translocation by the pokeweed antiviral protein and ricin. J Biol Chem. 1980;255:3251–3253. [PubMed] [Google Scholar]
- 5.Stirpe F, Battelli MG. Ribosome-inactivating proteins: progress and problems. Cell Mol Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajamohan F, Kurinov IV, Venkatachalam TK, Uckun FM. Deguanylation of human immunodeficiency virus (HIV-1) RNA by recombinant pokeweed antiviral protein. Biochem Biophys Res Commun. 1999;263:419–424. doi: 10.1006/bbrc.1999.1335. [DOI] [PubMed] [Google Scholar]
- 7.Hudak KA, Bauman JD, Tumer NE. Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA. 2002;8:1148–1159. doi: 10.1017/S1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh BA, Coetzer C, Tumer NE. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J Biol Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
- 9.Parikh BA, Tumer NE. Antiviral activity of ribosome inactivating proteins in medicine. Mini Rev Med Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- 10.Mansouri S, Choudhary G, Sarzala PM, Ratner L, Hudak KA. Suppression of human T-cell leukemia virus I gene expression by pokeweed antiviral protein. J Biol Chem. 2009;284:31453–31462. doi: 10.1074/jbc.M109.046235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Cruz OJ, Waurzyniakt B, Uckun FM. A 13-week subchronic intravaginal toxicity study of pokeweed antiviral protein in mice. Phytomedicine. 2004;11:342–351. doi: 10.1078/0944711041495209. [DOI] [PubMed] [Google Scholar]
- 12.Uckun FM, Rustamova L, Vassilev AO, Tibbles HE, Petkevich AS. CNS activity of pokeweed anti-viral protein (PAP) in mice infected with lymphocytic choriomeningitis virus (LCMV) BMC Infect Dis. 2005;5:9. doi: 10.1186/1471-2334-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapira A, Benhar I. Toxin-based therapeutic approaches. Toxins (Basel) 2010;2:2519–2583. doi: 10.3390/toxins2112519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benigni F, Canevari S, Gadina M, Adobati E, Ferreri AJ, Di Celle EF, Comolli R, Colnaghi MI. Preclinical evaluation of the ribosome-inactivating proteins PAP-1, PAP-S and RTA in mice. Int J Immunopharmacol. 1995;17:829–839. doi: 10.1016/0192-0561(95)00068-D. [DOI] [PubMed] [Google Scholar]
- 15.Monzingo AF, Collins EJ, Ernst SR, Irvin JD, Robertus JD. The 2.5 A structure of pokeweed antiviral protein. J Mol Biol. 1993;233:705–715. doi: 10.1006/jmbi.1993.1547. [DOI] [PubMed] [Google Scholar]
- 16.Kurinov IV, Myers DE, Irvin JD, Uckun FM. X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs. Protein Sci. 1999;8:1765–1772. doi: 10.1110/ps.8.9.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng ZH, He XL, Li HM, Hu Z, Wang DC. Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8 A resolution. J Struct Biol. 2003;141:171–178. doi: 10.1016/S1047-8477(02)00580-4. [DOI] [PubMed] [Google Scholar]
- 18.Hogg T, Kuta Smatanova I, Bezouska K, Ulbrich N, Hilgenfeld R. Sugar-mediated lattice contacts in crystals of a plant glycoprotein. Acta Crystallogr D Biol Crystallogr. 2002;58:1734–1739. doi: 10.1107/S0907444902014506. [DOI] [PubMed] [Google Scholar]
- 19.Brune DC. Alkylation of cysteine with acrylamide for protein sequence analysis. Anal Biochem. 1992;207:285–290. doi: 10.1016/0003-2697(92)90013-W. [DOI] [PubMed] [Google Scholar]
- 20.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst A. 1986;42:140–149. doi: 10.1107/S0108767386099622. [DOI] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Cryst A. 1976;32:922–923. doi: 10.1107/S0567739476001873. [DOI] [Google Scholar]
- 27.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, USA: 2002. http://www.pymol.org. [Google Scholar]
- 29.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honjo E, Dong D, Motoshima H, Watanabe K. Genomic clones encoding two isoforms of pokeweed antiviral protein in seeds (PAP-S1 and S2) and the N-glycosidase activities of their recombinant proteins on ribosomes and DNA in comparison with other isoforms. J Biochem. 2002;131:225–231. doi: 10.1093/oxfordjournals.jbchem.a003092. [DOI] [PubMed] [Google Scholar]
