Abstract
Background
Adipose-derived stromal cells (ASCs) represent a relatively abundant source of multipotent cells, with many potential applications in regenerative medicine. The present study sought to demonstrate the use of RNA sequencing (RNA-Seq) in identifying differentially expressed transcripts, particularly long non-coding RNAs (lncRNAs), associated with adipogenic differentiation to gain a clearer picture of the mechanisms responsible for directing ASC fate toward the adipogenic lineage.
Methods
Human ASCs were cultured in adipogenic differentiation media and RNA was harvested at Days 0, 1, 3, 5, and 7. Directional RNA-Seq libraries were prepared and sequenced. Paired-end reads were mapped to the human genome reference sequence hg19. Transcriptome assembly was performed and significantly differentially expressed transcripts identified. Gene Ontology (GO) term analysis was then performed to identify coding and non-coding transcripts of interest. Differential expression of several identified lncRNA was verified by quantitative real-time polymerase chain reaction.
Results
2868 significantly differentially expressed transcripts were identified; 207 were non-coding. Enriched GO terms among upregulated coding transcripts notably reflected differentiation toward the adipogenic lineage. Enriched GO terms among downregulated coding transcripts reflected growth arrest. Guilt-by-association analysis revealed non-coding RNA candidates with potential roles in the process of adipogenic differentiation.
Conclusions
The precise mechanisms that guide lineage-specific differentiation in multipotent cells are not yet fully understood. Defining lncRNAs associated with adipogenic differentiation allows for potential manipulation of regulatory pathways in novel ways. Thus, we present RNA-Seq as a powerful tool for expanding our understanding of ASCs and for development of novel applications employing these cells in regenerative medicine.
Keywords: Adipogenesis, adipogenic differentiation, adipose-derived stromal cell, non-coding RNA, transcriptome, RNA-Seq
Introduction
For most of the past several decades, the conventional view of molecular biologists has focused on protein-coding genes and the central dogma of DNA to mRNA to protein (1). In this paradigm, RNA was often regarded as a simple intermediary, transferring data encoded by DNA into functional proteins regulating cellular processes. However, with completion of the Human Genome Project, it became apparent that known genes encoding for proteins accounted for less than 2% of the information found in DNA. While the remaining 98% was largely thought to be filler, scientists have increasingly begun to appreciate the significance of these intergenic segments, identifying functional RNA molecules that are transcribed from DNA but not translated into protein (2-4). Known as non-coding RNA, well-characterized examples include transfer RNA, ribosomal RNA, and more recently described microRNA, short interfering RNA, and piwi-interacting RNA (Figure 1). Expansion of knowledge in this field has led to the understanding that non-coding RNAs may be as intimately involved in regulating cellular processes as proteins.
Figure 1.
Schematic of RNA classes. Transcription of DNA results in RNA, which may be translated into protein. Alternatively, RNA may remain un-translated and these non-coding RNA, including transfer RNA (tRNA), ribosomal RNA (rRNA), microRNA (miRNA), short interfering RNA (siRNA), piwi-interacting RNA (piRNA), and long non-coding RNA (lncRNA), among others, may participate in regulating cellular processes.
Importantly, over the last decade, large scale transcriptome analyses of mammalian genomes have identified another class of non-coding RNAs which are poorly conserved but found to be capable of influencing large transcriptional gene expression profiles (4, 5) (Figure 1). Named long non-coding RNAs (lncRNAs), this class of non-coding RNA is typically 200 nucleotides in length or greater (6, 7), and involvement of lncRNAs in diverse processes including cancer regulation (8, 9), X-chromosome inactivation (10), imprinting (11), and epidermal differentiation (12, 13) has already been described. Current transcriptome libraries suggest that lncRNAs may number in the tens of thousands in mammals, but despite accumulating evidence that most may be functional, only a small proportion have been delineated (6, 14). Nonetheless, given the ability of lncRNAs to perform widespread regulation and/or modulation of specific gene transcription profiles, the potential exists for identification and subsequent manipulation of lncRNAs to regulate cellular behavior.
