Summary
Many developmental genes are controlled by shadow enhancers, pairs of enhancers that drive overlapping expression patterns. We hypothesized that compensatory evolution can maintain the total expression of a gene while individual shadow enhancers diverge between species. To test this hypothesis, we analyzed expression driven by orthologous pairs of shadow enhancers from Drosophila melanogaster, Drosophila yakuba, and Drosophila pseudoobscura that control expression of Krüppel, a transcription factor that patterns the anterior-posterior axis of blastoderm embryos. We find that the expression driven by the pair of enhancers is conserved between these three species, but expression levels driven by the individual enhancers are not. Using sequence analysis and experimental perturbation, we show that each shadow enhancer is activated by different transcription factors. These results support the hypothesis that compensatory evolution can occur between shadow enhancers, which has implications for mechanistic and evolutionary studies of gene regulation.
Introduction
Compensatory evolution is the offset of a single deleterious mutation by a second mutation (Kimura, 1985). Searching for compensatory evolution within and between proteins has revealed physical interactions and led to global predictions of protein structure and function (Hopf et al., 2012; Suel et al., 2003; Hopf et al., 2014; Ovchinnikov et al., 2014). In studies of regulatory DNA, compensatory evolution has been observed at multiple scales. Within transcription factor (TF) binding sites, compensatory mutations of individual base pairs maintain the overall binding affinity (Mustonen et al., 2008). In enhancers, collections of TF binding sites that control tissue-specific gene expression, compensatory evolution has also been observed, though a slightly different definition is used: the function of the “whole” piece of regulatory DNA is conserved, while its constituent “parts” are not. For example, the whole even-skipped (eve) stripe 2 enhancer is functionally conserved between Drosophila species, but the 5’ and 3’ pieces of this enhancer are not conserved (Ludwig et al., 2000). This seminal study inspired the idea that enhancer function can be conserved without strict sequence conservation (Arnosti and Kulkarni, 2005; Weirauch and Hughes, 2010).
We hypothesize that compensatory evolution in regulatory DNA occurs at an even larger scale — between multiple enhancers controlling a single gene. Many developmental genes are controlled by pairs of “shadow” or “sibling” enhancers, enhancers that drive a gene in overlapping patterns of expression and can be important for driving robust gene expression patterns in the face of environmental and genetic perturbations (Hong et al., 2008; Perry et al., 2011; Frankel et al., 2010; Fujioka and Jaynes, 2012; Barolo, 2012; Frankel, 2012; Lam et al., 2015). Genome-scale studies support the hypothesis of compensatory evolution between enhancers. Gene expression levels are maintained between Drosophila species despite changes in TF binding (Paris et al., 2013). High-throughput measurements in cell culture suggest that compensatory evolution between enhancers is common, but measurements in cell culture do not allow the identification of bone fide shadow enhancers (Arnold et al., 2014). Here we test the hypothesis of compensatory evolution for a specific pair of defined shadow enhancers using spatially resolved measurements in the embryo. Testing this hypothesis using a specific pair may provide insights into how shadow enhancers together act on a single promoter to control a gene’s expression.
We looked for compensatory evolution in two shadow enhancers that control the expression of Krüppel (Kr), a transcription factor in the embryonic anterior-posterior patterning network of Drosophila. Kr is expressed as a single transverse stripe in the middle of the blastoderm embryo, and this expression domain is controlled by two shadow enhancers that drive nearly identical patterns (Hoch et al., 1990; Jacob et al., 1991; Perry et al., 2011). The Kr expression pattern is highly conserved between D. melanogaster (D. mel), D. yakuba (D. yak), and D. pseudoobscura (D. pse) (Fowlkes et al., 2011), despite widespread changes in regulatory DNA between these species (Clark et al., 2007). It is possible to measure the level and position of mRNA expression with high precision in Drosophila embryos (Luengo Hendriks et al., 2006; Wunderlich et al., 2014), making this pair of shadow enhancers an ideal case to test for compensatory evolution.
To assess how shadow enhancers evolve, we measured the expression driven by each of the two embryonic Kr enhancers singly and in combination from D. mel, D. yak, and D. pse. If compensatory evolution occurs between this pair, constructs that contain both Kr enhancers from a single species will drive similar expression, but the expression driven by individual enhancers will diverge between species, as will expression driven by interspecific chimeras.
