Abstract
Purpose
Aurora A kinase (AAK) is expressed exclusively during mitosis, and plays a critical role in centrosome duplication and spindle formation. Alisertib is a highly selective AAK inhibitor that has demonstrated marked clinical activity of alisertib across a spectrum of lymphomas, though particularly in patients with T-cell lymphoma (TCL). We sought to compare and contrast the activity of alisertib in preclinical models of B-cell lymphoma (BCL) and TCL, and identify combinations worthy of clinical study. High-throughput screening of pralatrexate, the proteasome inhibitor (ixazomib), and the histone deacetylase (HDAC) inhibitor (romidepsin) revealed that only romidepsin synergized with alisertib, and only in models of TCL. We discovered that the mechanism of synergy between AAK inhibitors and HDAC inhibitors appears to be mediated through cytokinesis failure.
Experimental Design
A high-throughput screening approach was used to identify drugs that were potentially synergistic in combination with alisertib. Live-cell imaging was used to explore the mechanistic basis for the drug: drug interaction between alisertib and romidepsin. An in vivo xenograft TCL model was used to confirm in vitro results.
Results
In vitro, alisertib exhibited concentration-dependent cytotoxicity in BCL and TCL cell lines. Alisertib was synergistic with romidepsin in a T-cell–specific fashion that was confirmed in vivo. Live-cell imaging demonstrated that the combination treatment resulted in profound cytokinesis failure.
Conclusions
These data strongly suggest that the combination of alisertib and romidepsin is highly synergistic in TCL through modulation of cytokinesis and merits clinical development.
Introduction
The T-cell lymphomas (TCL) are a heterogeneous subset of non-Hodgkin’s lymphoma that exhibit a poor prognosis. Present treatment options for patients with relapsed/refractory (RR) PTCL (peripheral T-cell lymphomas) are largely palliative. Recently, a retrospective study of patients (N = 153) with PTCL after first relapse or progression, who were not candidates for stem cell transplant, reported a median overall survival (OS) of only 5.5 months and a median progression-free survival (PFS) of only 3.1 months (1). These findings underscore the need to not only identify novel drugs active in PTCL, but to think about how these agents might be configured in rational combination regimens. Since 2009, the FDA has approved pralatrexate, two histone deacetylase (HDAC) inhibitors and the CD30-targeted immunoconjugate brentuximab vedotin (Bv) for patients with RR PTCL and anaplastic large T-cell lymphoma (ALCL; ref. 2). An intriguing feature of these drugs is their “apparent” lineage specific activity, as pralatrexate and the HDAC inhibitors have shown significant activity in patients with PTCL. Bv was approved in ALCL, the prototypical disease expressing CD30. Bv has also demonstrated activity in other malignancies known to express CD30, albeit not to the same extent seen in ALCL (3). Although the activity of these agents in heavily treated patients is impressive, their lineage-specific activity offers the prospect that they can form the basis of novel drug regimens with improved activity in PTCL.
Aurora kinases are a family of serine-threonine kinases (AAK, Aurora B and Aurora C kinases) that are highly expressed during mitosis, with very specific functions in cell signaling and mitotic division. AAK plays a critical role in chromosome maturation and separation as well as bipolar spindle assembly during G2–M phase of mitosis (4). High-level expression of AAK is associated with centrosome amplification, mitotic abnormalities, chromosomal instability, and malignant transformation (5). It has been shown that overexpression of AAK plays a role in the pathogenesis of various hematologic malignancies. Kanagal-Shamanna and colleagues (6) found that overexpression of AAK was detected in 68% of TCL cases, including ALK+ and ALK− ALCL, PTCL-not otherwise specified (PTCL-NOS), cutaneous T-cell lymphoma (CTCL), T-cell lymphoblastic lymphoma/leukemia, and T-cell prolymphocytic leukemia, providing support for its role in T-cell lymphomagenesis. AAK inhibitors have been shown to exhibit unique activity in TCL and do not produce the neurotoxicity seen with other M-phase–specific agents (7–9). Alisertib is a highly selective competitive inhibitor of the ATP-binding site on AAK. The inhibition of AAK causes a mitotic spindle defect that leads to abnormal mitosis, initiating an accumulation of cells in G2–M and the development of polyploidization. Preclinical studies of alisertib in models of B-cell lymphoma (BCL) have demonstrated that a dose of 20 and 30 mg/kg administered daily for 3 weeks exhibited 100% inhibition of tumor growth (10). Although these models support the broad activity of alisertib across many subtypes of BCL, virtually no preclinical data exist in TCL, where clinical development is largely focused.
We sought to systematically compare and contrast the activity of alisertib in panels of BCL and TCL, and screen for its potential synergy with other drugs active in PTCL, including ixazomib, pralatrexate, and romidepsin. We demonstrate that alisertib and romidepsin appear to exhibit a remarkably restricted pattern of synergy only in models of TCL, but not in BCL. To clarify the mechanistic basis for the synergy we developed a novel live suspension cell imaging technique to demonstrate that the combination of alisertib and romidepsin induces cytokinesis failure. These data report for the first time that this combination exhibits lineage-specific activity, which is confirmed in a novel cell–based in vitro imaging assay. We believe these findings may create prospects for biomarker discovery efforts in the clinic.
Materials and Methods
Cells and cell lines
H9, HH, C5MJ, J. Cam 1.6, SUP-T1, Tib152, and CCL119 are TCL cell lines purchased from the ATCC. SU-DHL6, SU-DHL2, Jeko-1, JVM-2, Z-138, Rec-1 are characterized BCL lines purchased from the ATCC. DND41 is a T-cell line purchased from Deutsche Sammlung von Mikroorganism und Zellkulturen GmbH (DSMZ; Braunschweig, Germany). OCI-LY10 and OCI-LY7 are BCL cell lines from DSMZ. All cell lines were grown in RPMI-1640 (H9, HH, J. Cam 1.6, SUP-T1, Tib-152, CCL119, DND41, Jeko-1, JVM-2, Z-138, Rec-1, SU-DHL6, and SU-DHL2) or IMDM media (C5MJ, OCI-LY10, and OCI-LY7) with 10% FBS (Life Technology) and maintained at a concentration of 0.3 × 106 cells/mL. All cell lines were authenticated from a hematopathologist, including verification of morphology and immunophenotype (11–13).
Materials
Reagents for Western blotting were obtained from Bio-Rad laboratories and Invitrogen Life Technologies. DMSO was obtained from Sigma-Aldrich. Drugs were obtained as follows: alisertib and ixazomib (MLN-2238) were provided by Takeda Pharmaceuticals, pralatrexate, and romidepsin were obtained from the institutional pharmacy. All reagents for cell-cycle and apoptosis analysis were obtained from Invitrogen Life Technologies.
Cytotoxicity assays
For all in vitro assays, cells were counted, incubated, and processed as previously described (11–15). Alisertib and ixazomib were diluted in DMSO to a final concentration of ≤0.01%. Romidepsin was diluted in 2 mL of 80% propylene glycol (USP) and 20% dehydrated alcohol (USP). Pralatrexate was diluted in 1 mL of diluent consisting of sodium chloride, sodium hydroxide, and hydrochloric acid to achieve an isotonic solution. For combination experiments, the final concentration of all drugs was selected to approximate the IC10–IC30. For all cytotoxicity experiments, Cell-Titer-Glo Reagent (Promega Corp.), a Synergy HT Multi-Detection Microplate Reader (Biotek Instruments, Inc.) were used as previously described (11–14, 16). Synergistic interactions were measured using excess over bliss (EOB) as previously described (16–18).
Flow cytometry
Cells were seeded at a density of 3 × 105 cells/mL and incubated for 72 hours with alisertib and romidepsin, alone or in combination at concentrations approximating the IC10–IC30. A minimum of 1 × 105 events were acquired for each sample. To quantitate apoptosis, cells were stained with Alex Fluor 488/Annexin V and propridium iodine (PI; Invitrogen #V13240) according to the manufacturer’s instruction. Flow cytometry was performed on a FACS Calibur System and the data were analyzed with Flowjo 8.8.6 software. Cells were considered in early apoptosis if annexin V positive but PI negative, late apoptotic if annexin V and PI positive, and dead if only PI positive.
