Abstract
Immunoglobulins M (IgMs) are gaining increasing attention as biopharmaceuticals since their multivalent mode of binding can give rise to high avidity. Furthermore, IgMs are potent activators of the complement system. However, they are frequently difficult to express recombinantly and can suffer from low conformational stability. Here, the broadly neutralizing anti-HIV-1 antibody 2G12 was class-switched to IgM and then further engineered by introduction of 17 germline residues. The impact of these changes on the structure and conformational stability of the antibody was then assessed using a range of biophysical techniques. We also investigated the effects of the class switch and germline substitutions on the ligand-binding properties of 2G12 and its capacity for HIV-1 neutralization. Our results demonstrate that the introduced germline residues improve the conformational and thermal stability of 2G12-IgM without altering its overall shape and ligand-binding properties. Interestingly, the engineered protein was found to exhibit much lower neutralization potency than its wild-type counterpart, indicating that potent antigen recognition is not solely responsible for IgM-mediated HIV-1 inactivation.
Keywords: Antibody engineering, Germline, HIV, IgM, Protein stability
Highlights
-
•
2G12-IgM is highly efficient in HIV-1 neutralization.
-
•
Framework engineering leads to 2G12-IgM with superior biophysical properties.
-
•
2G12-IgM tolerates 17 germline residues without loss of antigen binding.
-
•
Potent antigen recognition is not sufficient for IgM-mediated HIV-1 inactivation.
1. Introduction
Monoclonal antibodies (mAbs) are increasingly important therapeutics for the treatment of cancer as well as autoimmune and infectious diseases. The clinical success of mAbs is based on their potential to bind their antigens with high affinity and specificity, their long in vivo half-life and their effector functions such as antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity. The majority of currently approved mAbs are full-size IgGs (150 kDa), but smaller versions such as minibodies (80 kDa), Fab fragments (50 kDa) or scFv derivatives (27 kDa) are emerging as alternatives [1]. However, many of these new variants suffer from a short half-life or the absence of binding sites for ligands that trigger effector functions.
Another alternative to IgGs are IgMs. These large polymeric antibodies (~ 970 kDa in their pentameric form) are of increasing importance as therapeutics. IgMs are the first antibodies to be produced during a humoral immune response and thus tend to have low affinity, but their multivalent mode of binding allows for high avidity. Moreover, their complex structure makes them very effective in activating the complement system [2]. It has been demonstrated that IgMs can be employed in anti-cancer therapy [3–6], for combating microbial infections [7,8] or the treatment of graft-versus-host disease [9]. IgMs are considered to be difficult to produce in cell factories or other expression platforms [10,11], and the purified proteins frequently suffer from decreased conformational stability and heterogeneity in oligomeric structure. However, a human IgM has been recently produced in a commercially feasible scale [12].
Here we have investigated how the introduction of germline residues into a human immunodeficiency virus type 1 (HIV-1)-neutralizing IgM modifies its conformational and thermal stability as well as antigen binding and neutralization potency. The IgG version of the broadly neutralizing anti HIV-1 mAb 2G12 [13] exhibits a unique domain-swapped structure, which enables it to bind specifically to a highly conserved cluster of high-mannose N-glycans exposed on the viral envelope. A class switch from IgG to IgM improved virus neutralization substantially [14]. However, considerable heterogeneity was observed with respect to the oligomeric state of 2G12-IgM, which was attributed to a reduced conformational stability [15]. In the present study, a number of germline residues was selected by rational design and then incorporated into 2G12-IgM to improve the biochemical properties of the protein. Altogether, 17 changes were introduced into the framework of the variable domains of the heavy and light chains, aiming at an increase of protein stability while preserving antigen recognition. A thorough biophysical comparison of 2G12-IgG and its two IgM variants enabled us to determine the conformational and thermal stability of the three recombinant proteins and elucidate their unfolding pathways. Additionally, structural information was derived from small-angle X-ray scattering (SAXS) studies. Finally, the ligand-binding properties of wild-type and variant 2G12-IgMs were assessed by ELISA and neutralization assays.
2. Materials and methods
2.1. Cloning and establishment of cell lines
Highly purified 2G12-IgG was a gift from Polymun Scientific (Immunbiologische Forschung GmbH, Klosterneuburg, Austria). The generation of CHO lines producing wild-type 2G12-IgM (IgM-012) as well as a control antibody (IgM-617) was described previously [16]. The germline variant of 2G12-IgM (IgM-012_GL) was designed according to standard humanization strategies for monoclonal antibodies [17]. Briefly, the germline genes with the highest similarity to vH and vL of IgM-012 were identified as IGHV3-21/JH3 and IGKV1-5/JK1, respectively, using the online tool IMGT/V-QUEST [18,19] and aligned with the variable regions of IgM-012 using CLUSTAL 2.0.8. Suitable replacements were then chosen based on a superimposition of an IgM-012 model with the structure of 2G12-IgG (PDB: 1OM3) using Swiss-PDBViewer [20]. Finally, codon-optimized sequences (Life Technologies) were cloned into bi-cistronic pIRES vectors (Clontech, # 631621) and used for transfection of CHO DG44 cells (Invitrogen, # A10999-01) to establish a CHO cell line producing IgM-012_GL, as described previously [14,16].
Recombinant cell lines were routinely cultivated in chemically defined ProCHO5 medium (Lonza, # BE12-766Q) supplemented with 4 mM L-glutamine (Biochrom, # K0302), 15 μg/mL phenol red (Sigma-Aldrich, # P0290), 0.5 mg/mL G418 (PAA, # P11-012) and 100 nM methotrexate (Sigma-Aldrich, # M8407) [15].
2.2. Purification of IgMs
Collected cell-culture supernatants were concentrated by ultradiafiltration using a Millipore Labscale™ TFF System (Millipore, # XX42LSS12). In the case of IgM-617, the concentrate was stored overnight at 4 °C [21] followed by centrifugation at 8000 g for 30 min at 4 °C. Precipitated IgM-617 was dissolved in 0.2 M NaHCO3, 0.15 M NaCl (pH 8.5) containing 3 M urea and then dialyzed against 0.2 M NaHCO3, 0.15 M NaCl (pH 8.5).