Sensitive technologies, however, are needed in order to identify potential lncRNAs associated with specific cellular processes. Traditional hybridization-based techniques such as microarrays may provide a wealth of data, but are limited by the nature of printed probes attached to the slide, as they typically correspond to known genes. Modifications including tiling arrays have been made to detect lncRNAs (15, 16), but comprehensive transcriptome analysis using this approach would be cumbersome. In contrast, RNA sequencing (RNA-Seq) is a recently developed, high-throughput sequencing method that allows for the mapping and accurate quantification of entire transcriptomes (17). It offers several distinct advantages when compared to hybridization-based techniques, as RNA-Seq does not rely on an existing known genomic sequence, allowing for greater flexibility and applicability. Furthermore, RNA-Seq provides single base pair resolution with minimal background signal (17), and results are also highly reproducible and therefore comparable across studies (18, 19). Of note, other high-throughput sequencing-based methods for transcriptome analysis, including those relying on tag-based approaches such as serial analysis of gene expression (SAGE), can be relatively expensive and may be limited in their ability to analyze the entire transcriptome (17, 20).
While RNA-Seq is an exciting technology for the thorough interrogation of gene expression, its high-throughput nature has somewhat limited its practical applicability. Researchers must sift through an enormous amount of data generated by RNA-Seq in order to draw meaningful conclusions from experiments. Despite this hurdle, systematic evaluation of data may be performed to identify specific lncRNAs with potential significance. We describe such an approach to identify lncRNAs with potential involvement in the adipogenic differentiation of adipose-derived stromal cells (ASCs). Such cells hold significant promise for the treatment of various tissue deficiencies in regenerative medicine, and in contrast to bone marrow-derived mesenchymal stem cells (MSCs) and induced pluripotent stem cells (iPSCs), ASCs are a much more abundant and readily obtainable cell population with great potential for incorporation into cell-based therapies for soft tissue reconstruction (21). Importantly, while ASCs readily undergo adipogenic differentiation in vitro, efficiency of in vivo differentiation of ASCs for tissue engineering remains an elusive goal. By identifying lesser-known, more specialized regulatory transcripts such as lncRNAs associated with adipogenesis, we may be better able to understand how mature adipocytes may be generated from ASCs. Furthermore, manipulating lncRNAs associated with adipogenesis may provide a future avenue to alter large-scale transcriptional patterns to promote efficient differentiation for tissue engineering purposes. Thus, high-throughput assays such as RNA-Seq have great applicability not only in basic science research, where they are commonly employed, but in translational research as well.
Methods
ASC Harvest and Differentiation
After informed consent, lipoaspirate specimens were obtained from three healthy female human donors between 36-47 years of age, under a Stanford University Institutional Review Board approved protocol (#2188). Each lipoaspirate specimen was processed using standard techniques to isolate the stromal vascular fraction. Briefly, harvested fat was washed and treated with 0.075% collagenase type II (Sigma-Aldrich; St. Louis, MO) in Medium 199 (Cellgro; Manassas, VA) (22, 23). Digestion was performed for one hour at 37°C under gentle agitation. Centrifugation was then performed to yield a cellular pellet, which was then treated with red blood cell lysis buffer to remove red blood cells, and then plated onto 15 cm plates. Once subconfluent, cells were passaged onto six-well culture plates at equal densities. For adipogenic differentiation, cells were grown to 100% confluency and then treated with adipogenic differentiation media (ADM), which consisted of 10% FBS, 1% penicillin/streptomycin, 10 μg/mL insulin, 1 μM dexamethasone, 0.5 mM methylxanthine, and 200 μM indomethacin (22). Oil Red O (ORO) staining was performed on Days 1, 3, 5, and 7, verifying the successful differentiation of ASCs into adipocytes. Images were obtained using a Leica DC300 camera on a Leica DM IL inverted contrasting microscope at 10x, and quantification was performed by recording pixel-positive area per high power field using Image J (NIH; Bethesda, MD). Means and standard deviations were calculated from numerical data and error bars correspond to 1 SD. Statistical analysis was performed using a one-way analysis of variance test for multiple group comparison, and two-tailed Student's t-tests for direct comparisons between groups. A *p-value < 0.05 was considered statistically significant.