Cell-cycle analysis
Cells were seeded at a density of 3 × 105 cells/mL and incubated with alisertib and romidepsin, alone or in combination with concentrations approximating the IC10–IC30. After 24 hours of incubation, cells were harvested and washed twice with 1 mL of cold PBS. Cells were then fixed with 70% histology grade ethanol for 2 hours. After the incubation period, cells were resuspended in PBS and washed once. After cells were suspended in 1 mL of Triton 0.1x containing RNase A (Ambion #2286) and propridium iodide (Invitrogen #P3566) in a 1:50 dilution and kept at room temperature for 30 minutes. The fluorescence signal was acquired by FACS Calibur System and analyzed using Flowjo 8.8.6.
Live-cell imaging
Cells were plated onto 35-mm glass-bottom dishes (MatTek Corporation) and partially synchronized with 1 mmol/L nocodazole for 12 hours. Cells were then released from nocodazole and subjected to different drug treatments. Before live-cell imaging, cells were incubated with Hoechst 33342 (1 mg/mL) for 30 minutes. Time-lapse microscopic images were acquired every 10 minutes in a 37°C, 5% CO2 chamber for 48 hours using an inverted microscope (IX81; Olympus) with a 10×, NA = 0.6 dry objective lens (Olympus) and a monochrome CCD camera (Sensicam QE; Cooke), and processed using a Slidebook 5.5 software (Olympus).
Western blot analysis
Cells were incubated with the IC10–IC30 of each drug alone (alisertib, romidepsin) and in combination (alisertib plus romidepsin) under normal growth conditions for 72 hours. Proteins from total cell lysates were resolved on 4% to 20% tris-glycine gel (Invitrogen #EC6028BOX) and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked in TBS containing 0.2% Tween and 5% skim milk powder. Membranes were then incubated overnight with specific primary antibodies. Antibodies were detected as previously described (12). All monoclonal and polyclonal antibodies used were from cell signaling.
In vivo tumor model
In vivo experiments were performed as previously described (11, 12, 14). Five to 7-week-old SCID mice were injected with 5 × 106 cells/mL of HH, into the right flank subcutaneously with 50 μL of B.D. Matrigel (BD Biosciences). When the tumor volume reached an average of 50 to 100 mm3 mice were randomly divided into four cohorts of 10 mice each (control 0.01% DMSO, alisertib 20 mg/kg, romidepsin 1 mg/kg, alisertib 20 mg/kg + romidepsin 1 mg/kg). Tumor volumes were assessed as previously described (11). Control mice were given 0.09% saline i.p. on days 1, 9, 16. Romidepsin was diluted in 0.09% saline given i.p. on days 1, 9, 16. Alisertib was diluted in saline and administered daily for 21 days by oral gavage. In the combination cohort, romidepsin and alisertib were administered at the same dose and frequency as the single-agent cohorts. Control and drug treated mice received diluted DMSO concentrations of 0.01%.
Quantification of alisertib and romidepsin in mouse plasma and tumor tissue
To define and compare the pharmacokinetic (PK) profile of the two agents and their combination in blood, serial blood collections were performed at 30 minutes, 1, 2, and 6 hours in 2 mice per cohort (two time points were taken from each mouse) after day 16 of treatment. Tumor tissue was collected and harvested following 1 and 6 hours after treatment. Romidepsin and alisertib were quantified in serum and tissue by extraction using Acetonitrile followed by liquid chromatography tandem mass spectrometry (LC/MS-MS). Nine volumes of chilled acidified Acetonitrile (0.1% formic acid) were added to 100 μL of serum or 400 μL of aqueous tissue homogenate obtained by disrupting 100 mg of wet tissue using a tissue tearer homogenizer. The mixture was incubated at 4°C for 15 minutes followed by centrifugation at 13,000 × g for 10 minutes at 4°C. The supernatant was transferred to an LCMS vial and evaporated under nitrogen stream. The extracted compounds were resuspended in sample buffer (40% methanol) for further analysis. A calibration curve was prepared by spiking untreated mouse serum spanning a range between 50 pg/mL and 250 ng/mL and extracted same as the samples.
The method was developed on a platform comprising an Eksigent UPLC 100 integrated to API 4000 tandem mass spectrometer controlled by Analyst 1.6 software (all from AB Sciex). A 5 μL was injected into a Phenomenex Kinetex C18 column (50 × 2.1 mm, 1.7u, 100A) column preceded by a C18 guard column that was kept at 40°C. The flow rate was maintained at 200 μL/min. The initial flow conditions were 60% solvent A (H2O containing 0.1% Formic acid) and 40% solvent B (Methanol with 0.1% FA). After a 1 minute loading time, Solvent B was raised to 95% linearly over 7 minutes and held for 30 seconds and back to initial conditions by 8 minutes and held constant until 10 minutes for column equilibration and to avoid carry over. Romidepsin and Alisertib eluted at 4.37 and 6.09 minutes, respectively. Both compounds were detected in positive electrospray ionization (ESI+) and multiple reactions monitoring (MRM) mode with optimized mass spectrometer parameters for both compounds. Romidepsin and Alisertib were detected at mass to charge transitions of 541.2 > 423.9 and 519.0 > 328.1. Concentrations of romidepsin and alisertib in all samples were calculated by comparing integrated peak areas against calibration standards. Single-point standard addition method was used to assess the matrix effect of tumor tissue samples. Concentrations in tumor samples were corrected for any matrix effect.
Statistical analysis
The Kaplan–Meier survival functions were calculated for each cohort using the log-rank test to compare the median survival times among cohorts. Linear mixed model with random intercept was used to analyze log-transformed tumor burden in the in vivo experiment. Predictors in the model include time, treatment group, and interactions between time and group. Statistical significance is denoted by a ρ value of >0.05.
Results
Concentration:effect relationship of alisertib across a panel of BCL and TCL cell lines
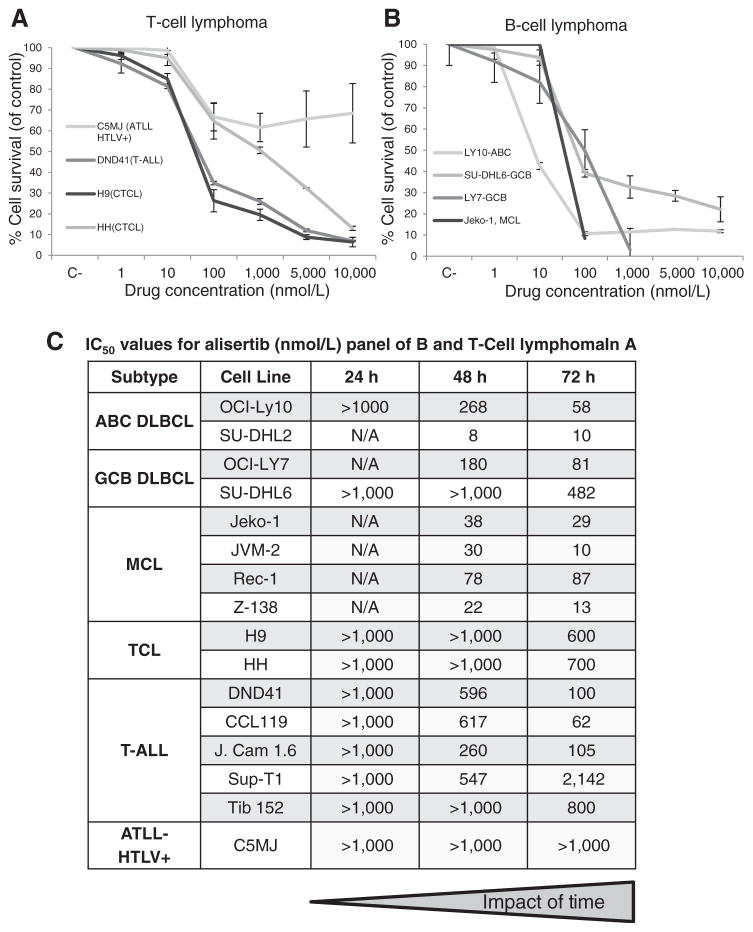
Previously published reports have demonstrated an IC50 of approximately 100 nmol/L for alisertib in select BCL (19). Figure 1 presents the concentration:effect relationships of alisertib across a broad panel of BCL (OCI-LY10, OCI-LY7, SU-DHL6, SU-DHL2, Jeko-1, JVM-2, Rec-1, and Z-138) and TCL (H9, HH, DND41, CCL119, J. Cam 1.6, SUP-T1, Tib 152, and C5MJ) cell lines (Fig. 1A and B). Alisertib exhibited a concentration and strong time dependent cytotoxic effect with the lowest IC50 values achieved at 72 hours in the range of 10 to 100 nmol/L and 60 to 1,000 nmol/L for BCL and TCL cell lines, respectively (Fig. 1C). In addition, continuous exposure to alisertib was more cytotoxic than a 1 and 3 hour pulse in two TCL cell lines (data not shown). The exquisite impact of time is consistent with the cell-cycle–dependent effects of an M-phase–specific drug, and suggests that area under the curve of exposure is critical in mediating cytotoxicity.