IgM-012 and IgM-012_GL concentrates were subjected to affinity chromatography using IgM CaptureSelect Affinity Matrix (Life Technologies, # 289010). 0.1 M glycine (pH 3.0) was used as elution buffer. Eluted IgMs were immediately neutralized to pH 7.0 using Tris–HCl (pH 9.5). Finally, dialysis against 0.2 M NaHCO3, 0.15 M NaCl (pH 8.5) was performed.
Purified protein samples were loaded onto NuPage® gradient 3–12% Bis–Tris gels (Life Technologies, # BN1001BOX) and run at 200 V for 60 min in Tris-Acetate SDS buffer (Life Technologies, # LA0041). Gels were stained either with silver [22] or Sypro® Ruby (Bio-Rad Laboratories, # 170-3126) [15,23]. NativeMark™ unstained protein standard (Life Technologies, # LC0725) was used to estimate the molecular mass of the IgM bands. Densitometric analysis of silver-stained gels was done using Quantity One (Bio-Rad).
2.3. Electronic circular dichroism spectroscopy
Overall secondary structure composition as well as temperature-mediated unfolding was investigated by electronic circular dichroism (ECD) spectroscopy (Chirascan, Applied Photophysics). The instrument was equipped with a Peltier element for temperature control. Temperature-mediated denaturation was monitored between 20 °C and 90 °C with a heating rate of 1.0 °C min− 1. For overall secondary structure determination, wavelength scans between 190 and 260 nm were performed. Temperature-mediated unfolding was studied at 218 nm (spectral bandwidth 3.0 nm; scan time per point: 12.5 s). Samples were analyzed in 0.2 M NaHCO3, 0.15 M NaCl (pH 8.5).
2.4. Differential scanning calorimetry
Differential scanning calorimetry (DSC) was performed on a Microcal VP-capillary DSC microcalorimeter (GE Healthcare) equipped with a 96-well plate autosampler. Samples (cell volume: 137 μL) underwent programmed heating, using a scan rate of 60 °C h− 1 over the temperature range of 20 °C to 110 °C. Collected DSC data were corrected for the buffer baseline and normalized for protein concentration. Heat capacity (Cp) was expressed in kcal mol− 1 K− 1 (1 cal = 4.184 J). Data points were fitted to non-two-state unfolding models. Samples were measured in 0.2 M NaHCO3, 0.15 M NaCl (pH 8.5).
2.5. Fluorescence spectroscopy
Chemical unfolding in the presence of increasing concentrations of urea (Sigma-Aldrich, # U5378) was followed by fluorescence spectroscopy on an F-7000 Fluorescence Spectrophotometer (Hitachi) by monitoring changes in the intrinsic fluorescence of tryptophan residues (excitation wavelength: 295 nm; emission wavelength: 300 – 450 nm; scan speed: 60 nm min− 1; PMT voltage: 700 V; response: 0.5 s).
2.6. Small-angle X-ray scattering
All small-angle X-ray scattering (SAXS) data were recorded on an in-house instrument (SAXSess mc2, Anton Paar) equipped with a Kratky camera, a sealed X-ray tube source and a two-dimensional Princeton Instruments PI•SCX:4300 CCD detector (Roper Scientific). The scattering patterns were measured with 180 min exposure times (1080 frames, each 10 s) for several solute concentrations in the range of 1–10 mg/mL. Radiation damage was excluded based on a comparison of individual frames of the 180 min exposures. A range of momentum transfer of 0.012 < s < 0.63 Å− 1 was covered (s = 4π sin(θ) / λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength). All SAXS data were analyzed with the package ATSAS (version 2.5). The data were processed with the SAXSQuant software (version 3.9), and desmeared using the programs GNOM [24] and GIFT [25]. Forward scattering intensity (I (0)), radius of gyration (Rg), maximum dimension (Dmax) and the inter-atomic distance distribution function (P(R)) were computed with the program GNOM. The masses of the solutes were evaluated by comparison of the forward scattering intensity with that of a human serum albumin reference solution of known concentration (molecular mass 66.5 kDa) and using Porod's law. To generate ab initio shape models, a total number of 50 models were calculated using the program DAMMIF [26] and aligned, and averaged using the program DAMAVER [27] assuming five-fold symmetry.
2.7. ELISA
96-well plates were coated with 2 μg/mL (100 ng/well) of sheep anti-gp120 antibody (Aalto, # D7324) and incubated overnight at 4 °C. After washing and blocking with 4% non-fat dry milk in PBS (containing 0.05% Tween-20) for 1 h at 37 °C, plates were further incubated with 50 ng per well of soluble HIV-1 BG505 SOSIP.664 gp140 [28] for 2 h at 37 °C. Plates were then washed and subsequently incubated with serially diluted (1:4) antibody samples (starting at 10 μg/mL). After 1 h at 37 °C, the plates were washed, and bound antibody was detected with a goat anti-human IgG + IgA + IgM-peroxidase conjugate (Jackson ImmunoResearch, # 109035064). Finally, plates were washed, developed with 3,3′,5,5′-tetramethylbenzidine as substrate, and analyzed with a BioTek plate reader at 450 nm. EC50 values were calculated using GraphPad Prism 5.0 software for Windows (GraphPad Software).
2.8. Neutralization assays
Pseudotyped virions were generated as described previously [29]. In brief, 5 × 105 human embryonic kidney 293T cells (ATCC, # CRL-3216) were co-transfected with 4 μg of the HIV-1 envelope glycoprotein (Env)-deleted backbone plasmid pSG3ΔEnv (NIHARRRP; contributed by J. Kappes and X. Wu) and 2 μg of the respective HIV-1 Env complementation kiplasmid using polyethyleneimine (18 μg) as a transfection reagent. Cell culture supernatants were harvested 48 h after transfection, cleared by centrifugation at 4000 g for 10 min, and then used for single-round infectivity assays as described elsewhere [30]. Briefly, pseudotyped virus was added at a 1:1 volume ratio to serially diluted (1:3) mAbs (starting at 40 μg/mL) and incubated at 37 °C. After 1 h TZM-bl reporter cells (NIH AIDS Reagent Program, # 8129) were added (1:1 by volume) at 1 × 104 cells/well, supplemented with 10 μg/mL DEAE-dextran and then incubated for a further 48 h at 37 °C. Next, the cells were washed, lysed, and developed with luciferase assay reagent according to the manufacturer's instructions (Promega). Relative light units were then measured using a microplate luminometer (BioTek, Synergy 2 luminescence microplate reader). All experiments were performed at least in duplicate. The extent of virus neutralization in the presence of antibody was determined as the 50% inhibitory concentration (IC50) as compared to samples treated without mAb.