Library Preparation, Sequencing, and Alignment
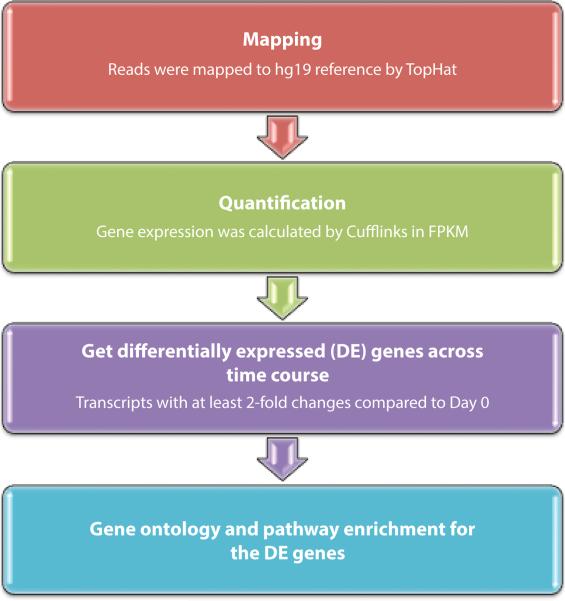
On Days 0, 1, 3, 5, and 7, total RNA was harvested and phase separated using Trizol (Invitrogen; Carlsbad, CA), and isolated using the RNeasy Mini Kit (Qiagen; Hilden, Germany). RNA samples were treated with Turbo DNAse (Ambion; Carlsbad, CA), to eliminate DNA from the samples, and then purified. An Agilent Bioanalyzer was used to determine RNA Integrity Number (RIN; median RIN score = 10). Directional RNA-Seq libraries were prepared using the Mondrian™ SP+ Workstation and the Encore SP+ Complete DR Multiplex System (NuGEN; San Carlos, CA), analyzed again with an Agilent Bioanalyzer to ensure successful library creation, and then sequenced with the Illumina HiSeq 2500 System (Illumina, Inc.; San Diego, CA). Paired-end reads were mapped to the human genome reference sequence hg19 using TopHat (24) (Figure 2).
Figure 2.
Schematic of RNA-Seq data processing. RNA-Seq reads were mapped to the hg19 reference genome using TopHat. Gene expression was calculated based on FPKM using Cufflinks. Greater than two-fold change in expression was used to define differentially expressed (DE) loci for subsequent evaluation. Finally, Gene Ontology term analysis was performed for differentially expressed genes.
Quantification of Gene Expression
Transcriptome assembly was performed with Cufflinks, and transcriptomes were subsequently merged (Figure 2). Fragments per kilobase of transcript per million mapped reads (FPKM) for known transcripts were calculated, which normalizes expression level data to gene length and library size to allow for comparison between different samples. Expression levels across each time point were merged over the three patient samples, and significantly differentially expressed (DE) transcripts across the time course were identified. Differentially expressed genes were defined as genes that underwent a minimum two-fold unidirectional change compared to Day 0 expression levels.
Gene Ontology Term Analysis
Gene Ontology (GO) term analysis was performed on our DE coding transcripts to yield enriched biological themes. Long non-coding RNAs were then determined using NBCI annotations, and a “guilt-by-association” analysis was performed to predict functions of DE non-coding RNA transcripts. GO terms were assigned to non-coding transcripts based on the GO terms associated with their co-expressed coding transcripts.
Quantitative Real-Time Polymerase Chain Reaction Analysis
Reverse transcription was performed on the RNA samples from Days 0, 1, 3, 5, and 7 using TaqMan Reverse Transcription Reagents (Invitrogen), to obtain the corresponding cDNA. Primers for several DE lncRNAs from the RNA-Seq and subsequent guilt-by-association analyses were designed using GenScript (Piscataway, NJ), and were assessed for size, specificity, and linear amplification. Primer sequences are listed in Supplemental digital content 1, Table showing Primer sequences for qRT-PCR analysis, INSERT LINK. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using an ABI Prism 7900HT Sequence Detection System with Power SYBR Green PCR Master Mix (Applied Biosystems; Foster City, CA) as the reporter. All expression data were normalized to GAPDH expression levels as the housekeeping gene, and then calibrated to Day 0 baseline expression to generate relative expression levels.