Figure 1.
Alisertib is cytotoxic in TCL and BCL. Cytotoxicity curves were generated for a panel of TCL and BCL cell lines following 24, 48, 72 hours of treatment. A, concentration:effect of alisertib following 72 hours of treatment in four representative TCL cell lines (C5MJ, DND41, H9, and HH). B, concentration:effect of alisertib following 72 hours of treatment in four representative BCL cell lines (LY10, SU-DHL6, LY7, and Jeko-1). C, IC50 values were generated for alisertib following 24, 48, 72 hours of treatment in a panel of TCL and BCL cell lines.
Alisertib is not synergistic with pralatrexate or ixazomib in BCL and TCL cell lines
Single-agent concentration:effect relationship curves were generated for pralatrexate and the proteasome inhibitor ixazomib (MLN-2238) in a panel of B- and T-cell lines. The 72 hour IC50 values across a panel of BCLs and TCLs for pralatrexate and ixazomib were in a range from 1 to 5 nmol/L and 2 to 40 nmol/L (ixazomib). Using the IC10–IC20 of pralatrexate (0.75 and 1.5 nmol/L) or ixazomib (10 and 15 nmol/L) and the IC10–IC30 of alisertib (50, 100, 1,000 nmol/L), we evaluated the drug:drug interactions in a schedule (drug A before B, drug B before drug A, and simultaneous exposure of drug A and drug B) and time-dependent manner. The combinations of alisertib and pralatrexate in both BCL and TCL generated additive effects on cytotoxicity defined by excess over bliss (EOB) values less than or equal to 13 (Supplementary Fig. S1A and S1B). On the other hand, when alisertib was combined with ixazomib, an antagonistic effect was observed across of the BCL and TCL cell lines, with the EOB values in the range of −5 to 5 (Supplementary Fig. S1C and S1D). These data establish that in these models the combination of alisertib with pralatrexate or ixazomib does not produce a synergistic interaction in either BCL or TCL cell lines.
Alisertib is markedly synergistic with romidepsin in in vitro TCL models but not in in vitro BCL models
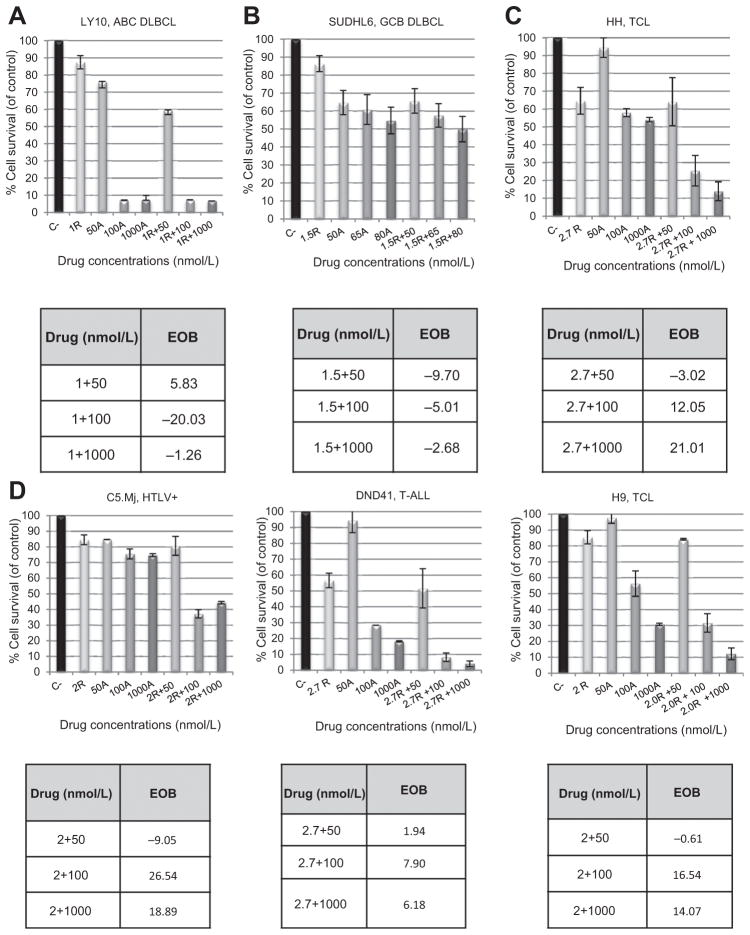
Using the IC10–IC30 of alisertib, we evaluated the synergistic interaction using the IC10–IC20 (approximately 1–2.7 nmol/L) of romidepsin across a panel of in vitro T- and B-cell lines following 24, 48, and 72 hours of simultaneous drug exposure. The IC10–IC20 of romidepsin was determined for all cell lines based on the single-agent concentration:effect relationship curves. When alisertib was combined with romidepsin in BCL, there was an additive to antagonistic interaction across all BCL cell lines evaluated (4 DLBCL and 4 MCL). Figure 2A and B depict two representative diffuse large B-cell lymphoma (DLBCL) cell lines (1 ABC and 1 GCB), indicating the additive to antagonistic interaction between alisertib and romidepsin following 72 hours of treatment.
Figure 2.
Alisertib in combination with romidepsin is synergistic in TCL but not in BCL. The IC10–20 of romidepsin (R) was used in combination with 50, 100, 1,000 nmol/L (IC10–30) of alisertib (A) to evaluate drug:drug interactions for up to 72 hours of treatment. A and B, following 72 hours of simultaneous drug exposure, synergy was not observed in a representative panel of BCL cell lines. EOB values were in the range of −20.03 to 5.83, signifying an antagonistic drug:drug interaction. C and D, following 72 hours of simultaneous drug exposure, synergy was observed in a panel of TCL cell lines with EOB values ranging from −9.05 to 21.01.
Interestingly, there was a highly synergistic interaction of alisertib and romidepsin in TCL following 72 hours of drug exposure. Notably, the greatest synergistic interaction was observed in C5MJ, an alisertib resistant ATLL HTLV-1 Tax+ cell line (Fig. 2C and D). The EOB values for all 8 TCL cell lines evaluated were greatest after 72 hours of drug exposure.