3. Results
3.1. Design and production of engineered 2G12-IgM
The rational design of IgM-012_GL was based on the primary sequence of IgM-012. Ten mutations were introduced into the framework of IgM-012 vH and 7 mutations into that of IgM-012 vL (Tables 1a and 1b). In essence, all residues were changed into their germline counterparts except those which are (i) known to be important for antigen binding (as identified by previous studies), (ii) located in Vernier zones, and (iii) responsible for the unique domain swap of 2G12 (Fig. 1, Tables 1a and 1b). While the percentage of identities between IgM-012 and the corresponding germline sequences is 68% (vH) and 84% (vL), these values increase to 78% and 91% in the case of IgM-012_GL, leading to an improvement of the germinality index [31] from 0.8 (IgM-012) to 0.9 (IgM-012_GL). However, the natural IgM-617 displays an even higher extent of homology with its germline counterpart (vH: 95%; vL: 98%). It should be noted that the mutations introduced into IgM-012 did not lead to substantial changes in hydrophobicity (IgM-012: 244 hydrophobic residues; IgM-012_GL: 243 hydrophobic residues; total number of residues in both cases: 789) and isoelectric point (IgM-012: 6.72; IgM-012_GL: 6.81).
Table 1a.
Germline IGHV3-21/JH3 residues introduced into the variable regions of the heavy chain of IgM-012 leading to IgM-012_GL. Amino acids previously identified as structurally or functionally important [32–37] were kept unchanged.
| Position | 2G12 | IGHV3/JH3 | Change | Reasoning |
|---|---|---|---|---|
| 14 | A | P | No | Crucial for domain exchange [34] |
| 19 | I | R | No | Hydrophobic patch in VH/VH′ interface [33] |
| 23 | G | A | G > A | |
| 24 | V | A | V > A | |
| 26 | N | G | No | N might contribute to conformation |
| 28 | R | T | No | Vernier zone [37] |
| 29 | I | F | No | Vernier zone [37] |
| 31 | A | S | No | Contact with antigen [33,34] |
| 32 | H | Y | No | Contact with antigen [33,34] |
| 33 | T | S | No | Contact with antigen [33,34] |
| 39 | R | Q | No | Crucial for domain exchange [34] |
| 40 | V | A | V > A | |
| 43 | G | K | No | Positioned in VH/VL interface [34] |
| 49 | A | S | No | Vernier zone [37] |
| 52A | T | S | No | Contact with antigen [33] |
| 55 | T | S | No | Contact with antigen [34] |
| 57 | R | I | No | Important in VH/VH′ interface [33] |
| 58 | D | Y | No | Important according to [33] |
| 62 | A | S | No | Both residues small and uncharged |
| 69 | V | I | No | Vernier zone [37] |
| 73 | D | N | No | Vernier zone [37] |
| 74 | L | A | L > A | |
| 75 | E | K | No | Crucial for domain exchange [34] |
| 76 | D | N | D > N | |
| 77 | F | S | No | Hydrophobic patch in VH/VH′ interface [33] |
| 78 | V | L | No | Vernier zone |
| 82A | H | N | H > N | |
| 82B | K | S | K > S | |
| 82C | M | L | M > L | |
| 84 | V | A | No | V stabilizes structure [33] |
| 89 | I | V | I > V | |
| 105 | P | Q | No | Unique structural properties of P |
| 108 | V | M | V > M | |
| 113 | P | S | No | Important for domain swap [33] |
Table 1b.
Germline IGKV1-5/JK1 residues introduced into the variable regions of the light chain of IgM-012 leading to IgM-012_GL. Amino acids previously identified as structurally or functionally important [32–37] were kept unchanged.
| Position | 2G12 | IGKV1-5/JK1 | Change | Reasoning |
|---|---|---|---|---|
| 2 | V | I | I > V | |
| 3 | V | Q | Q > V | |
| 18 | T | R | T > R | |
| 19 | I | V | I > V | |
| 30 | E | S | No | Vernier zone [37] |
| 31 | T | S | No | Canonical residue [37] |
| 53 | T | S | No | Specificity-determining residue [37] |
| 55 | K | E | No | Specificity-determining residue [37] |
| 56 | T | S | No | Specificity-determining residue [37] |
| 77 | G | S | G > S | |
| 80 | F | P | F > P | |
| 87 | H | Y | No | H unusual at this position |
| 90 | H | Q | No | Canonical residue [37] |
| 92 | A | N | No | Specificity-determining residue [37] |
| 93 | G | S | No | Contact with antigen [33] |
| 96 | A | W | No | Specificity-determining residue [37] |
| 103 | R | K | R > K |
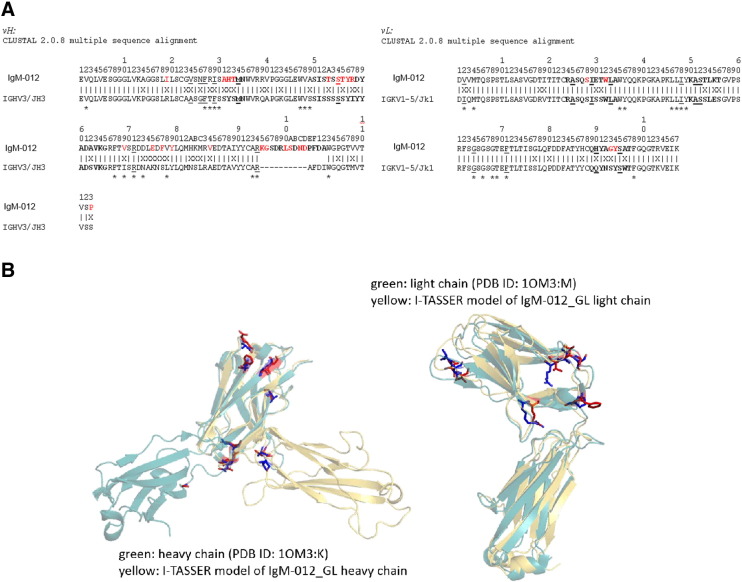
Fig. 1.