Results
Staining and Transcriptome Analysis of Adipogenesis
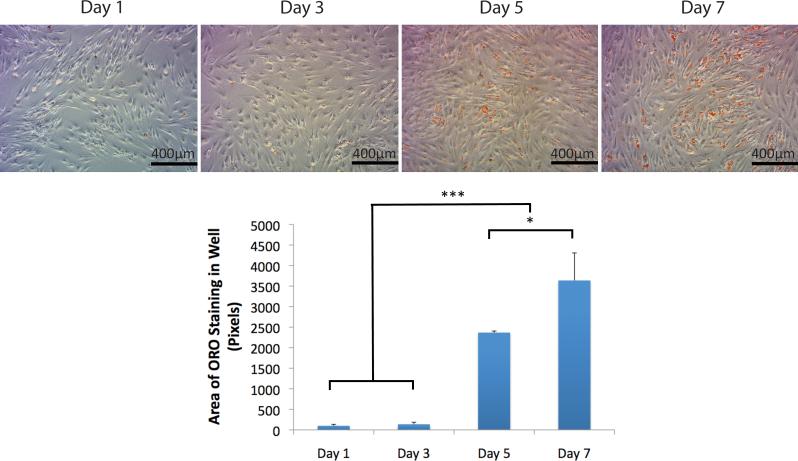
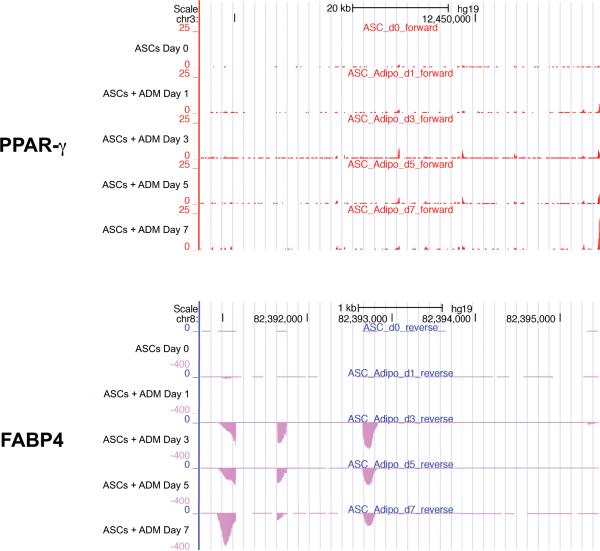
ASCs cultured in ADM were stained with ORO on Days 1, 3, 5, and 7. As seen in Figure 3A, progressive accumulation of triglyceride and lipids could be appreciated. Quantification was also performed, demonstrating significantly increased ORO staining at Days 5 and 7 (Figure 3B). Lipid droplet staining thus confirmed adipogenic differentiation by ASCs. Total RNA was then harvested from similarly treated ASCs for transcriptome analysis using RNA-Seq. Of the 2868 total transcripts significantly differentially expressed, 2661 were protein-coding and 207 were non-coding. To visualize genome-mapped data from RNA-Seq, results were uploaded to the UCSC Genome Browser. Importantly, peroxisome proliferator-activated receptor gamma (PPAR-γ) and fatty acid binding protein 4 (FABP4), two genes widely known to play integral roles in adipogenic differentiation, were found to significantly increase over the time course of ADM treatment. Reads within the PPAR-γ locus on the forward strand (Figure 4A) and FABP4 locus on the reverse strand (Figure 4B) were both observed to progressively rise with culture time in ADM, lending validity to data generated by RNA-Seq.
Figure 3.
Oil Red O staining and quantification. A) ASCs cultured in adipogenic differentiation medium for 1, 3, 5, and 7 days were stained with Oil Red O. Scale bar corresponds to 400 μm. B) Quantification of staining demonstrated a significant increase in triglyceride and lipid accumulation at Days 5 and 7 (*p < 0.05, ***p < 0.001).
Figure 4.
Visualization of RNA-Seq data with UCSC tracks. A) Reads at the PPAR-γ locus on the forward strand progressively increased the longer ASCs were cultured in adipogenic differentiation medium, as represented by higher peaks along the Y-axis. Similarly, reads for FABP4 (B), which resides on the reverse strand, also increased with prolonged culture in adipogenic differentiation medium, as represented by longer bars along the Y-axis.