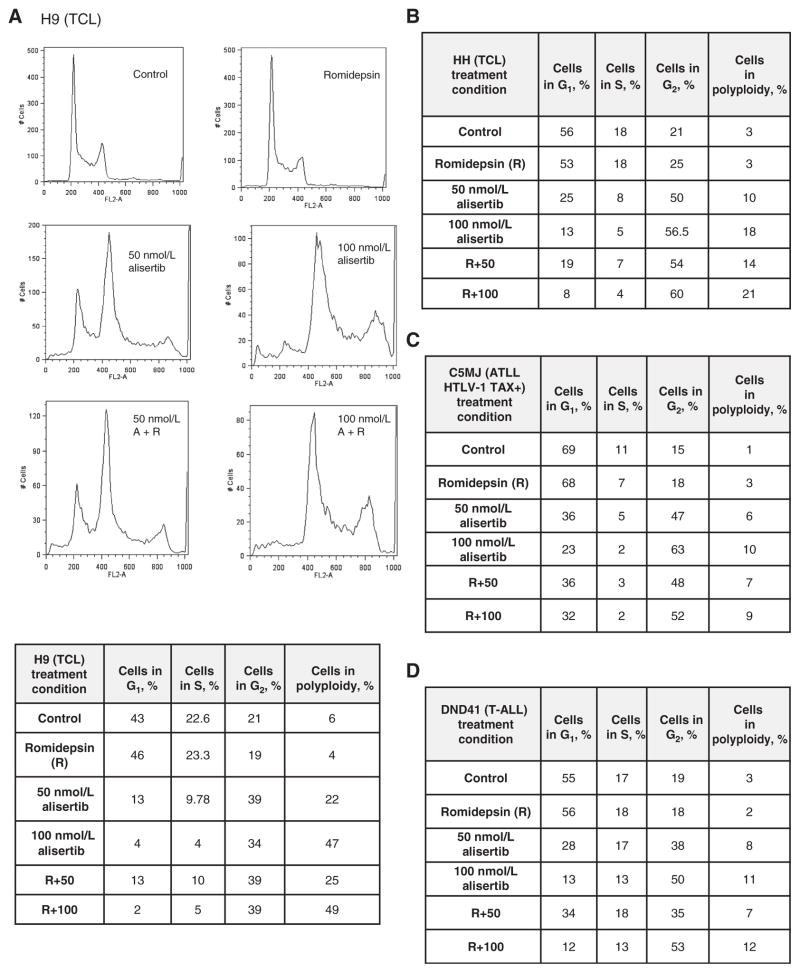
Alisertib in combination with romidepsin induces polyploidy in TCL cell lines
Romidepsin treatment has been shown to cause a G1 arrest through the upregulation of p21cip1/waf1 leading to the activation of the G1–S cell-cycle checkpoint, whereas alisertib treatment inhibits AAK, leading to a defect in spindle formation and a G2–M arrest (20–22). We evaluated cell-cycle arrest upon treatment of alisertib and romidepsin as single agents and in combination in 4 TCL cell lines (H9, HH, C5MJ, and DND41) following 24 hours of treatment. In all cell lines evaluated, romidepsin induced a modest increase (≤3.2%) in the percentage of cells in G1 compared with control, whereas alisertib demonstrated a dose-dependent increase in the percentage of cells in G2–M arrest following 50 and 100 nmol/L of drug (in the range of 13%–48%) when compared with control. The combination of alisertib and romidepsin induced a marked increase in polyploidy (in the range 10% to 42%), while inducing a significant decrease of cells in G1 relative to the control. The combination of alisertib and romidepsin at the IC10 and IC20, respectively, was the most potent in inducing polyploidy (up to 42%; Fig. 3A–D). Following 48 hours of treatment, virtually all cells were polyploid with little distinction among the other stages of cell cycle (data not shown).
Figure 3.
Combined treatment of alisertib and romidepsin induces polyploidy in TCL cell lines following 24 hours of treatment. Cells were stained with PI following 24 hours of treatment with an IC10–20 of romidepsin (R) and 50 or 100 nmol/L of alisertib as a single agent or in combination. A, H9, TCL cell line demonstrates the percentage of cells in G1, S, G2–M, or polyploid; B, the HH TCL cell line demonstrates the percentage of cells in G1, S, G2–M, or polyploid; C, C5MJ, ATLL HTLV-1 cell line demonstrates the percentage of cells in G1, S, G2–M, or polyploid; D, DND41, T-ALL cell line demonstrates the percentage of cells in G1, S, G2–M, or polyploid.
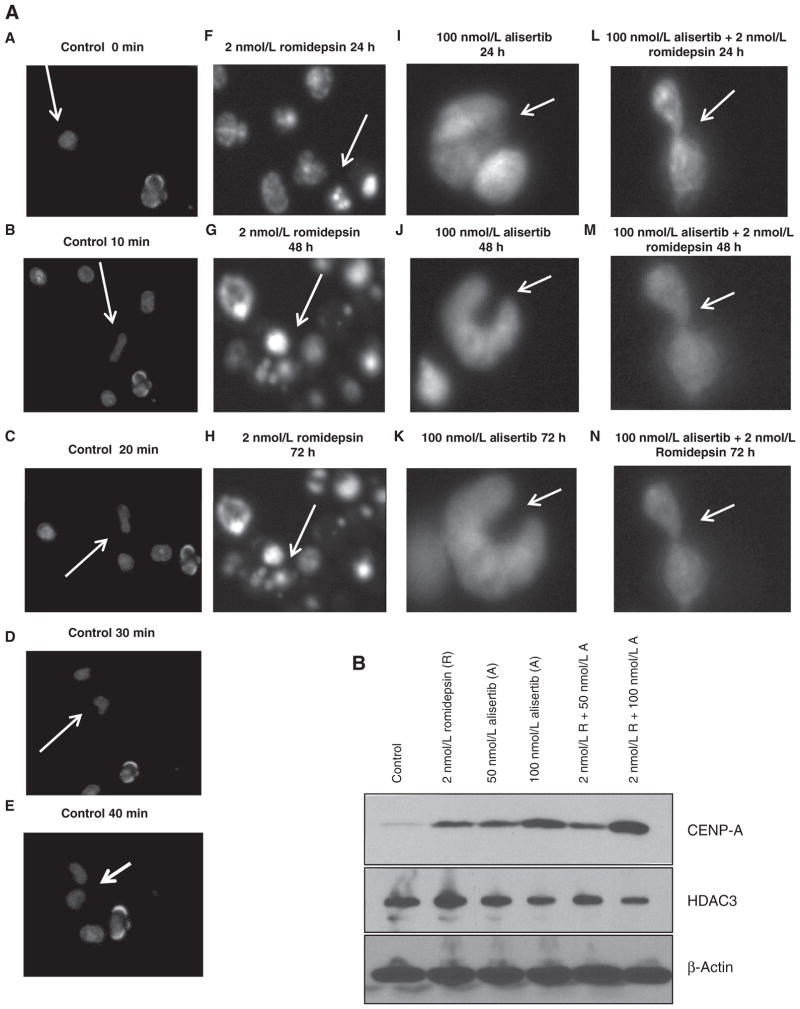
Live-cell imaging of TCL confirms failure of cytokinesis with alisertib and romidepsin in TCL
To explore the mechanism of action in more detail, we developed a live-cell imaging method for suspension cells. Figure 4A depicts single-cell images of all treatment conditions (control, 2 nmol/L romidepsin, 100 nmol/L alisertib, 100 nmol/L alisertib +2 nmol/L romidepsin) after synchronization with nocodazole in the H9 cell line. The control cells underwent normal mitosis in approximately 40 minutes as depicted by images in Fig. 4A–E. Following treatment with romidepsin, increased apoptosis was observed within 24 hours of treatment (Fig. 4F). Following treatment with alisertib, an accumulation of cells in G2–M was observed in about four hours (Fig. 4I). These results remained consistent following 48 to 72 hours of treatment with either romidepsin (Fig. 4G and H) or alisertib (Fig. 4J and K) and support the cell-cycle analysis data presented above. Interestingly, when alisertib was combined with romidepsin a spindle defect was observed inducing cytokinesis failure as soon as 15 hours following treatment (Fig. 4L–N). Supplementary Fig. S2 demonstrates the live-cell imaging time-lapse video from 0 to 72 hours for all treatment conditions [control (Supplementary Fig. S2A), romidepsin (Supplementary Fig. S2B), alisertib (Supplementary Fig. S2C), combination (Supplementary Fig. S2D), respectively]. Cytokinesis failure was confirmed after a corresponding increase in CENP-A protein levels following 72 hours of treatment with 100 nmol/L of alisertib, which was augmented with the combination treatment (Fig. 4B). CENP-A is a chromatin-associated protein that is histone h3 variant and plays a role in the final stages of cytokinesis. HDAC3 protein levels were evaluated following 72 hours of treatment with 50 or 100 nmol/L of alisertib and 2 nmol/L of romidepsin following both single agent and combination treatment (Fig. 4B). HDAC 3 is known to deacetylate AAK preventing AAK from proteolytic degradation (23). A slight decrease in HDAC3 levels following 100 nmol/L of alisertib and the combination treatment was appreciated in H9 cell line. This finding raises the prospect that the combination down regulates HDAC3, which has been shown to contribute to cytokinesis defects (24).