Sequence alignment and structural model of 2G12-IgM. A) Alignment of IgM-012 and the respective germline sequences IGHV3-21/JH3 and IGKV1-5/JK1 using Clustal 2.0.8. Sequences are numbered according to the Kabat system. Complementarity-determining regions are in bold letters. Canonical residues are underlined and Vernier residues are marked by asterisks. Residues previously identified as structurally and functionally important [32,34] are displayed in red. (B) Superimposition of 2G12-IgG (PDB identifier: 1OM3) with models of IgM-012_GL heavy and light chains. The introduced germline residues and their wild-type counterparts are displayed in blue and red, respectively. Note that in the model of the IgM-012_GL heavy chain, the CH1 domain does not overlap with that of 2G12-IgG due to differences in the orientation of a flexible loop.
Bi-cistronic pIRES vectors containing the engineered sequences were used for transfection of CHO DG44 cells [12,14], giving rise to a stable CHO cell line producing IgM-012_GL. IgM-012 and IgM-012_GL could be purified in good yields and were found to be highly soluble (> 10 mg/mL). SDS-PAGE combined with densitometric analysis revealed that purified IgM-012 consists of 68 ± 5% pentamers, 26 ± 3% hexamers and 5 ± 3% dimers (n = 3). Despite the replacement of 17 residues, assembly of IgM-012_GL into pentamers (58 ± 6%) and hexamers (23 ± 1%) is remarkably efficient, with only a moderate increase in dimers amounting to 19 ± 5% (n = 4; Fig. 2). However, it cannot be completely ruled out that the observed small differences in oligomer distribution might exert subtle effects on the biophysical and functional properties of the proteins.
Fig. 2.
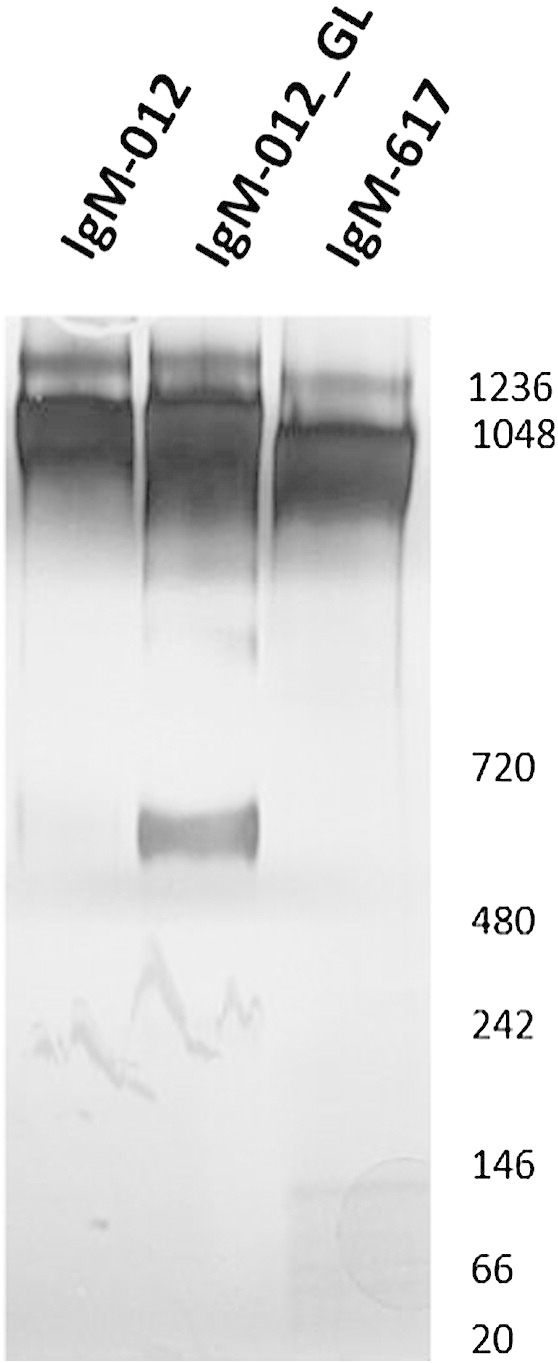
SDS-PAGE of 2G12-IgM. Purified IgM-012, IgM-012_GL and IgM-617 (control) were separated on 3–12% gradient NuPage® Bis-Tris gels and detected by silver staining.
3.2. Thermal and chemical stability
Next we probed the overall secondary structure and thermal stability of 2G12-IgG, IgM-012, IgM-012_GL and a natural IgM (IgM-617) by ECD spectroscopy. The far-UV spectra of these proteins were similar, exhibiting a minimum at 218 nm, which is typical for protein structures dominated by β-sheets (Fig. 3A left panel). Upon increasing the temperature to > 80 °C, the proteins unfolded irreversibly leading to far-UV spectra of increased ellipticity with a broad minimum between 205 and 220 nm (dashed lines in Fig. 3A, left panel). This indicates the residual presence of undefined structural elements after thermal denaturation of the antibodies, which agrees well with the observation that protein precipitation did not occur. Similar results were previously reported for antibodies of different isotypes [32].