GO Term Analysis of Coding Genes
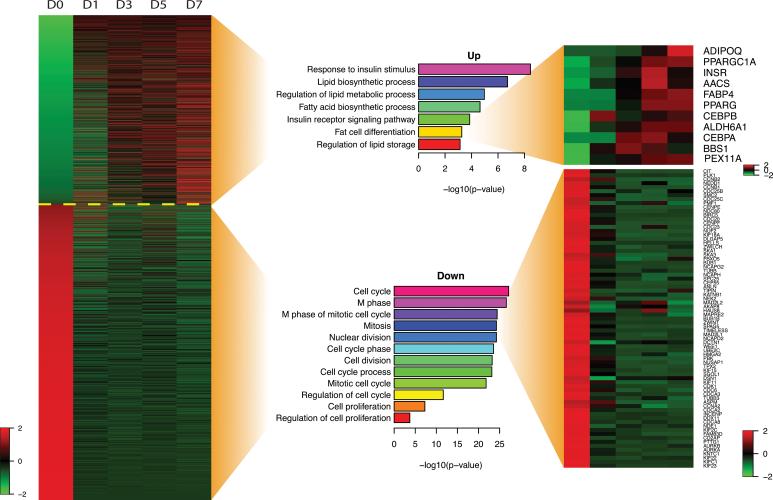
Among the 2868 transcripts found to be differentially expressed, 1146 transcripts were upregulated and 1722 were downregulated during adipogenic differentiation of ASCs (Figure 5A). Focusing first on the upregulated loci, as expected, GO term analysis of coding genes identified many processes related to adipocyte differentiation and function. Along with fat cell differentiation, other GO terms with some of the greatest association to upregulated coding genes included lipid biosynthetic process, regulation of lipid metabolic process, response to insulin stimulus, and regulation of lipid storage (Figure 5B and Supplemental Digital content 1, Figure shows GO term analysis for upregulated and downregulated loci. A) The 30 most highly associated GO terms with upregulated transcripts. Longer bars correspond to greater number of loci associated with specific GO term. B) The 30 most highly associated GO terms with downregulated transcripts. Longer bars correspond to greater number of loci associated with specific GO term, INSERT LINK). Looking specifically at the GO term fat cell differentiation, upregulated loci in our sequencing data included PPAR-γ and FABP4, as mentioned above, as well as other known fat-related genes such as adiponectin (ADIPOQ), Acetoacetyl-CoA Synthase (AACS), and CCAAT/enhancer binding protein (CEBPA) (Figure 5B).
Figure 5.
Analysis of differentially expressed transcripts. A) 2868 differentially expressed transcripts were identified through RNA-Seq. 1146 loci were upregulated in ASCs cultured with adipogenic differentiation medium (above dotted yellow line) and 1722 loci were downregulated in ASCs cultured with adipogenic differentiation medium (below dotted yellow line). Red signifies increased expression and green signifies decreased expression. B) GO term analysis revealed many upregulated coding genes to be associated with fat-related processes. In particular, 11 upregulated genes were associated with the GO term fat cell differentiation. C) GO term analysis also revealed many downregulated coding genes to be associated with cell cycle and cell proliferation.
In contrast to upregulated genes, of the 1722 loci with decreasing level of expression, some of the most frequent GO terms associated with these downregulated genes were those broadly related to regulation of cell cycle. These included cell cycle, cell division, mitotic cell cycle, nuclear division, and cell proliferation, among others (Figure 5C and Supplemental Digital Content 2, INSERT LINK). Collectively, these findings suggest arrest of ASC proliferation in response to culture in ADM and are consistent with more differentiated adipocytes.