Figure 4.
Combined treatment of alisertib and romidepsin induces cytokinesis failure. A, live-cell imaging was used following 24 hours of treatment with 2 nmol/L of romidepsin and 100 nmol/L of alisertib both as a single agent and in combination in the H9, TCL cell line. Cells were imaged from 0 to 72 hours. A to E, images from control H9 sample demonstrate an H9 cell going through the stages of mitosis (prophase, metaphase, anaphase, telophase, and cytokinesis). Each stage is depicted every 10 minutes, for a total of 40 minutes. F, following 24 hours of 2 nmol/L romidepsin treatement H9 cells begin to display apoptotic effects depicted by the fragmented cells. G and H, following 48 and 72 hours of romidepsin treatment (respectively), there was an increase in fragmented cells; however, no mitosis defect was observed. I, following 24 hours of 100 nmol/L alisertib treatment, H9 cells demonstrate a multinucleated form depicted by the arrow. J and K, following 48 and 72 hours of alisertib treatment (respectively), the cell was shown to be arrested, no longer entering mitosis. L, following 24 hours of combination treatment (100 nmol/L alisertib + 2 nmol/L romidepsin), the H9 cell demonstrated cytokinesis failure. M and N, cytokinesis failure was observed following 48 and 72 hours of treatment, respectively. B, evaluation of CENP-A protein levels following 72 hours of treatment with 2 nmol/L of romidepsin(R), 50 nmol/L of alisertib(A), 100 nmol/L of alisertib (A) as a single agent of in combination. Evaluation of HDAC3 protein levels following 72 hours of treatment with 2 nmol/L of romidepsin, 50 nmol/L of alisertib, 100 nmol/L of alisertib as a single agent of in combination.
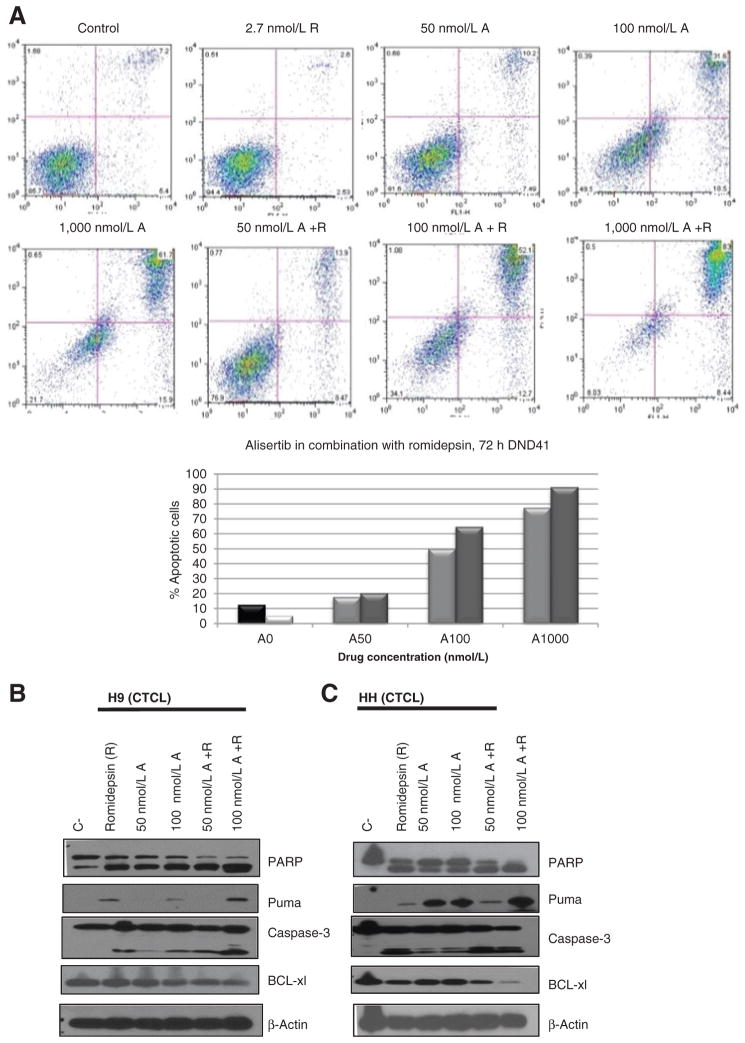
Alisertib in combination with romidepsin induces apoptosis in TCL cell lines
Apoptosis as measured by positive annexin V staining was analyzed in a TCL cell line following 72 hours of treatment with the IC10 of romidepsin and IC10–30 of alisertib both as a single agent or in combination (Fig. 5A). Clearly, there was an increase in apoptosis as a function of the alisertib concentration, with approximately 13% and 52% of apoptotic cells being observed following treatment with 2.7 nmol/L of romidepsin and 50 nmol/L of alisertib and 2.7 nmol/L of romidepsin and 100 nmol/L of alisertib, respectively. The induction in apoptosis corresponded with an increase in caspase-3 activation and PARP cleavage, as well as an increase in the proapoptotic protein PUMA as well as a decrease in antiapoptotic protein Bcl-xL (Fig. 5B and C).
Figure 5.
Alisertib in combination with romidepsin induces apoptosis following 72 hours of treatment. A, following 72 hours of 2.7 nmol/L romidepsin (R), 50 nmol/L alisertib (A), 100 nmol/L alisertib (A), and combination 50 nmol/L A+R, 100 nmol/L A+R, we analyzed apoptosis by staining for annexin V/PI as measured by FACS. B, Western blot analysis was used to confirm apoptosis in H9, TCL cell line. Following 72 hours of R, 50 nmol/L A, 100 nmol/L A, 50 nmol/L A+R, 100 nmol/L A+R. C, Western blot analysis was used to confirm apoptosis in the HH, TCL cell line. Following 72 hours of R, 50 nmol/L A, 100 nmol/L A, 50 nmol/L A+R, 100 nmol/L A+R.