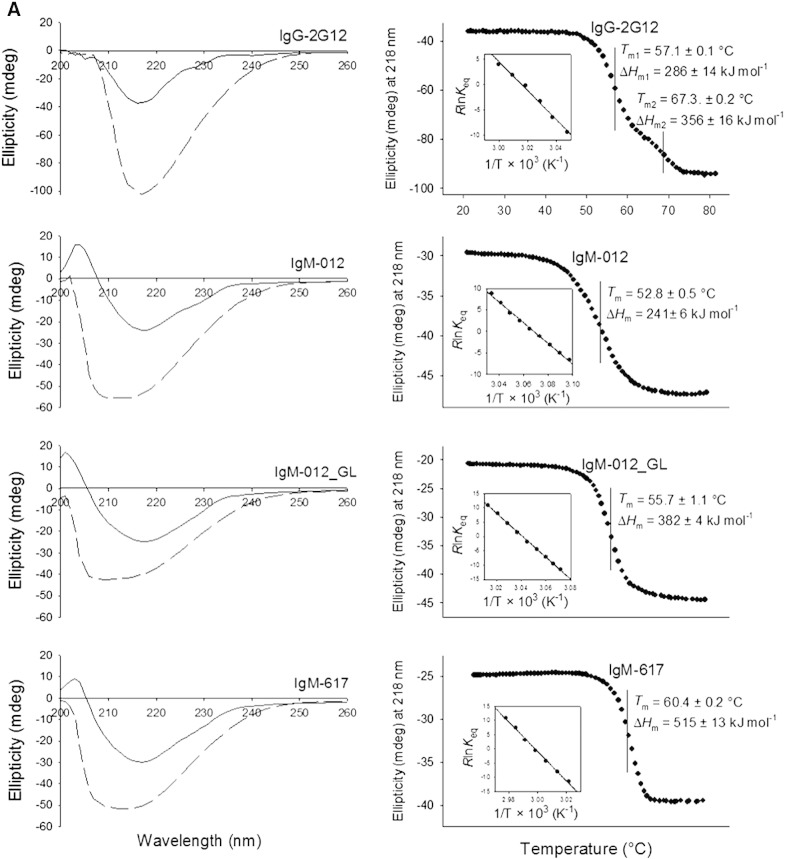
Fig. 3.
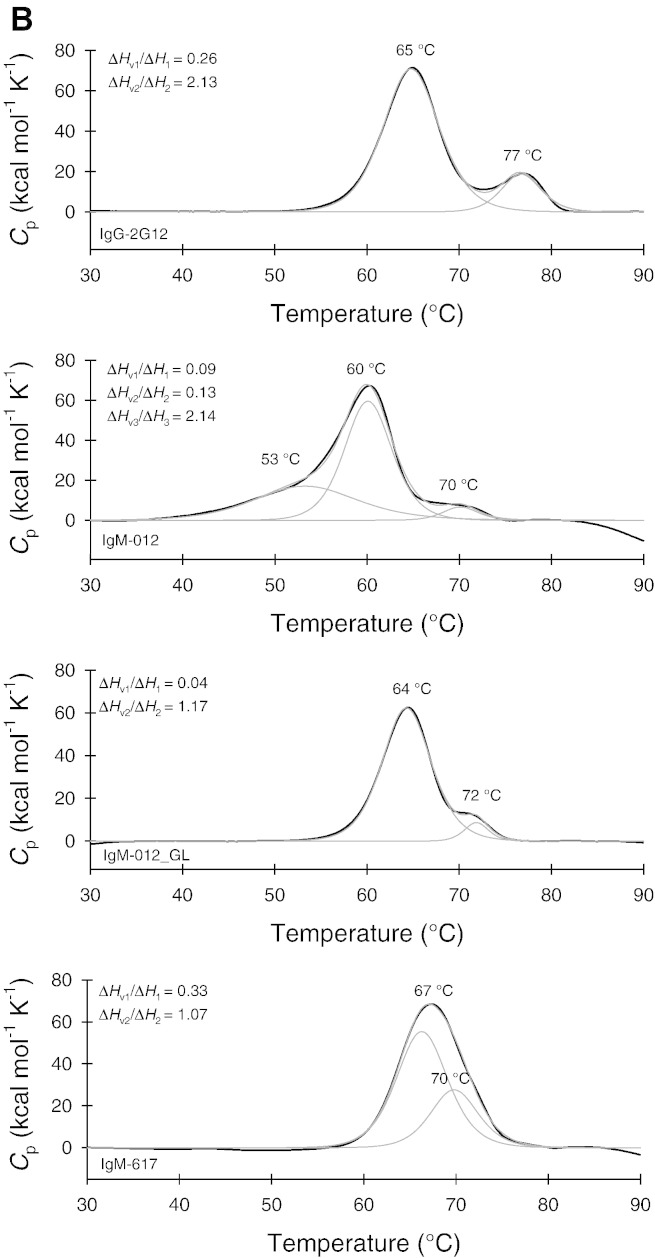
Temperature-mediated unfolding of 2G12 variants. A) Electronic circular dichroism spectroscopy. Far UV-spectra of 2G12-IgG, IgM-012, IgM-012_GL, and IgM-617 (control) in the native state (continuous lines) and in the completely unfolded state (at 80 °C; dashed lines). Temperature-mediated unfolding was followed at 218 nm. Insets display the corresponding van't Hoff plots. B) Differential scanning calorimetry. Thermograms are depicted with fits (red line) based on the assumption of non-two-state (i.e. multi-step) transitions. The ratio ΔHv/ΔH gives information about the cooperativity of a transition (ΔHv/ΔH < 1, non-cooperative; ΔHv/ΔH > 1, cooperative unfolding).
2G12-IgG clearly follows a non-two-state unfolding pathway with Tm values of 57.1 ± 0.1 °C (Tm1) and 67.3 ± 0.2 °C (Tm2). The corresponding van't Hoff enthalpies were calculated to be 382 ± 4 kJ mol− 1 and 356 ± 16 kJ mol− 1, respectively (Fig. 3A right panel). By contrast, its class-switched counterpart IgM-012 unfolds with a broad transition range between ~ 40 °C and ~ 65 °C (Tm 52.8 ± 0.5 °C, van't Hoff enthalpy 241 ± 6 kJ mol− 1). Closer inspection shows the occurrence of an unfolding intermediate (which is even more evident upon analysis with DSC as demonstrated below). In the case of IgM-012_GL, ECD suggests that the unfolding pathway follows a two-state transition within a smaller temperature range, pointing to a mode of cooperative unfolding (Tm 55.7 ± 1.1 °C, van't Hoff enthalpy 383 ± 4 kJ mol− 1). The natural IgM-617 used for comparison was found to exhibit a Tm value of 60.4 ± 0.2 °C and a van't Hoff enthalpy of about 515 kJ mol− 1 (Fig. 3A right panel).