Results and Discussion
Kruppel’s expression levels are conserved between species
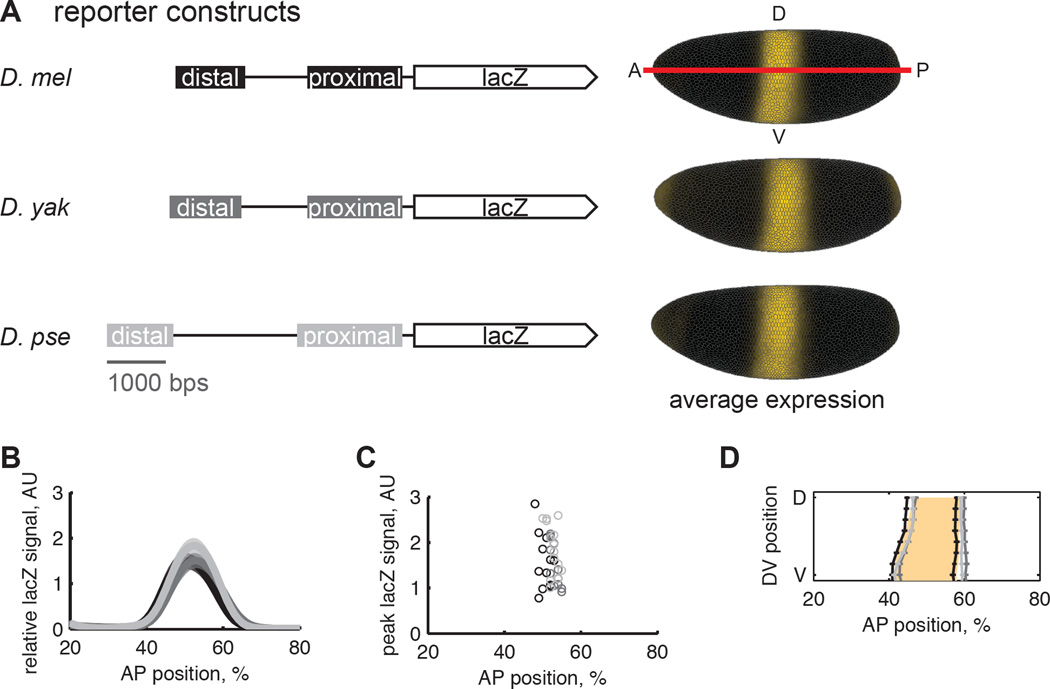
We first verified that the level and position of mRNA expression driven by the pair of shadow enhancers is quantitatively conserved between D. mel, D. yak, and D. pse. The endogenous Kr pattern in the three species is spatially conserved with respect to the expression pattern of its regulators (Fowlkes et al., 2011). Compared to other TFs involved in embryonic patterning, RNA-seq measurements show that the overall levels of Kr expression are among the most strongly conserved between the three species (Paris et al., 2013). We directly measured shadow enhancer function using transgenic reporter lines in D. mel, where the trans environment, promoter, and other variables affecting expression are identical. By normalizing levels of mRNA with a co-stain, we can compare both expression patterns and levels between reporter constructs (Wunderlich et al., 2014).
We found that the pair of Kr shadow enhancers from D. mel, D. yak, and D. pse drive indistinguishable gene expression levels and highly similar expression patterns (Figure 1B). The median expression levels are not statistically different between reporter lines (Figure 1C; p-values> 0.3, pair wise-rank sum tests with Benferonni correction). The boundaries of the expression patterns are similar, but the D. mel reporter line drives a pattern that is shifted to the anterior by 1% or ~1 cell width (Figure 1D).
Figure 1.
The pair of shadow enhancers from D. mel, D. yak, and D. pse drive similar levels of gene expression. (A) We created transgenic D. mel lines that contain lacZ reporters for the Kr distal and proximal enhancers with endogenous intervening sequence from D. mel, D. yak, and D. pse. All reporter constructs in this study were integrated into the same site in the genome. The “virtual” embryos show the average lacZ expression pattern in yellow and are oriented with anterior left, posterior right, dorsal up and ventral down. We used in situ hybridization with a co-stain to detect lacZ expression. We find that the spatial pattern and level of lacZ expression driven by these three constructs are nearly identical (B, C, D). (B) Average lacZ signal from each reporter line is plotted as a function of anterior-posterior position along the lateral side of the embryo, with shaded regions showing the standard error of the mean. D. mel is in black; D. yak in dark gray, and D. pse in light gray. (C)The position and magnitude of peak lacZ expression is plotted for individual embryos. Using a rank-sum test with a Bonferroni multiple comparison correction, we find the median peak expression levels between to the three reporter lines is not statistically different, p-value>0.3. (D) The position of the lacZ expression boundaries is plotted with the standard error of the mean. The anterior expression boundary does not significantly vary between lines, but the posterior boundary is shifted to the anterior in the D. mel reporter line by 1% of the anterior-posterior axis, about 1 cell width. The shaded orange region is the endogenous D. mel Kr expression pattern. The D. mel reporter matches endogenous expression to within a cell width, indicating that most of Kr’s spatial regulatory information is captured by the reporter.