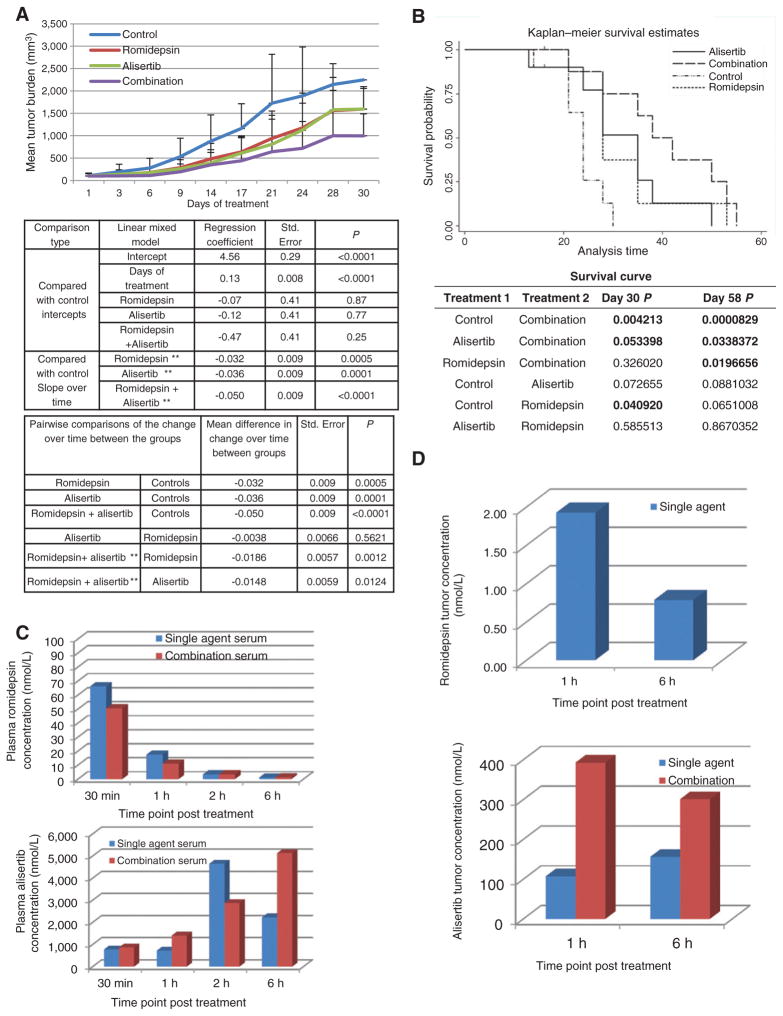
Alisertib in combination with romidepsin is synergistic in an in vivo xenograft model of TCL
The in vivo activity of alisertib and romidepsin as single agents or in combination were evaluated in an in vivo xenograft model of TCL using the HH cell line. Figure 6A demonstrates that the combination cohort was statistically superior compared with the single-agent treatment and the control cohort (P < 0.05.) over time. Figure 6B establishes that the survival of the combination cohort was also statistically significant (P < 0.05) when compared with the single-agent treatment and control arms (by day 58). PK analyses were used to determine the concentration of romidepsin and alisertib on day 16, 30 minutes, 1, 2, and 6 hours after treatment (one sample collected/timepoint/treatment cohort). Figure 6C (top histogram) demonstrates that the single-agent romidepsin concentration in plasma samples was approximately 60 nmol/L at 30 minutes after treatment, and then decreased to 3 and 1 nmol/L at 2 and 6 hours after treatment, respectively. At 6 hours after treatment, the single-agent romidepsin plasma concentration approximated the IC5, which is obviously less than the IC10–20 concentration used in vitro. It is important to note that the PK profile of romidepsin was nearly identical in both single agent and combination cohorts. These data are consistent with previous PK analyses done in our laboratory with romidepsin in NOG mice (16). The alisertib concentration in plasma was in the range of 770 to 4,600 nmol/L in both the single agent and combination plasma samples and the concentration time pattern of the single agent was similar to that described earlier (Fig. 6C, bottom histogram; refs. 10, 25). These concentrations approximated the IC70–80 in our in vitro studies. Interestingly, the plasma concentration of alisertib is still lower than that achieved in patients at the MTD. A 50 mg twice a day dose of alisertib produces a concentration of approximately 192 μmol/L. Figure 6D presents the PK data evaluating the concentration of romidepsin (top histogram) and alisertib (bottom histogram) in tumor tissue on day 16 following 1 and 6 hours of treatment. The intratumor concentration of romidepsin was approximately 2 and 1 nmol/L at 1 and 6 hour after treatment, respectively. These data are concordant with the single-agent romidepsin concentration in plasma samples as well as our in vitro combination data and our previous PK analyses (16). Interestingly, the concentration of romidepsin at 1 and 6 hours was equivalent in plasma and tumor samples supporting rapid distribution of drug in vivo. In contrast, the combination tumor samples revealed the romidepsin concentration was at the lower detection limit of 0.3 nmol/l per gram of tumor, with an average tumor size of 200 mg. The single-agent alisertib tumor concentration was 100 and 150 nmol/L at 1 and 6 hours after treatment, respectively. These concentrations are approximate the IC35–40 at 72 hours in our in vitro analyses. Even though, the romidepsin concentration was at the detection limit of 0.3 nmol/l per gram of tumor, the alisertib concentration in the combination tumor samples increased from 100 to 400 nmol/L (1 hour after treatment) and 150 to 300 nmol/L (6 hours after treatment) when compared with the single-agent alisertib tumor samples. These data support the synergistic cytotoxicity of alisertib and romidepsin.
Figure 6.
Alisertib in combination with romidepsin is synergistic in a xenograft model of cutaneous TCL. The HH, TCL cell line was injected into SCID mice. Control mice were injected i.p. with 0.01% DMSO (N = 10). Romidepsin 1 mg/kg was given i.p. day 1, 9, 16 (N = 10). Alisertib 20 mg/kg was given orally once a day from days 1 to 21 (N = 10). Combination mice followed the same treatment schedule as single agents (N = 10). Cycle 1 ended on day 21, and cycle 2 began on day 23, and ended on day 42. A, combination mice showed a statistically significant (P < 0.05) log-transformed tumor burden when compared with the control cohort over time (top table) and single agents over time (bottom table). B, Kaplan–Meier survival curve demonstrates that combination mice surpassed survival of all other cohorts and demonstrates statistical significance when compared with control and single agents (P < 0.05). C, PK analysis on plasma samples was performed following 30 minutes, 1, 2, and 6 hours of treatment. The top histogram depicts romidepsin (R) concentration for all timepoints evaluated whereas bottom histogram depicts the alisertib (A) concentration. D, PK analysis on tumor samples was performed on samples following 1 and 6 hours of treatment. Romidepsin intratumor concentration is depicted in the top histogram whereas alisertib intratumor concentration is depicted in the bottom histogram.
Discussion
AAK is a serine threonine kinase that autophosphorylates at threonine 288. AAK plays a major role in the regulation of mitosis, including targeting G2–M transition and DNA content. Although this role is well known, the exact mechanism of action of how AAK regulates mitosis is still not clear. Marumoto and colleagues (26) have demonstrated that AAK phosphorylates histone 2b and histone 3 while maintaining its maximal kinase activity during M-phase. The activation of AAK is associated with activity of cyclin B-associated kinase, which may suggest that AAK interacts with cyclin B1 to facilitate entry into mitosis. In addition, AAK inactivation occurs through DNA damage induced at the end of G2. However, if AAK is overexpressed the G2 checkpoint will be abrogated and cellular proliferation will occur. Although AAK plays a major role in G2–M, it is well known that HDAC’s play a role in inducing G1–S. HDAC inhibitors have been shown to alter kinetochore assembly through hyperacetylation of pericentromeric histones (23). Park and colleagues (27) have demonstrated that HDAC inhibitors induce degradation of AAK and Aurora B kinase. In addition to the cytokinesis failure demonstrated here, the data from Park and colleagues suggest that HDAC inhibitor–mediated degradation of AAK could further complement the activity of the combination seen with an AAK inhibitor. The finding that the AAK inhibitor, alisertib, and an HDAC inhibitor were selectively active in the T-cell lineage over B-cell was unexpected. One potential explanation relates to the practical observation that HDAC inhibitors are highly active drugs in TCL with minimal activity in BCL. With three different HDAC inhibitors approved for clinical use all in essentially the same disease, there is irrefutable evidence that TCL are sensitive to this class of drugs. Another explanation could be related to data from Kretzner and colleagues (28), suggesting that vorinostat in combination with a pan-aurora kinase inhibitor is synergistic in BCL due to a down-regulation of c-MYC. The C-MYC protein is a likely downstream target of aurora B kinase. Therefore, it could be hypothesized that aurora B kinase would need to be inhibited leading to down-regulation of c-MYC to see synergy in BCL. Despite this, in these model systems, the combination of alisertib and romidepsin consistently demonstrated substantially more activity in cell lines derived from T-cell malignancies compared with those derived from B-cell malignancies. What remains elusive, however, is that despite nearly 2 decades of preclinical and clinical research into these drugs, there are still no good predictive biomarkers of activity in these diseases, likely owing to their pleiotropic properties.