To further investigate these observed differences in thermal stability, DSC was performed (Fig. 3B). 2G12-IgG clearly shows two main transitions at 65 °C (Tm1) and 77 °C (Tm2), whereas the endotherm of IgM-012 could be best fitted by three transitions with Tm values of 53 °C, 60 °C, and 77 °C. By introduction of the germline residues, both the cooperativity and thermal stability of IgM-012_GL increased significantly (Tm1 = 64 °C, Tm2 = 72 °C) and became similar to that of IgM-617 (Tm1 = 67 °C, Tm2 = 70 °C; Fig. 3B). With both IgM-012_GL and IgM-617 the ratio of van't Hoff enthalpy and calorimetric enthalpy calculated from DSC experiments suggested a non-cooperative unfolding event for the first transition (ΔHv/ΔH < 1) and a cooperative unfolding for the second transition (ΔHv/ΔH > 1) (Table 2). Typically, Tm values obtained from DSC were higher compared to those derived from ECD. The latter allows direct temperature control in the cuvette, whereas in DSC measurements higher scan rates may result in overestimation of Tm values. Importantly, the hierarchy of Tm values of investigated proteins is the same for both methods.
Table 2.
Calorimetric enthalpies and van't Hoff enthalpies derived from differential scanning calorimetric measurements (see Fig. 3B).
| Tm (°C) | ΔH (cal mol− 1) | ΔHv (cal mol− 1) | ΔHv/ΔH | |
|---|---|---|---|---|
| IgG-2G12 | 64.8 | 4.3 × 105 | 1.1 × 105 | 0.26 |
| 76.7 | 8.0 × 104 | 1.7 × 105 | 2.13 | |
| IgM-012 | 53.4 | 6.4 × 105 | 5.7 × 104 | 0.09 |
| 60.1 | 1.0 × 106 | 1.3 × 105 | 0.13 | |
| 70.1 | 8.4 × 104 | 1.8 × 105 | 2.14 | |
| IgM-012_GL | 64.3 | 3.3 × 106 | 1.3 × 105 | 0.04 |
| 71.8 | 2.3 × 105 | 2.7 × 105 | 1.17 | |
| IgM-617 | 66.7 | 4.0 × 105 | 1.3 × 105 | 0.33 |
| 70.2 | 1.5 × 106 | 1.6 × 105 | 1.07 |
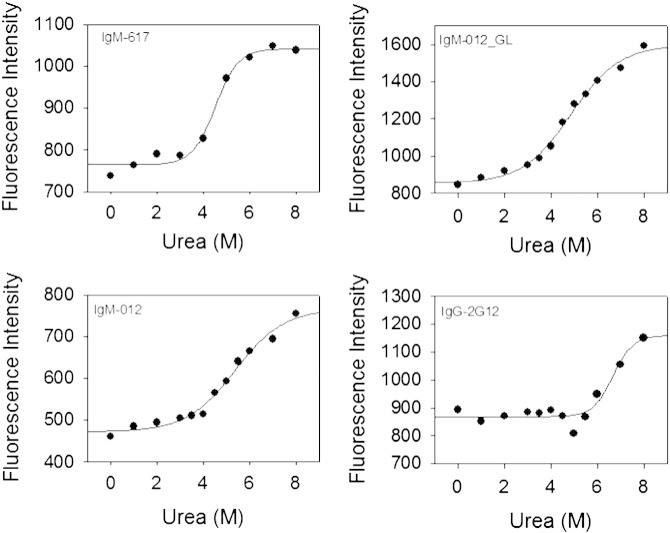
Finally, the conformational stability was probed by chemical denaturation of the four recombinant antibodies using increasing concentrations of urea (0–8 M). The change in tryptophan fluorescence intensity was monitored spectrofluorimetrically. Fig. 4 depicts the corresponding curves demonstrating that 2G12-IgG is stable up to 5 M urea. Unfolding starts at 5.5 M urea, but does not reach completion even at the highest urea concentration used (8 M). Its IgM counterpart IgM-012 already shows some minor unfolding between 1 and 4 M urea, but this is again followed by a steady increase in fluorescence without achieving an upper plateau. The presence of germline residues in IgM-012_GL only slightly increased its chemical stability, with the first (minor) transition being less pronounced than in the case of its wild-type counterpart. These data are comparable to natural IgM-617 that also follows a non-two-state transition with a first (minor) unfolding event below 3 M and a second (major) one within the range of 4–7 M urea (Fig. 4).
Fig. 4.
Urea-mediated unfolding of 2G12 variants. Change in intrinsic fluorescence upon addition of increasing concentrations of urea. Note that only the control IgM (IgM-617) could be completely unfolded. For the other mAbs, the upper plateau of the unfolding curve could not be determined. Hence, comparative estimates for the conformational stability of the antibodies could not be deduced.
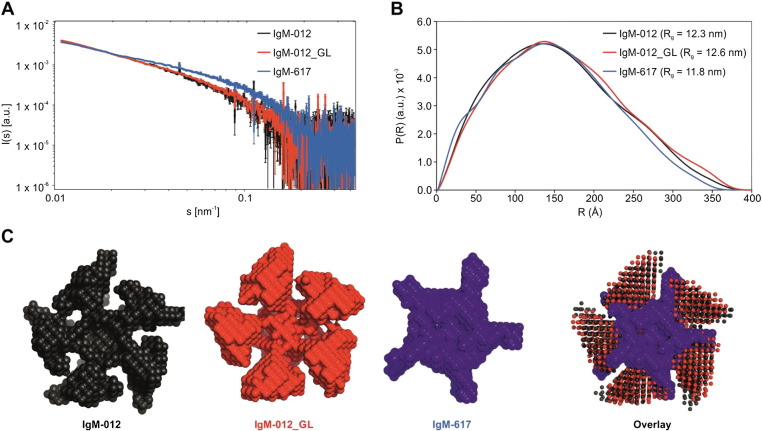
3.3. SAXS studies
For further structural evaluation small-angle X-ray scattering (SAXS) was employed. The raw data (Fig. 5A) already allows to clearly discriminate between IgM-617 and the two 2G12-IgM variants, for which almost identical scattering curves were obtained. This is further supported by the calculated gyration radius (Rg) values showing that IgM-012 and IgM-012_GL adopt a less compact fold than IgM-617 (Fig. 5B). Notably, the determined Rg values are in good agreement with those of previous SAXS experiments on IgMs [33,34]. A five-fold symmetry was imposed for low-resolution shape modeling of IgM-012 and IgM-617, and a total of 50 structures were used to generate density models. In agreement with the scattering curves and the pair distance distribution function, the superimposition of the shape models confirms that IgM-617 adopts a more compact conformation than IgM-012 and IgM-012_GL (Fig. 5C).
Fig. 5.