- 31.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 33.Rajamohan F, Pugmire MJ, Kurinov IV, Uckun FM. Modeling and alanine scanning mutagenesis studies of recombinant pokeweed antiviral protein. J Biol Chem. 2000;275:3382–3390. doi: 10.1074/jbc.275.5.3382. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Hogg T, Hilgenfeld R. Protein crystallography in drug discovery. In: Taylor JB, Triggle DJ, Kubinyi H, editors. Comprehensive Medicinal Chemistry II. 2nd edition. Vol. 3. Elsevier; Amsterdam: 2007. pp. 875–900. [DOI] [Google Scholar]
- 36.Ho BK, Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Struct Biol. 2005;5:14. doi: 10.1186/1472-6807-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hase S, Fujimura K, Kanoh M, Ikenaka T. Studies on heterogeneity of Taka-amylase A: isolation of an amylase having one N-acetylglucosamine residue as the sugar chain. J Biochem. 1982;92:265–270. doi: 10.1093/oxfordjournals.jbchem.a133922. [DOI] [PubMed] [Google Scholar]
- 38.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci USA. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YC, Jahren N, Stone MD, Udeshi ND, Markowski TW, Witthuhn BA, Shabanowitz J, Hunt DF, Olszewski NE. Identification and origin of N-linked β-D-N-acetylglucosamine monosaccharide modifications on Arabidopsis proteins. Plant Physiol. 2013;161:455–464. doi: 10.1104/pp.112.208900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Islam MR, Kung SS, Kimura Y, Funatsu G. N-acetyl-D-glu cosamine-asparagine structure in ribosome-inactivating proteins from the seeds of Luffa cylindrica and Phytolacca americana. Agric Biol Chem. 1991;55:1375–1381. doi: 10.1271/bbb1961.55.1375. [DOI] [PubMed] [Google Scholar]
- 42.Wells L, Whelan SA, Hart GW. O-GlcNAc: a regulatory post-translational modification. Biochem Biophys Res Commun. 2003;302:435–441. doi: 10.1016/S0006-291X(03)00175-X. [DOI] [PubMed] [Google Scholar]
- 43.Khalkhall Z, Marshall RD. Glycosylation of ribonuclease A catalysed by rabbit liver extracts. Biochem J. 1975;146:299–307. doi: 10.1042/bj1460299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khalkhali Z, Marshall RD. UDP-N-acetyl-D-glucosamine-as paragine sequon N-acetyl-b-D-glucosaminyl-transferase-activity in human serum. Carbohydr Res. 1976;49:455–473. doi: 10.1016/S0008-6215(00)83163-8. [DOI] [PubMed] [Google Scholar]
- 45.Khalkhali Z, Marshall RD, Reuvers F, Habets-Willems C, Boer P. Glycosylation in vitro of an asparagine sequon catalysed by preparations of yeast cell membranes. Biochem J. 1976;160:37–41. doi: 10.1042/bj1600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajamohan F, Mao C, Uckun FM. Binding interactions between the active center cleft of recombinant pokeweed antiviral protein and the alpha-sarcin/ricin stem loop of ribosomal RNA. J Biol Chem. 2001;276:24075–24081. doi: 10.1074/jbc.M011406200. [DOI] [PubMed] [Google Scholar]
- 47.Bonness MS, Ready MP, Irvin JD, Mabry TJ. Pokeweed antiviral protein inactivates pokeweed ribosomes; implications for the antiviral mechanism. Plant J. 1994;5:173–183. doi: 10.1046/j.1365-313X.1994.05020173.x. [DOI] [PubMed] [Google Scholar]
- 48.Poyet JL, Hoeveler A. cDNA cloning and expression of pokeweed antiviral protein from seeds in Escherichia coli and its inhibition of protein synthesis in vitro. FEBS Lett. 1997;406:97–100. doi: 10.1016/S0014-5793(97)00250-0. [DOI] [PubMed] [Google Scholar]
- 49.Barbieri L, Battelli MG, Stirpe F. Ribosome-inactivating proteins from plants. Biochem Biophys Acta. 1993;1154:237–282. doi: 10.1016/0304-4157(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 50.Gross V, Heinrich PC, vom Berq D, Steube K, Andus T, Tran-Thi TA, Decker K, Gerok W. Involvement of various organs in the initial plasma clearance of differently glycosylated rat liver secretory proteins. Eur J Biochem. 1988;173:653–659. doi: 10.1111/j.1432-1033.1988.tb14048.x. [DOI] [PubMed] [Google Scholar]