What has been established is that HDAC inhibitors do cause chromosome segregation defects through pericentromeric heterochromatin. Taddei and colleagues (24) demonstrated that prolonged exposure to low concentrations HDAC inhibitors leads to relocation and alteration within the pericentromeric heterochromatin. Corroborating these findings, Ishii and colleagues (29) demonstrated that HDAC3 uniquely localizes to the mitotic spindle during mitotic progression. When HDAC3 was knocked-down in HELA cells, the cells were unable to maintain a proper chromosome alignment due to defects within the mitotic spindles and kinetochore assembly (30). Using a unique live suspension cell imaging system, which we developed, we demonstrated that the combination of alisertib and romidepsin induces cytokinesis failure following 15 hours of treatment. This result is supportive of the Taddei and colleagues finding that longer exposure to low concentrations of HDAC inhibitors alters the pericentromeric heterochromatin, leading to improved therapeutic effects (24, 31). These findings are supported by the downregulation of HDAC3 and an increase in CENP-A. CENP-A is a chromatin-associated protein that contains a histone H3–related fold domain that is regulated through AAK phosphorylation. CENP-A is required for the recruitment of centromeric proteins, proper kinetochore assembly, and chromosome segregation. After DNA replication and cytokinesis, CENP-A accumulates in the nucleosome region of replicated centromeres to maintain homeostasis between CENP-A protein levels and the epigenetic mechanism of chromatin assembly of the newly replicated centromeres (32, 33). If there is a defect in the completion of cytokinesis, then CENP-A will not accumulate in the nucleosome region of the newly replicated centromeres and will accumulate in the cytosol as free CENP-A protein. Our data strongly support this as the primary mechanism of action for these two drugs, as cytokinesis failure results in an increase of CENP-A protein levels following treatment with alisertib and romidepsin.
Our in vivo experiment demonstrates that combination treatment is statistically beneficial when compared with single agents in log transformed tumor burden and survival analysis. Interestingly, we found that the intratumor concentration of romidepsin following combination treatment was very low. There are a few reasons for this. First, the concentration of romidepsin was found to be at the lower detection limit of romidepsin is 0.3 nmol/L per gram of tumor with an average tumor size of 200 mg. It is possible that due to low concentration of intratumor romidepsin, the detection of the intratumor romidepsin following combination treatments was too low to quantify. Second, previous work by our laboratory and Amiri-Kordestani and colleagues (34) demonstrated that romidepsin is a multidrug-resistant (MDR) substrate and can induce the MDR, increasing the efflux of the drug. Although the basis for the increase in intratumor alisertib following combination treatment is unlikely correlated with the inhibition of the MDR because romidepsin is an inducer of the MDR, the data may suggest that romidepsin can selectively induce an influx pump pathway. This induction may lead to an increase in intratumor alisertib concentration following combination treatment when compared with the intratumor single-agent alisertib, which is observed in our PK analysis. It is important to note that even with low concentrations of romidepsin there was broad marked synergy seen in the combination cohort.
Although our data demonstrate a selective synergy with an HDAC inhibitor, it establishes an additive or worse interaction with pralatrexate and ixazomib. The antagonism with pralatrexate is not that unexpected, given that exposure to a drug that induces G1–S arrest would preempt cells from entering mitosis, and thus nullify the impact of an M-phase–specific AAK inhibitor. The antagonism observed when alisertib was combined with ixazomib was a little surprising, given that proteasome inhibitors have been shown to synergize with so many different classes of drugs, and are also known to induce mitotic catastrophe (11, 12, 35–37). Cha and colleagues have demonstrated that panobinostat induced degradation of AAK and ABK (presumably through acetylation) through the ubiquitin–proteasome pathway (UPP) by directly targeting HDAC3 and HDAC6. Interestingly, when Cha and colleagues treated HELA cells with the proteasome inhibitor MG132, there was marked suppression of panobinostat induced AAK and ABK depletion. The authors suggested that inhibition of the UPP led to accumulation of AAK, which could be a mechanism of resistance to MG132. Alternatively, it was been well established that proteasome inhibitors increase CDK inhibitors like p21 and p27, inducing a G1–S arrest. It is possible that some combination of these events could account for the observed lack of synergism.
Presently, there is a randomized phase III clinical trial with alisertib in TCL, versus dealers choice (pralatrexate, romidepsin, or gemcitabine), and a phase 1 study of alisertib and romidepsin (NCT01482962 and NCT01897012). Future preclinical studies will be focused on the biochemical effects of these drugs and determine whether there are other mitotic agents that can more efficiently modulate mitotic proteins.
Supplementary Material
Translational Relevance.
Aurora A kinase (AAK) overexpression has been demonstrated in hematologic malignancies, including TCL. Recently, selective inhibitors of AAK have been studied in the clinic and have been shown to be active across a spectrum of lymphomas, particularly in peripheral T-cell lymphomas (PTCL). Capitalizing on the activity in PTCL requires a systematic evaluation of alisertib’s activity with other T-cell–active drugs, given none of these agents will be used exclusively as single agents in the future. Using a high-throughput screening approach, we demonstrated a highly selective synergistic interaction between alisertib and romidepsin in preclinical and clinical models of TCL. We show that the combination of alisertib and romidepsin synergize through induction of complete cytokinesis failure, generating a compelling rationale for the clinical evaluation of this combination in patients with PTCL.
Acknowledgments
The authors thank the Columbia University Lymphoma Research Fund for support. The authors thank University of Tennessee/West Cancer Center for research support for this project.
Grant Support
This publication was supported by the National Center for Advancing Translational Sciences, NIH, through grant number UL1TR000040, and in part by a grant from Takeda/Millennium.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
O.A. O’Connor reports receiving a commercial research grant from Takeda/Millennium Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: K.M. Zullo, L. Cooke, S. Cremers, D. Mahadevan, O.A. O’Connor
Development of methodology: K.M. Zullo, Y. Guo, L. Cooke, Y. Mao, R. Nandakumar, S. Cremers, D. Mahadevan, O.A. O’Connor
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): K.M. Zullo, Y. Guo, L. Cooke, X. Jirau-Serrano, R. Nandakumar, S. Cremers
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): K.M. Zullo, L. Cooke, X. Jirau-Serrano, M. Mangone, L. Scotto, J.E. Amengual, Y. Mao, R. Nandakumar, S. Cremers, J. Duong, O.A. O’Connor
Writing, review, and/or revision of the manuscript: K.M. Zullo, Y. Guo, L. Cooke, M. Mangone, L. Scotto, J.E. Amengual, Y. Mao, D. Mahadevan, O.A. O’Connor
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K.M. Zullo, Y. Guo, L. Cooke, X. Jirau-Serrano, M. Mangone, D. Mahadevan, O.A. O’Connor
Study supervision: L. Scotto, J.E. Amengual, D. Mahadevan, O.A. O’Connor
References
- 1.Mak V, Hamm J, Chhanabhai M, Shenkier T, Klasa R, Sehn LH, et al. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J Clin Oncol. 2013;31:1970–6. doi: 10.1200/JCO.2012.44.7524. [DOI] [PubMed] [Google Scholar]