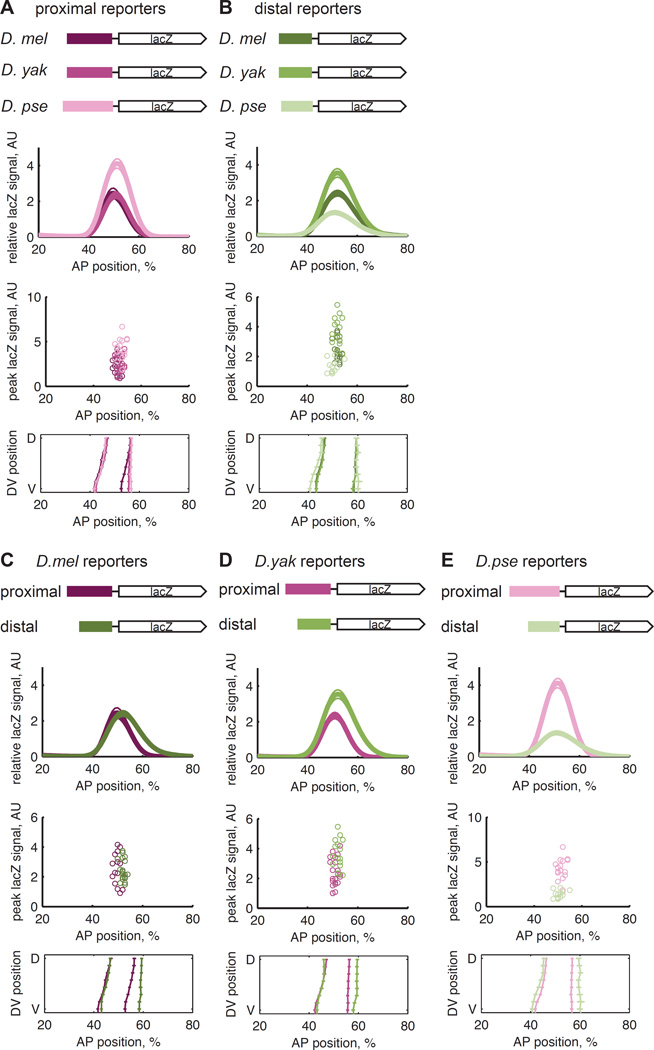
Individual enhancers diverge in expression levels between species
If compensatory evolution occurs between a pair of shadow enhancers, expression driven by each individual enhancer may differ between species. We therefore measured the mRNA expression driven by all six individual enhancers comprising the three orthologous Kr pairs. In this setting, the signature of compensatory evolution is that the function of the combined construct is conserved, while the function of individual enhancers may diverge. For this reason, we focus on expression level, which is conserved in the combined constructs, and not the spatial pattern, which differs slightly between species (Figure 1D). However, we note that the differences in spatial expression patterns are more dramatic in constructs driven by individual enhancers (Figure 2) compared to the combined constructs (Figure 1), suggesting that compensatory evolution may also stabilize the spatial pattern.
Figure 2.
The spatial expression patterns and levels of mRNA driven by individual Kr enhancers vary between species. We measured the expression driven by six additional reporter lines containing the proximal and distal enhancers from D. mel, D. yak, and D. pse. (A) The proximal enhancers do not drive conserved expression patterns or levels. The median peak expression levels between the D. mel and D. pse and the D. yak and D. pse lines are statistically different (rank sum test, p-values <= 0.002). (B) The distal enhancers do not drive conserved expression patterns or levels (rank sum test of median peak levels, p-values <= 0.002). In (C), (D), and (E), the data from (A) and (B) is represented to compare the enhancers from each species. The D. mel enhancers drive similar levels of expression (rank sum, p-value = 0.81) but different patterns, while the D. yak and D. pse enhancers drive different patterns and levels of mRNA (rank sum, p-values = 0.0017 and 8e-6, respectively).
The expression levels driven by the isolated enhancers differ between species. The D. pse proximal enhancer drives higher levels of mRNA expression than the D. mel and D. yak proximal enhancers (Figure 2A, p-values <= 0.002, rank sum test with Bonferroni correction). In contrast, the D. mel and D. yak distal enhancers drive higher levels of mRNA expression than the D. pse distal enhancer (Figure 2B, p-values <= 0.002, rank sum test with Bonferroni correction). These differences are consistent with the phylogenetic distances between the species, which are ~25 million years between D. mel and D. pse, and ~10 million years between D. mel and D. yak. Thus, while the expression level driven by the pair of Kr shadow enhancers is conserved between these three species, the levels of mRNA expression driven by each individual enhancer are not, supporting our hypothesis that the shadow enhancers are subject to compensatory evolution.