Global shape of 2G12-IgM. (A) Experimental X-ray scattering data of IgMs. The s axis is shown in a logarithmic representation. (B) SAXS data showing a comparison of the experimental radial density distributions of IgMs. Calculated Rg values are shown. (C) Models of IgM-012, IgM-012_GL and IgM-617 (control) proteins and their overlay. Models were calculated by DAMMIF using an average of 50 structures, with P5 symmetry being imposed.
3.4. Ligand-binding properties and neutralization activity of 2G12-IgM
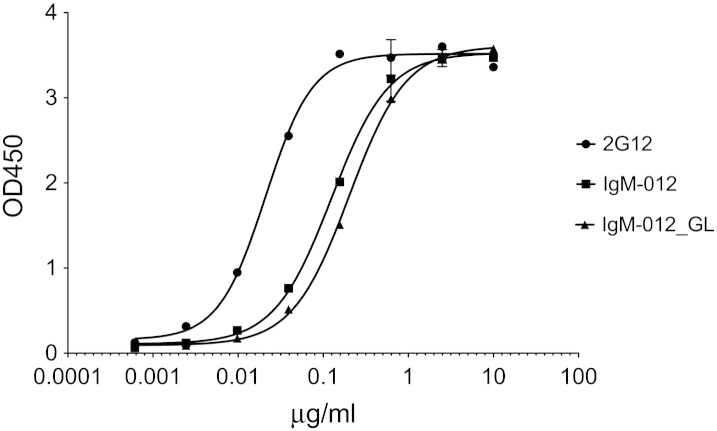
Next we compared the ligand-binding properties of 2G12-IgG, IgM-012 and IgM-012_GL. Binding to trimeric HIV-1 envelope glycoprotein BG505 SOSIP.664 gp140 (which has an architecture very similar to that of native envelope spikes on HIV-1 virions) was assessed by ELISA. As documented by the binding curves depicted in Fig. 6, IgM-012_GL is still able to bind efficiently to HIV-1 gp140. There is a 1.6-fold difference in EC50 values between IgM-012 (EC50 = 0.12 μg/mL) and IgM-012_GL (EC50 = 0.20 μg/mL), suggesting only a minimal loss in affinity. Based on protein concentration, the EC50 of 2G12-IgG for HIV-1 gp140 is considerably lower (0.02 μg/mL) than that of either IgM. However, it is possible that due to steric hindrance only one binding site per IgM molecule is reactive in our ELISA system. In terms of molarity, the ligand-binding capacity of the IgM and IgG molecules would then be comparable, as the molar mass of IgM-012 and IgM-012_GL is > 5 times higher than that of 2G12-IgG.
Fig. 6.
Ligand-binding activity of 2G12 variants. Interaction of 2G12-IgG and 2G12-IgM with trimeric BG505 SOSIP.664 HIV-1 gp140 as determined by ELISA.
Finally, the neutralization activity and breadth of the 2G12 mAbs were evaluated using a panel of pseudotyped HIV-1 isolates from clades B, C and E. IgM-012 was able to neutralize all tested clade B isolates at a median IC50 of 0.1 μg/mL. 2G12-IgG, which neutralized all clade B viruses except ADA, revealed a 10-fold higher median IC50 value (1.0 μg/mL). By contrast, IgM-012_GL neutralized only one clade B isolate (TRO.11) with high potency. None of the above mentioned mAbs neutralized HIV-1 strains from clades C and E (Table 3).
Table 3.
Neutralization potency of IgM and IgG variants of 2G12 towards pseudotyped HIV-1 isolates.
| HIV-1 isolate (clade) | IC50 [μg antibody/mL] |
||
|---|---|---|---|
| IgM-012 | IgM-012_GL | 2G12 | |
| JR-FL (B) | 0.22 | 8.91 | 0.64 |
| JR-CSF (B) | 0.10 | > 10 | 0.95 |
| SF162 (B) | 0.07 | > 10 | 1.55 |
| HxB2 (B) | 0.08 | > 10 | 0.19 |
| ADA (B) | 0.31 | > 10 | > 10 |
| PVO.4 (B) | 0.10 | > 10 | 1.88 |
| TRO.11 (B) | 0.09 | 0.38 | 0.44 |
| ZM53M (C) | > 10 | > 10 | > 10 |
| ZM214M (C) | > 10 | > 10 | > 10 |
| 93TH966.8 (E) | > 10 | > 10 | > 10 |
4. Discussion
In addition to the previously established CHO cell lines for production of the mAbs IgM-012 and IgM-617 [16], in the present work a CHO cell line was newly generated that expresses the germline variant IgM-012_GL. Engineering of IgM-012 followed a rational approach to avoid changes at amino-acid positions of importance either for antigen binding or for structural integrity [35–39]. The attempt was to (i) increase its conformational and thermal stability, (ii) preserve antibody specificity and (iii) decrease the immunogenicity of IgM-012_GL compared to the wild-type protein [39].
Since biogenesis of secretory IgMs is a stringently controlled process involving several quality control checkpoints [40], the slightly increased heterogeneity of IgM-012_GL with respect to its oligomerization status is more likely the consequence of undesirable fragmentation upon or during secretion than of imperfect assembly in the endoplasmic reticulum. Contamination with undesired proteolytic activity is a widespread problem in mAb production [41], and it has been shown that reduced conformational stability and elevated structural flexibility can give rise to a higher tendency of fragmentation [42].
The original IgG version of 2G12 is well characterized and was therefore included in this study for comparative purposes. Its antigen-binding regions are intertwined on the top of the molecule due to a domain swap, resulting in the formation of a structurally unique Fab dimer. In the gp140-bound state, the two Fab arms of 2G12-IgG were shown to be also locked above the Fc suggesting no substantial change upon ligand binding [43]. Temperature-mediated unfolding of 2G12-IgG as monitored by both ECD and DSC followed a clear non-two-state unfolding pathway with two domains of different sensitivity, representing the Fab and Fc fragments of the IgG molecule [32]. By contrast, urea-mediated unfolding monitored by fluorescence spectroscopy suggested only one main unfolding event. Upon class switch to IgM, both the thermal and chemical stability of 2G12 decreased significantly and showed at least three transitions. This could indicate that IgM sequences interfere with the swapping of the VH domains, since this unique structural feature is known to contribute to the rigidity of the 2G12 molecule [35]. However, SAXS analysis of IgM-012 and IgM-012_GL provided no evidence for inefficient domain-swapping during the assembly of 2G12-IgMs.