- 2.O’Connor OA, Pro B, Pinter-Brown L, Bartlett N, Popplewell L, Coiffier B, et al. Pralatrexate in patients with relapsed or refractory peripheral T-cell lymphoma: results from the pivotal PROPEL study. J Clin Oncol. 2011;29:1182–9. doi: 10.1200/JCO.2010.29.9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz SM, Advani RH, Bartlett NL, Jacobsen ED, Sharman JP, O’Connor OA, et al. Objective responses in relapsed T-cell lymphomas with single-agent brentuximab vedotin. Blood. 2014;123:3095–100. doi: 10.1182/blood-2013-12-542142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Camacho E, Bea S, Salaverria I, Lopez-Guillermo A, Puig X, Benavente Y, et al. Analysis of Aurora-A and hMPS1 mitotic kinases in mantle cell lymphoma. Int J Cancer. 2006;118:357–63. doi: 10.1002/ijc.21370. [DOI] [PubMed] [Google Scholar]
- 6.Kanagal-Shamanna R, Lehman NL, O’Donnell JP, Lim MS, Schultz DS, Chitale DA, et al. Differential expression of aurora-A kinase in T-cell lymphomas. Mod Pathol. 2013;26:640–7. doi: 10.1038/modpathol.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg JW, Mahadevan D, Cebula E, Persky D, Lossos I, Agarwal AB, et al. Phase II study of alisertib, a selective Aurora A kinase inhibitor, in relapsed and refractory aggressive B- and T-cell non-Hodgkin lymphomas. J Clin Oncol. 2014;32:44–50. doi: 10.1200/JCO.2012.46.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita M, Mori N. Aurora A selective inhibitor MLN8237 suppresses the growth and survival of HTLV-1-infected T cells in vitro. Cancer Sci. 2010;101:1204–11. doi: 10.1111/j.1349-7006.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi W, Spier C, Liu X, Agarwal A, Cooke LS, Persky DO, et al. Alisertib (MLN8237) an investigational agent suppresses Aurora A and B activity, inhibits proliferation, promotes endo-reduplication and induces apoptosis in T-NHL cell lines supporting its importance in PTCL treatment. Leuk Res. 2013;37:434–9. doi: 10.1016/j.leukres.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, et al. Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res. 2011;17:7614–24. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 11.Marchi E, Paoluzzi L, Scotto L, Seshan VE, Zain JM, Zinzani PL, et al. Pralatrexate is synergistic with the proteasome inhibitor bortezomib in in vitro and in vivo models of T-cell lymphoid malignancies. Clin Cancer Res. 2010;16:3648–58. doi: 10.1158/1078-0432.CCR-10-0671. [DOI] [PubMed] [Google Scholar]
- 12.Paoluzzi L, Scotto L, Marchi E, Zain J, Seshan VE, O’Connor OA. Romidepsin and belinostat synergize the antineoplastic effect of bortezomib in mantle cell lymphoma. Clin Cancer Res. 2010;16:554–65. doi: 10.1158/1078-0432.CCR-09-1937. [DOI] [PubMed] [Google Scholar]
- 13.Amengual JE, Clark-Garvey S, Kalac M, Scotto L, Marchi E, Neylon E, et al. Sirtuin and pan-class I/II deacetylase (DAC) inhibition is synergistic in preclinical models and clinical studies of lymphoma. Blood. 2013;122:2104–13. doi: 10.1182/blood-2013-02-485441. [DOI] [PubMed] [Google Scholar]
- 14.Kalac M, Scotto L, Marchi E, Amengual J, Seshan VE, Bhagat G, et al. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood. 2011;118:5506–16. doi: 10.1182/blood-2011-02-336891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng C, Lipstein M, Rodriguez R, Serrano XO, McIntosh C, Tsai WY, et al. The novel IKK2 inhibitor LY2409881 potently synergizes with histone deacetylase inhibitors in preclinical models of lymphoma through the downregulation of NF-kB. Clin Cancer Res. 2015;21:134–45. doi: 10.1158/1078-0432.CCR-14-0384. [DOI] [PubMed] [Google Scholar]
- 16.Jain S, Jirau-Serrano X, Zullo K, Scotto L, Palermo CF, Satstra SA, et al. A multimodality imaging approach confirms marked activity of pralatrexate and romidepsin in combination in a bioluminescent murine model of human T-cell lymphoma. Clin Cancer Res. 2015 Feb 12; doi: 10.1158/1078-0432.CCR-14-2249. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–8. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 18.Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–82. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahadevan D, Morales C, Cooke LS, Manziello A, Mount DW, Persky DO, et al. Alisertib added to rituximab and vincristine is synthetic lethal and potentially curative in mice with aggressive DLBCL co-overexpressing MYC and BCL2. PLoS ONE. 2014;9:e95184. doi: 10.1371/journal.pone.0095184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–33. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 21.Burgess AJ, Pavey S, Warrener R, Hunter LJ, Piva TJ, Musgrove EA, et al. Upregulation of p21(WAF1/CIP1) by histone deacetylase inhibitors reduces their cytotoxicity. Mol Pharmacol. 2001;60:828–37. [PubMed] [Google Scholar]
- 22.Estevam J, Danaee H, Liu R, Ecsedy J, Trepicchio WL, Wyant T. Validation of a flow cytometry based G(2)M delay cell cycle assay for use in evaluating the pharmacodynamic response to Aurora A inhibition. J Immunol Methods. 2011;363:135–42. doi: 10.1016/j.jim.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Cha TL, Chuang MJ, Wu ST, Sun GH, Chang SY, Yu DS, et al. Dual degradation of aurora A and B kinases by the histone deacetylase inhibitor LBH589 induces G2-M arrest and apoptosis of renal cancer cells. Clin Cancer Res. 2009;15:840–50. doi: 10.1158/1078-0432.CCR-08-1918. [DOI] [PubMed] [Google Scholar]
- 24.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–20. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 25.Palani S, Patel M, Huck J, Zhang M, Balani SK, Yang J, et al. Preclinical pharmacokinetic/pharmacodynamic/efficacy relationships for alisertib, an investigational small-molecule inhibitor of Aurora A kinase. Cancer Chemother Pharmacol. 2013;72:1255–64. doi: 10.1007/s00280-013-2305-8. [DOI] [PubMed] [Google Scholar]
- 26.Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–82. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 27.Park JH, Jong HS, Kim SG, Jung Y, Lee KW, Lee JH, et al. Inhibitors of histone deacetylases induce tumor-selective cytotoxicity through modulating Aurora-A kinase. J Mol Med. 2008;86:117–28. doi: 10.1007/s00109-007-0260-8. [DOI] [PubMed] [Google Scholar]
- 28.Kretzner L, Scuto A, Dino PM, Kowolik CM, Wu J, Ventura P, et al. Combining histone deacetylase inhibitor vorinostat with aurora kinase inhibitors enhances lymphoma cell killing with repression of c-Myc, hTERT, and microRNA levels. Cancer Res. 2011;71:3912–20. doi: 10.1158/0008-5472.CAN-10-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii S, Kurasawa Y, Wong J, Yu-Lee LY. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc Natl Acad Sci U S A. 2008;105:4179–84. doi: 10.1073/pnas.0710140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnaghi-Jaulin L, Eot-Houllier G, Fulcrand G, Jaulin C. Histone deacetylase inhibitors induce premature sister chromatid separation and override the mitotic spindle assembly checkpoint. Cancer Res. 2007;67:6360–7. doi: 10.1158/0008-5472.CAN-06-3012. [DOI] [PubMed] [Google Scholar]
- 31.Grewal C, Hickmott J, Rentas S, Karagiannis J. A conserved histone deacetylase with a role in the regulation of cytokinesis in Schizosaccharomyces pombe. Cell Div. 2012;7:13. doi: 10.1186/1747-1028-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155:1147–57. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdivia MM, Hamdouch K, Ortiz M, Astola A. CENPA a genomic marker for centromere activity and human diseases. Curr Genomics. 2009;10:326–35. doi: 10.2174/138920209788920985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amiri-Kordestani L, Luchenko V, Peer CJ, Ghafourian K, Reynolds J, Draper D, et al. Phase I trial of a new schedule of romidepsin in patients with advanced cancers. Clin Cancer Res. 2013;19:4499–507. doi: 10.1158/1078-0432.CCR-13-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Au WY, Wong KY, Shimizu N, Tsuchiyama J, Kwong YL, et al. Cell death by bortezomib-induced mitotic catastrophe in natural killer lymphoma cells. Mol Cancer Ther. 2008;7:3807–15. doi: 10.1158/1535-7163.MCT-08-0641. [DOI] [PubMed] [Google Scholar]
- 36.Paoluzzi L, Gonen M, Bhagat G, Furman RR, Gardner JR, Scotto L, et al. The BH3-only mimetic ABT-737 synergizes the antineoplastic activity of proteasome inhibitors in lymphoid malignancies. Blood. 2008;112:2906–16. doi: 10.1182/blood-2007-12-130781. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor OA, Smith EA, Toner LE, Teruya-Feldstein J, Frankel S, Rolfe M, et al. The combination of the proteasome inhibitor bortezomib and the bcl-2 antisense molecule oblimersen sensitizes human B-cell lymphomas to cyclophosphamide. Clin Cancer Res. 2006;12:2902–11. doi: 10.1158/1078-0432.CCR-05-0308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.