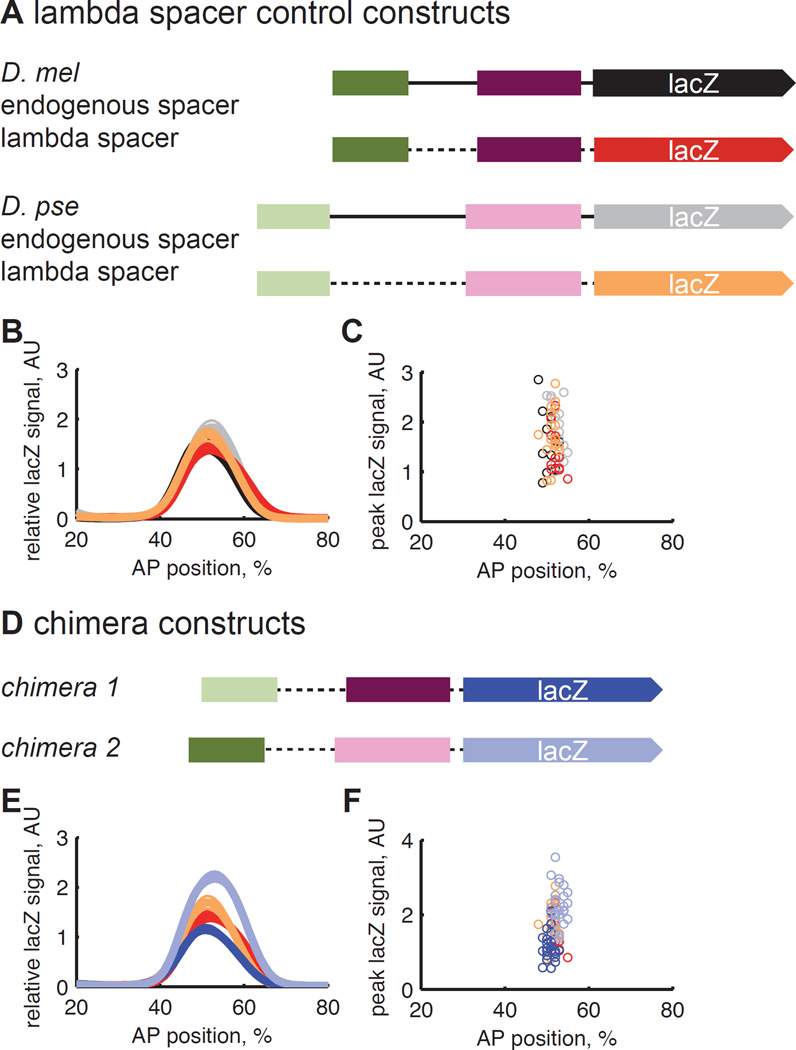
We also made chimeric constructs that combine the proximal and distal enhancers from different species. We hypothesized that these chimeras would not drive the same expression levels as constructs containing pairs of shadow enhancers from a single species. Because the endogenous sequence between the enhancers is unalignable between D. mel and D. pse, we replaced this sequence with an artificial spacer from the lambda phage genome. We confirmed that the single species constructs with lambda spacer drive the same expression levels as those with endogenous spacers (Figure 3A; p-values > 0.5, paired rank sum tests with Bonferroni correction). We then generated chimeras with combinations of the D. mel and D. pse enhancers and found that these chimeras drive significantly different levels of gene expression than each other; they also drive significantly different levels of gene expression than the single species constructs, with the exception of chimera 1 and the D. mel lambda spacer construct (Figure 3B; p-values < 0.05, paired rank sum test with Bonferroni correction). These results together suggest that compensatory evolution acts to maintain overall Kr expression levels and, more broadly, that selection simultaneously works on all the regulatory DNA controlling a gene, in line with our previous work and the work of others (Wunderlich et al., 2012; Arnold et al., 2014; Villar et al., 2015).
Figure 3.
Chimeric enhancer constructs drive different levels of gene expression. (A) To generate chimeric constructs with proximal and distal enhancers from D. mel and D. pse, we first made constructs in which we replaced the endogenous sequence between the two enhancers with a spacer taken from the lambda phage genome. These constructs, shown in orange, do not drive expression levels that are significantly different from the constructs with the endogenous spacer, shown in black and gray (rank sum, p-values > 0.5). (B) We then made chimeric enhancer constructs, which contained the proximal and distal enhancers from different species. These constructs drive significantly different levels of expression from each other (rank sum, p-value = 2.9e-8) and generally drive different levels of expression from the lambda spacer control constructs (rank sum with Bonferroni correction, p-values < 0.02), with the exception of chimera 1 and the D. mel lambda spacer construct (rank sum with Bonferroni correction, p-value = 0.17).
Each Kr shadow enhancer is controlled by different activators
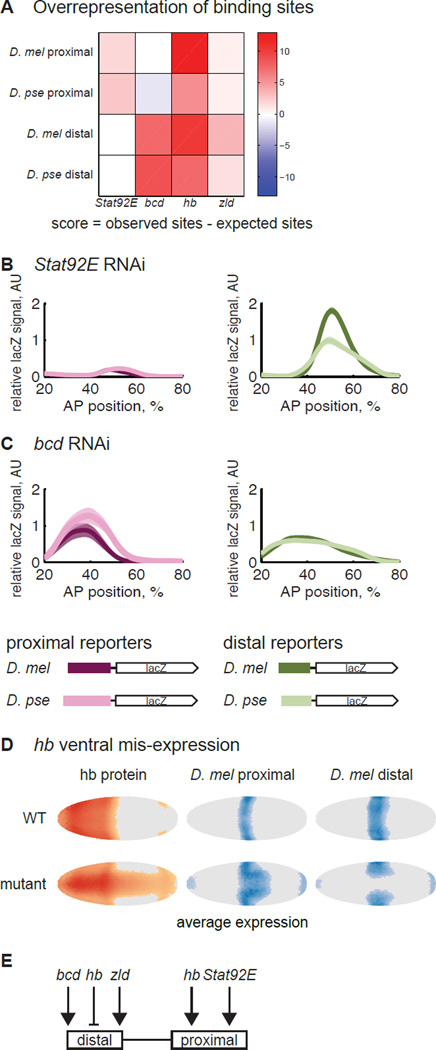
To uncover which DNA sequence changes are responsible for compensatory evolution of expression level, we looked at the binding site content of each enhancer. We focused on activators because at this stage in development, the spatial pattern of Kr’s expression is set by repressors while the level of mRNA is set by activators (Jaeger, 2011). Genetic studies have shown that Kr is activated by bicoid (bcd) (Hoch et al., 1990; Jacob et al., 1991), Stat92E (Tsurumi et al., 2011), zelda (zld) (Nien et al., 2011), and hunchback (hb) (Struhl et al., 1992; Schulz and Tautz, 1994). To map these genetic interactions to the individual shadow enhancers, we looked for TFs whose binding sites are overrepresented in each enhancer sequence compared to the genomic background. bcd and zld sites were overrepresented in the D. mel and D. pse distal enhancers and Stat92E sites were overrepresented in the D. mel and D. pse proximal enhancers (Figure 4A; Figure S1). hb sites were overrepresented in the proximal and distal enhancers from both species. We therefore hypothesized that the proximal and distal enhancers are activated by different TFs.