lncRNA Analysis
Along with the 2661 protein-coding genes identified by RNA-Seq to be differentially expressed, changes to transcriptional patterns at 207 non-coding loci were also noted. Of these 207 lncRNAs identified, 109 were upregulated and 98 were downregulated (Figure 6A). To determine potential roles some of these lncRNAs may play, a guilt-by-association analysis was performed. Coding genes with similar changes to transcriptional profiles as lncRNA were first identified (Supplemental Digital Content 3, Figure shows Guilt-by-association GO term analysis for lncRNA. Number of associated coding genes assigned to each lncRNA corresponding to more GO terms assigned to each specific lncRNA, INSERT LINK). GO terms associated with these coding genes were then determined and assigned to each lncRNA (Figure 6B). Based on this approach, multiple potential lncRNAs were found with a potential role in adipogenesis. Looking specifically at seven fat-related GO terms (lipid biosynthetic process, regulation of lipid metabolic process, response to insulin stimulus, regulation of lipid storage, insulin receptor signaling pathway, fatty acid biosynthetic process, and fat cell differentiation), 26 lncRNAs were identified, all of which were upregulated (Figure 6C).
Figure 6.
Analysis of differentially expressed lncRNA. A) Of the 207 lncRNAs differentially expressed, 109 were upregulated (red, above dotted yellow line) and 98 were downregulated (green, below dotted yellow line). B) Schematic of “guilt-by-association” GO term analysis of lncRNA. Expression patterns of every lncRNA (triangle) were associated with similarly expressed coding genes (circles). GO terms of associated coding genes were then assigned to each lncRNA. C) 26 lncRNAs were identified that could be assigned fat-related GO terms based on guilt-by-association analysis, of which all were upregulated (red).
qRT-PCR Validation of Differential Expression
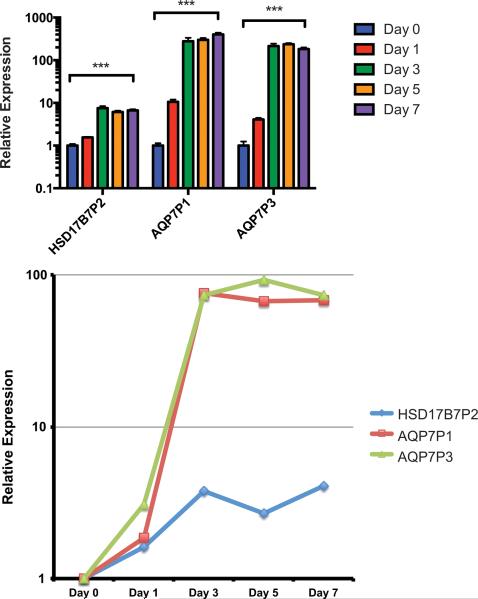
Of the 26 lncRNAs identified, expression levels of three significantly upregulated lncRNAs (HSD17B7P2, AQP7P1, and AQP7P3) were evaluated at each time point with qRT-PCR in order to validate the results from the RNA-Seq analysis. As expected, qRT-PCR analysis demonstrated statistically significant differential expression of each transcript over time (each ***p < 0.001), with increased expression of HSD17B7P2, AQP7P1, and AQP7P3 over time (Figure 7A). Notably, these relative changes in expression levels, as measured by qRT-PCR, closely followed the changes in expression in FPKM from the earlier RNA-Seq analysis (Figure 7B).
Figure 7.
qRT-PCR analysis of identified lncRNAs. A) Transcript levels of HSD17B7P2, AQP7P1, and AQP7P3 at Days 0, 1, 3, 5, and 7, as measured by quantitative real-time polymerase chain reaction, demonstrated statistically significant differential expression (***p < 0.001). Expression levels are calibrated to Day 0. B) Relative gene expression levels in FPKM as measured through RNA-Seq analysis, calibrated to Day 0 baseline expression.
Discussion
When compared to other members of the transcriptome, including other non-coding RNA species, lncRNAs are vastly less understood and therefore represent a relatively untapped pool of potential targets for regulation of cell fate and differentiation. The reported functions of lncRNAs thus far range widely across diverse cellular processes and tissue types, though a common thread of participation in cell differentiation has emerged over the past decade (25). Long non-coding RNAs have been shown to act at multiple levels of gene regulation, from epigenetic modification, to transcription, to post-transcription (26). Therefore, it has been suggested that lncRNAs are a precise marker of functionally significant biological events, such as the specific cell state in differentiation (27).