Figure 4.
The proximal and distal enhancers are not activated by the same transcription factors. (A) We compared the observed number of binding sites for Kr’s known activators, Stat92E, bcd, hb and zld, to a background distribution of binding sites, derived from the DNAse-sensitive regions of the genome at the blastoderm stage. The p-values associated with these scores are in Figure S1. The analysis suggests that the proximal enhancers are controlled by Stat92E and hb and distal enhancers by bcd, hb and zld. To verify the prediction, we crossed the single enhancer constructs into Stat92E (B) and bcd RNAi lines (C) and measured lacZ expression in these RNAi embryos. As expected, the proximal enhancers drive weak expression in the Stat92E RNAi embryos, and the distal enhancers drive weak expression in the bcd RNAi embryos. In bcd RNAi, the proximal enhancers are also lowly-expressed; this is likely due to indirect effects of bcd RNAi on hb levels (Struhl et al., 1989). The expression pattern is also shifted to the anterior and widened, as has been previously observed with the Kr endogenous pattern in bcd RNAi embryos (Staller et al., 2015a). We cannot compare the expression levels in RNAi embryos to the WT embryos in Figure 2 because we do not know the effect of bcd and Stat92E RNAi on our co-stain. But the unequal expression levels of D. mel enhancers in bcd and Stat92E RNAi as compared to the equal levels in WT demonstrates the differential sensitivity of these enhancers to the perturbation. (D) To test each enhancer’s hb sensitivity, we mis-expressed it in the ventral part of the embryo. In the left column, we show the average pattern of hb protein expression in WT and hb mis-expression embryos at mid-blastoderm stage, taken from (Staller et al., 2015b). We thresholded cells with expression levels greater than the mode + 0.5 standard deviation as “on” and colored the rest of the cells gray as “off”. Deeper colors indicate higher expression. The middle and right columns show the average lacZ expression pattern in each genetic background. The data were thresholded at the mode + 1 standard deviation. The expansion of the proximal enhancer’s pattern is consistent with hb activation, while the retreat of the distal enhancer’s pattern suggests repression by hb. The expression in the poles of the embryos is an unused hkb co-stain. (E) Our evidence suggests that the distal enhancer is activated by bcd and zld and repressed by hb. The proximal enhancer is activated by Stat92E and hb.
To test this hypothesis for bcd and Stat92E, we measured the activity of each enhancer reporter line in embryos depleted for each gene separately (Staller et al., 2013). In support of our hypothesis, the proximal enhancers drive very low expression levels in Stat92E RNAi embryos (Figure 4B). In bcd RNAi embryos, the distal enhancers drive weaker expression than the proximal enhancers (Figure 4C).
To test the role of zld in regulating these two enhancers, we focused on the onset of expression, because zld regulates timing of transcription in blastoderm embryos (Nien et al., 2011; Li et al., 2014). We therefore hypothesized the zld-sensitive distal enhancers drive transcription earlier than proximal enhancers. We counted the fraction of embryos expressing lacZ during stage 4, the stage of development immediately preceding the blastoderm stage. The D. mel distal enhancer reporter line drove robust lacZ expression in all stage 4 embryos (n = 8), compared with none from the D. mel proximal line (n = 13). The same trend holds true for the D. pse distal (83%, n = 18) and proximal (0%, n = 14) reporter lines.
To test how hb regulates the Kr shadow enhancers, we measured the D. mel enhancers in embryos with ventrally mis-expressed hb (Clyde et al., 2003). In ventral region, the proximal enhancer’s expression pattern expands to the posterior, while the distal enhancer’s pattern retreats, suggesting that hb activates the proximal enhancer and represses the distal (Figure 4D). This is consistent with genetic evidence that hb both activates and represses Kr (Zuo et al., 1991; Struhl et al., 1992; Schulz and Tautz, 1994).
Because these experiments are trans-perturbations, we cannot assess whether each TF is directly acting on each enhancer; testing direct interaction would require mutation of the corresponding binding sites in each enhancer. Though possible, this can be challenging (Struffi et al., 2011). We favor the hypothesis that these interactions are direct because of our binding site analysis and existing ChIP-seq data, which shows differential binding of bcd, zld, and hb activators to the Kr shadow enhancers (Figure S1). However, even if these interactions are not direct, the different responses of the proximal and distal enhancers to perturbation show that these enhancers use different regulatory logic, contradicting the initial picture of shadow enhancers as binding to the same set of TFs (Hong et al., 2008; Barolo, 2012). Our emerging picture is that shadow enhancers each build the same pattern in different ways; previous studies have shown that the snail shadow enhancers respond differently to a single repressor, (Dunipace et al., 2011), and in the eve locus, hb activates one shadow enhancer for stripe 7 and represses the other (Staller et al., 2015b). Here, we find an even more dramatic difference in the regulatory logic of the Kr shadow enhancers: each is activated by a non-overlapping set of TFs.
Our motivation for identifying the activators of the two Kr enhancers was to uncover the changes in regulatory DNA that lead to compensation in expression levels. But because the pair uses different TFs, this is impossible in a small dataset. If both enhancers were controlled by the same TF, we could look for anti-correlation in TF binding sites, such as gain of Bcd sites in one enhancer with compensatory loss in the other. But because the two enhancers are controlled by different sets of activators (Figure 4E), a large number of sequence differences could explain expression level changes between species, e.g. changes in the number, strength or arrangement of any of the activator binding sites. This is insufficiently constrained by measurements of three pairs of orthologous shadow enhancers. However, with measurements from a much larger set of orthologs, it may be possible to discern the sequence changes underlying compensatory evolution of expression levels.
Limitations
We assessed compensatory evolution between shadow enhancers using reporter constructs that contain the eve basal promoter (Experimental Procedures), which come with inherent strengths and weaknesses. The reporter constructs are highly controlled: when integrated in a site-specific manner, any significant differences between transgenic reporter lines can be ascribed to the differences in the enhancer sequences driving the reporter. Ludwig, et al. showed the power of this approach in their study of compensatory evolution within the eve stripe 2 enhancer (Ludwig et al., 2000). However, in the intact animal, promoters, UTRs, and other factors also affect expression, and therefore, reporter constructs do not fully recapitulate evolution in the natural setting. The advent of genome editing tools will make experiments in the intact locus more feasible (Gratz et al., 2015; Housden et al., 2014), but interpretation of these experiments will be complicated by the feedback mechanisms present in many transcriptional circuits (Jaeger, 2011; Schier and Gehring, 1992), which will make it hard to discern the direct effects of changes in regulatory DNA.
Implications
The observation of compensatory evolution within enhancers led to the influential idea that stabilizing selection acts on entire enhancers rather than on individual TF binding sites (Ludwig et al., 2000). This allows for flexible constraints on TF binding site organization within an enhancer (Arnosti and Kulkarni, 2005; Weirauch and Hughes, 2010). Our results support the hypothesis that there is also stabilizing selection between shadow enhancers and therefore flexibility in how expression is controlled by multiple enhancers in a locus. The fact that there are multiple ways to get the same gene expression pattern, with flexibility both in individual pieces of regulatory DNA and how information is allocated between them, leads to a vast neutral sequence space of regulatory DNA. This genetic variation is the substrate upon which evolution can act. Within a species, this large sequence space allows the species to be mutationally close to a wide range of novel phenotypes, allowing for a large number of evolutionary paths to novelty (Huynen, 1996).
The list of genes controlled by shadow enhancers, super enhancers, and other large constellations of regulatory DNA is rapidly growing, and these genes are often involved in key developmental programs or disease progression (Hnisz et al., 2015; Adam et al., 2015). Deciphering how these pieces of regulatory DNA work together to control expression, and by extension how they are constrained during evolution, is an important step towards decoding transcriptional regulation in animals.
Experimental Procedures
Transgenic fly line creation
We used the D. mel proximal and distal enhancers used in (Perry et al., 2011); the coordinates for the proximal enhancer are chr2R:21112355-21113940 and for the distal enhancer are chr2R:21110141-21111300 (BDGP R5/dm3 assembly). We identified orthologous pieces in D. yak and D. pse using the UCSC Genome Browser’s liftOver tool (https://genome.ucsc.edu/cgibin/hgLiftOver). Supplemental File 1 contains the exact sequences. We confirmed that there was not significant sequence conservation outside of these regions (Figure S2). Using Gibson assembly, we cloned the combined construct and the individual enhancers into the pBΦY vector, which contains the eve basal promoter driving lacZ, the Amp and mini-white marker genes, and an attB site for site-specific integration (Hare et al., 2008; Groth et al., 2004). Each construct was injected into white118 flies carrying the attP2 integration site by Genetic Services and Best Gene. Flies were homozygoused using the mini-white marker.