Studies of the Hox genes, clustered in four loci in the mammalian genome and responsible for cell differentiation along the anterior-posterior anatomic axis, have revealed several lncRNAs with critical functions in transcriptional regulation of these genes. HOTAIR, a lncRNA found within the HoxC locus, represses genes in the HoxD cluster (15). Concurrent changes in the chromatin remodeling complex PRC2 suggest that HOTAIR acts by inducing a repressive chromatin state. Furthermore, HOTAIR has also been shown to be overexpressed in breast and colorectal cancer (28), where it appears to participate in the repression of metastasis-suppressing genes through chromatin modification (9, 29). Meanwhile, HOTTIP, a lncRNA in the HoxA cluster, appears to be involved in the transcriptional activation of several other HoxA genes (30). Silencing of HOTTIP reduces expression levels of these genes and is associated with specific changes in the chromatin state as well. These findings suggest an important role of lncRNAs in the regulation of cell fate. However, identification and assignment of function for the vast majority of lncRNAs remains in its infancy, and substantial progress in this area can be made through application of high-throughput sequencing methods. It is with this very application that use of RNA-Seq may be most powerful.
High-throughput sequencing has also helped to identify other lncRNA transcripts with roles in regulation and maintenance of cell differentiation. Using analogous guilt-by-association analyses, Kretz et al. described an 855 bp lncRNA, named anti-differentiation ncRNA (ANCR), which was dramatically downregulated in differentiated keratinocytes, adipocytes, and osteoblasts (12). Depletion of ANCR in progenitor cell populations led to rapid induction of differentiation genes. In contrast, the 3.7 kb lncRNA terminal differentiation-induced ncRNA (TINCR) was found to be one of the most highly induced transcripts during keratinocyte differentiation (13). TINCR-deficient epidermis was shown to lack ultrastructure consistent with terminal differentiation, thus highlighting the importance of this lncRNA in guiding somatic tissue differentiation (13).
Aside from involvement in differentiation, lncRNAs have also been implicated in potential regulation of pluripotency. In particular, the lncRNA RoR has been shown to be enriched in iPSCs after reprogramming, and RoR is a direct target of the key pluripotency-regulating transcription factors Oct4, Sox2, and Nanog (31). Notably, over-expression of RoR leads to more efficient reprogramming, while inhibition reduces the ability to generate iPSCs.
Other techniques for transcriptome analysis include hybridization- and tag-based sequencing techniques. While these techniques are more than adequate to analyze expression within the coding genome, traditional techniques are more limited in their ability to analyze the entire transcriptome with high detail. Newer developments such as the high-density genomic tiling microarray allow for higher resolution mapping of the transcriptome (both coding and non-coding), using overlapping oligonucleotide probes to determine length of RNAs. However, while resolution has improved, tiling microarrays still do not provide efficient, comprehensive transcriptome analysis capable with RNA-Seq.
Using RNA-Seq, we have identified 2868 total transcripts of potential interest in the adipogenic differentiation of ASCs. GO term functional analysis of both up- and down-regulated differentially expressed transcripts revealed biological processes concordant with adipogenesis and increased cell maturity, elucidating the functions of the DE genes identified. Additionally, as expected, the increase in well-described transcription factors such as PPAR-γ and FABP4 also demonstrates adipogenic differentiation. Of the 207 lncRNAs identified to be differentially expressed, guilt-by-association analysis allowed for assignment of fat-related GO terms to 26 loci. Furthermore, confirmation of differential expression by qRT-PCR validates the findings provided by RNA-Seq analysis, affirming RNA-Seq as a powerful and accurate high-throughput technology of great potential utility. These 26 lncRNAs may thus be promising novel targets for future studies aimed at understanding and promoting adipogenesis with ASCs, which would benefit from further analysis in order to more definitively elucidate functional significance. Regardless, as adipocyte differentiation is a complex process which we do not yet fully understand, the use of RNA-Seq provides new capabilities to identify and quantify significant potential molecular targets or signals in the adipogenic differentiation of ASCs. Our study provides an example of how to harness this technology to provide a clearer picture of the mechanisms responsible for directing cell fate towards the adipogenic lineage in vitro and in vivo, and demonstrates the potential role of lncRNAs in the regulation of adipogenesis.
Conclusions
The precise mechanisms that guide lineage-specific differentiation in multipotent stem cells are intently investigated but not yet fully understood. With a global, temporal picture of the ASC transcriptome in response to adipogenic differentiation cues, the potential exists to identify and harness new molecular targets for the augmentation of ASC use in soft tissue reconstruction. Long non-coding RNAs are especially exciting candidates for use in manipulating regulatory networks in previously unexplored ways and, as we have demonstrated, may be identified using RNA-Seq. As such, RNA-Seq represents a technology with powerful capabilities for mapping and quantifying the entire transcriptome, and which we believe holds great promise in aiding the development of effective new translational therapies by both plastic surgeons and researchers in regenerative medicine.
Supplementary Material
Acknowledgments
We thank the surgeons and medical staff at Plastic Surgery Center Palo Alto for their assistance providing biological samples for these experiments.
We also thank AccuBay Corporation for the professional bioinformatics analysis.
M.T.L. was supported by NIH grants U01 HL099776, R01 DE021683-03, R21 DE024230-02, the Oak Foundation, Hagey Laboratory for Pediatric Regenerative Medicine, and the Gunn/Olivier Fund. D.C.W. was supported by the ACS Franklin H. Martin Faculty Research Fellowship, NIH grant 1K08 DE024269-01, the Hagey Laboratory for Pediatric Regenerative Medicine, and the Stanford University Child Health Research Institute Faculty Scholar Award.
Footnotes
Data presented in this manuscript have previously been presented at the Stanford University Plastic Surgery Research Conference.
Financial Disclosure and Products:
None of the authors has a financial interest in any of the products, devices, or drugs mentioned in this manuscript.
Author Roles:
Anna Luan, M.S. – contributed to project design, acquisition and analysis of data, drafting and revision of the manuscript
Kevin J. Paik, A.B. – contributed to project design, acquisition and analysis of data, drafting and revision of the manuscript
Jiang Li, M.S. – contributed to project design, acquisition and analysis of data
Elizabeth R. Zielins, M.D. – contributed to drafting of the manuscript
David A. Atashroo, M.D. – contributed to analysis of data
Andrew Spencley, B.S. – contributed to project design
Arash Momeni, M.D. – contributed to revision of the manuscript
Michael T. Longaker, M.D., M.B.A. – contributed to project conception and design, interpretation of data, revision of the article, and approval of the final version
Kevin C. Wang, M.D., Ph.D. – contributed to project conception and design, interpretation of data, revision of the article, and approval of the final version
Derrick C. Wan, M.D. – contributed to project conception and design, interpretation of data, revision of the article, and approval of the final version
References
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Wan DC, Wang KC. Long noncoding RNA: significance and potential in skin biology. Cold Spring Harbor perspectives in medicine. 2014:4. doi: 10.1101/cshperspect.a015404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in genetics : TIG. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harbor symposia on quantitative biology. 2010;75:325–331. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagano T, Fraser P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mammalian genome : official journal of the International Mammalian Genome Society. 2009;20:557–562. doi: 10.1007/s00335-009-9218-1. [DOI] [PubMed] [Google Scholar]
- 12.Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Briefings in functional genomics & proteomics. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 15.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cloonan N, Forrest AR, Kolle G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 20.Gerhard DS, Wagner L, Feingold EA, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome research. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuk P. Adipose-Derived Stem Cells in Tissue Regeneration: A Review. ISRN Stem Cells. 2013;2013:35. [Google Scholar]
- 22.Levi B, James AW, Glotzbach JP, Wan DC, Commons GW, Longaker MT. Depot-specific variation in the osteogenic and adipogenic potential of human adipose-derived stromal cells. Plast Reconstr Surg. 2010;126:822–834. doi: 10.1097/PRS.0b013e3181e5f892. [DOI] [PubMed] [Google Scholar]
- 23.Levi B, James AW, Nelson ER, et al. Human adipose derived stromal cells heal critical size mouse calvarial defects. PLoS One. 2010;5:e11177. doi: 10.1371/journal.pone.0011177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNASeq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO reports. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Liu N, Wang JP, et al. Regulatory long non-coding RNA and its functions. Journal of physiology and biochemistry. 2012;68:611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA biology. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 30.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.