RNAi and hb ventral mis-expression
To measure reporter expression in RNAi backgrounds, we first crossed virgin females with a maternal-tubulin-Gal4 driver to males with a UAS-shRNA construct. The maternal-tubulin-Gal4 is homozygous for two insertions of a construct containing the alphaTub67C promoter and the 3’ UTR from alphaTub84B (Staller et al., 2013). The UAS-shRNA-bcd line is Bloomington Drosophila Stock Center line number 35478 (Transgenic RNAi Project construct TRiP.GL00407), the Stat92E line is number 33637 (TRiP.HMS00035). We then collected virgin female offspring and crossed these flies to reporter line males. The resulting embryos were collected as described below. The shRNA against bcd was validated in (Staller et al., 2015a). Using qPCR, we confirmed the Stat92E shRNA knocked down Stat92E mRNA expression by ~90% (Figure S1). As in (Staller et al., 2015a), we removed embryos with a weak knockdown from the data set based on the expression pattern of fushi-tarazu (ftz). Ftz is normally expressed in seven transverse stripes along the anterior-posterior axis, and its pattern is altered in a stereotypical manner in response to bcd and Stat92E knockdown. Bcd RNAi embryos with a strong knockdown have only 6 ftz stripes, so we removed embryos that had 7 ftz stripes. To curate the Stat92E RNAi embryos, we removed embryos that did not have thick ftz stripes 6 and 7 and weak ftz stripe 4. We mis-expressed hb in the ventral part of the embryo using a construct driving hb with the snail promoter as described in (Clyde et al., 2003), kindly provided by Steve Small.
In situ hybridization
The in situ hybridization was done as described in (Luengo Hendriks et al., 2006). We collected and fixed 0–4 hour old embryos at 25°C. To stain the embryos, we incubated the embryos at 56°C for two days with DNP-labeled probes for lacZ and hkb and DIG-labeled probes for ftz. The hkb probes as a standard for normalizing expression levels between different reporter lines. We sequentially detected the probes with anti-DIG-HRP antibody (Roche, Indianapolis, IN) and coumarin-tyramide color reaction (Perkin-Elmer, Waltham, MA) and anti-DNP-HRP (Perkin-Elmer) antibody and Cy3-tyramide color reaction (Perkin-Elmer). We treated the embryos with RNAseA and then stained the nuclei with Sytox Green (Life Technologies, Grand Island, NY). We mounted the embryos in DePex (Electron Microscopy Sciences, Hatfield, PA), using a bridge of #1 coverslips to preserve embryo morphology.
Image acquisition, processing, and analysis
Using 2-photon laser scanning microscopy, we acquired z-stacks of each embryo on a Zeiss LSM 710 with a plan-apochromat 20X 0.8 NA objective. Using the software described in (Luengo Hendriks et al., 2006), each stack was converted into a PointCloud, a text file that includes the location and levels of gene expression for each nucleus. We imaged embryos in the early blastoderm stage (4–10% membrane invagination) for the figures in the main text, but we have included the results for older embryos (26–100% membrane invagination) in the Figure S3.
To normalize the lacZ levels, we identified the 95% quantile of hkb expression in the posterior 10% of each embryo and divided the lacZ signal by that amount (Wunderlich et al., 2014). Within a genotype, we expect the lacZ and hkb levels to be correlated with each other. To verify this, we ran a regression of the 99% quantile lacZ value from each embryo with the 95% quantile hkb value. We discarded influential outliers using Cook’s distance (Cook, 1977), as described in (Wunderlich et al., 2014). We show the numbers of embryos in Table S1. Importantly, we only compare lacZ levels in embryos stained in a single batch in the same genetic background to avoid extraneous sources of noise in the normalization.
To generate the line traces of embryos, we used the extractpattern command in the PointCloud toolbox (http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=analysis). This divides the embryo into 16 strips along the anterior-posterior (AP) axis of the embryo, and for each strip, calculates the mean expression level in 100 bins along the AP axis. We averaged the strips along the right and left lateral side of the embryos, averaged them, and subtracted the minimum value along the axis to remove background noise. The boundaries of the expression pattern were defined as the inflection point of the lacZ expression levels.
Transcription factor binding site analysis
To calculate the background rate of TF binding site motif matches, we used accessible regions of the genome during the blastoderm stage, as identified by DNAse sensitivity (Thomas et al., 2011). For each TF of interest, we calculated the number of motif matches to the background DNA using PATSER (http://ural.wustl.edu/software.html), with a p-value = 0.001. The binding motifs are from FlyFactorSurvey (http://pgfe.umassmed.edu/ffs/) (Noyes et al., 2008), and we used a pseudocount of 0.1 and a GC content of 0.406 when generating position weight matrices from these count matrices (Supplemental File 2). The scores shown in Figure 4A are the difference between the observed number of TF binding sites and the expected number of binding sites based on the background distribution.
Supplementary Material
Highlights.
-
*
Expression levels driven by a pair of Kr enhancers conserved between Drosophilids
-
*
Expression levels driven by the individual enhancers diverge between species
-
*
Compensatory evolution acts to maintain overall Kr expression levels
-
*
Each shadow enhancer is controlled by different activating TFs
Acknowledgements
We thank Max Staller for his assistance with designing the RNAi experiments and noticing the difference in expression onset time in the Kr enhancers. We thank Francheska Lopez Rivera for assistance in setting up the RNAi crosses, Clarissa Scholes for her thorough comments on the manuscript, and all the DePace lab members for valuable discussions. We thank the TRiP at Harvard Medical School (NIH R01 GM084947) for providing transgenic RNAi fly stocks used in this study. Research reported in this publication was supported by the NIH Grant K99/R00 HD073191 (to ZW), NIH Grant U01 GM103804-01A1 (to AHD), the David Baltimore/Broad Foundation Endowment fellowship (to JW), and the Novartis Fellowship (to JE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ZW, BJV, and AHD designed the study, ZW, MDJB, BJV, and JAW collected the data, ZW and JE processed the data, ZW analyzed the data, and ZW and AHD wrote the paper, with input from all authors.
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, Fuchs E. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 2014;46:685–692. doi: 10.1038/ng.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays. 2012;34:135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Clyde DE, Corado MS, Wu X, Pare A, Papatsenko D, Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- Cook RD. Detection of Influential Observation in Linear Regression. Technometrics. 1977;19:15–18. [Google Scholar]
- Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes CC, Eckenrode KB, Bragdon MD, Meyer M, Wunderlich Z, Simirenko L, Luengo Hendriks CL, Keranen SV, Henriquez C, Knowles DW, Biggin MD, Eisen MB, DePace AH. A conserved developmental patterning network produces quantitatively different output in multiple species of Drosophila. PLoS Genet. 2011;7:e1002346. doi: 10.1371/journal.pgen.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N. Multiple layers of complexity in cis-regulatory regions of developmental genes. Dev Dyn. 2012;241:1857–1866. doi: 10.1002/dvdy.23871. [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB. Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev Biol. 2012;362:309–319. doi: 10.1016/j.ydbio.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Harrison MM, Wildonger J, O’Connor-Giles KM. Precise Genome Editing of Drosophila with CRISPR RNA-Guided Cas9. Methods Mol Biol. 2015;1311:335–348. doi: 10.1007/978-1-4939-2687-9_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Schroder C, Seifert E, Jackle H. cis-acting control elements for Kruppel expression in the Drosophila embryo. EMBO J. 1990;9:2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Scharfe CP, Rodrigues JP, Green AG, Kohlbacher O, Sander C, Bonvin AM, Marks DS. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3:e03430. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden BE, Lin S, Perrimon N. Cas9-based genome editing in Drosophila. Methods Enzymol. 2014;546:415–439. doi: 10.1016/B978-0-12-801185-0.00019-2. [DOI] [PubMed] [Google Scholar]
- Huynen MA. Exploring phenotype space through neutral evolution. J Mol Evol. 1996;43:165–169. doi: 10.1007/BF02338823. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Sather S, Martin JR, Ollo R. Analysis of Krüppel control elements reveals that localized expression results from the interaction of multiple subelements. Proc Natl Acad Sci U S A. 1991;88:5912–5916. doi: 10.1073/pnas.88.13.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. The gap gene network. Cell Mol Life Sci. 2011;68:243–274. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The role of compensatory neutral mutations in molecular evolution. Journal of Genetics. 1985;64:7–19. [Google Scholar]
- Lam DD, de Souza FS, Nasif S, Yamashita M, López-Leal R, Otero-Corchon V, Meece K, Sampath H, Mercer AJ, Wardlaw SL, Rubinstein M, Low MJ. Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold. PLoS Genet. 2015;11:e1004935. doi: 10.1371/journal.pgen.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife. 2014;3:e03737. doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Luengo Hendriks CL, Keranen SV, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, Malik J, Sudar D, Biggin MD, Knowles DW. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V, Kinney J, Callan CG, Lässig M. Energy-dependent fitness: a quantitative model for the evolution of yeast transcription factor binding sites. Proc Natl Acad Sci U S A. 2008;105:12376–12381. doi: 10.1073/pnas.0805909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris M, Kaplan T, Li XY, Villalta JE, Lott SE, Eisen MB. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet. 2013;9:e1003748. doi: 10.1371/journal.pgen.1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- Staller MV, Fowlkes CC, Bragdon MD, Wunderlich Z, Estrada J, DePace AH. A gene expression atlas of a bicoid-depleted Drosophila embryo reveals early canalization of cell fate. Development. 2015a;142:587–596. doi: 10.1242/dev.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Vincent BJ, Bragdon MD, Wunderlich Z, Estrada J, DePace AH. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc Natl Acad Sci U S A. 2015b;112:785–790. doi: 10.1073/pnas.1413877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, Depace AH, Perrimon N. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193:51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development. 2011;138:4291–4299. doi: 10.1242/dev.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, Biggin MD, Stamatoyannopoulos JA. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi A, Xia F, Li J, Larson K, LaFrance R, Li WX. STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLoS Genet. 2011;7:e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, Turner JM, Bertelsen MF, Murchison EP, Flicek P, Odom DT. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z, Bragdon MD, DePace AH. Comparing mRNA levels using in situ hybridization of a target gene and co-stain. Methods. 2014;68:233–241. doi: 10.1016/j.ymeth.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Bragdon MD, Eckenrode KB, Lydiard-Martin T, Pearl-Waserman S, DePace AH. Dissecting sources of quantitative gene expression pattern divergence between Drosophila species. Mol Syst Biol. 2012;8:604. doi: 10.1038/msb.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley JL. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 1991;5:254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.