When compared with 2G12-IgG, a reduction was observed for the ligand-binding capacity of IgM-012, with the EC50 value for binding of HIV-1 gp140 increasing from 0.02 μg/mL to 0.12 μg/mL. This could either reflect an unanticipated steric hindrance affecting the antigen-binding sites of 2G12 in the IgM format or be a direct consequence of the reduced conformational stability of the protein. By introduction of germline residues, the thermal stability and cooperativity of unfolding of 2G12-IgM could be improved significantly. The observation that affinity-matured antibodies exhibit a lower stability than their germline precursors has already been reported for other mAbs [44,45]. Importantly, IgM-012_GL still shows good gp140 recognition, with its EC50 being only ~ 1.6-fold higher than that of IgM-012, thus demonstrating that ligand binding was only slightly compromised by the introduction of the germline residues. This is in good agreement with SAXS experiments indicating that the overall structure of IgM-012 and IgM-012_GL is very similar.
The present study also established a higher HIV-1 neutralization potency of IgM-012 as compared to 2G12-IgG, as evident from a 10-fold difference in the median IC50 value for the 2G12-sensitive viral isolates tested. With a median IC50 of 0.1 μg/mL, the neutralization potency of IgM-012 is even better than that of the latest generation of broadly neutralizing anti-HIV IgGs such as PG9 (median IC50 0.4 μg/mL) or VRC01 (median IC50 0.2 μg/mL). It has been observed previously that the immunological performance of 2G12 improves upon class switch from IgG to IgM [14]. Although the effector functions of IgM-012 still remain to be tested in detail, our findings highlight the anti-viral potential of mAbs based on the IgM format. Despite being barely compromised in its gp140-binding capacity, the stabilized variant IgM-012_GL has lost most of its ability to neutralize HIV-1. Future studies will therefore aim at identifying the introduced residues which account for this selective reduction in neutralization potency.
Transparency document
Transparency document.
Acknowledgments
This work was performed with support of the PhD program BioToP—Biomolecular Technology of Proteins funded by the Austrian Science Fund (FWF Project W1224). Further support came from the Bavarian Ministry of Sciences, Research and the Arts (Bavarian Molecular Biosystems Research Network, to T.M.), the Austrian Academy of Sciences (APART fellowship, to T.M.) and the German Research Foundation (Emmy Noether program MA 5703/1-1, to T.M.). The authors acknowledge the Leibniz Supercomputing Centre (LRZ, www.lrz.de) for providing computing time on the Linux Cluster. Additionally, we would like to thank John P. Moore and Albert Cupo for providing B6505 SOSIP.664 gp140. We also thank Polymun Scientific for the supply with 2G12-IgG.
Footnotes
The Transparency document associated with this article can be found, in the online version.
Contributor Information
Christian Obinger, Email: christian.obinger@boku.ac.at.
Renate Kunert, Email: renate.kunert@boku.ac.at.
References
- 1.Holliger P., Hudson P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 2.Thomas H.I., Morgan-Capner P. The avidity of specific IgM detected in primary rubella and reinfection. Epidemiol. Infect. 1990;104:489–497. doi: 10.1017/s095026880004749x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollmers H.P., Brändlein S. Natural human immunoglobulins in cancer immunotherapy. Immunotherapy. 2009;1:241–248. doi: 10.2217/1750743X.1.2.241. [DOI] [PubMed] [Google Scholar]
- 4.Irie R.F., Ollila D.W., O'Day S., Morton D.L. Phase I pilot clinical trial of human IgM monoclonal antibody to ganglioside GM3 in patients with metastatic melanoma. Cancer Immunol. Immunother. 2004;53:110–117. doi: 10.1007/s00262-003-0436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasche L., Duell J., Morgner C., Chatterjee M., Hensel F., Rosenwald A., Einsele H., Topp M.S., Brändlein S. The natural human IgM antibody PAT-SM6 induces apoptosis in primary human multiple myeloma cells by targeting heat shock protein GRP78. PLoS One. 2013;8:e63414. doi: 10.1371/journal.pone.0063414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensel F., Eckstein M., Rosenwald A., Brädlein S. Early development of PAT-SM6 for the treatment of melanoma. Melanoma Res. 2013;23:264–275. doi: 10.1097/CMR.0b013e328362cbc8. [DOI] [PubMed] [Google Scholar]
- 7.Derkx B., Wittes J., McCloskey R. Randomized, placebo-controlled trial of HA-1A, a human monoclonal antibody to endotoxin, in children with meningococcal septic shock. European Pediatric Meningococcal Septic Shock Trial Study Group. Clin. Infect. Dis. 1999;28:770–777. doi: 10.1086/515184. [DOI] [PubMed] [Google Scholar]
- 8.Horn M.P., Zuercher A.W., Imboden M.A., Rudolf M.P., Lazar H., Wu H., Hoiby N., Fas S.C., Lang A.B. Preclinical in vitro and in vivo characterization of the fully human monoclonal IgM antibody KBPA101 specific for Pseudomonas aeruginosa serotype IATS-O11. Antimicrob. Agents Chemother. 2010;54:2338–2344. doi: 10.1128/AAC.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macmillan M.L., Couriel D., Weisdorf D.J., Schwab G., Havrilla N., Fleming T.R., Huang S., Roskos L., Slavin S., Shadduck R.K., Dipersio J., Territo M., Pavletic S., Linker C., Heslop H.E., Deeg H.J., Blazar B.R. A phase 2/3 multicenter randomized clinical trial of ABX-CBL versus ATG as secondary therapy for steroid-resistant acute graft-versus-host disease. Blood. 2007;109:2657–2662. doi: 10.1182/blood-2006-08-013995. [DOI] [PubMed] [Google Scholar]
- 10.Mader A., Chromikova V., Kunert R. Recombinant IgM expression in mammalian cells: a target protein challenging biotechnological production. Adv. Biosci. Biotechnol. 2013;4:7. [Google Scholar]
- 11.Loos A., Gruber C., Altmann F., Mehofer U., Hensel F., Grandits M., Oostenbrink C., Stadlmayr G., Furtmüller P.G., Steinkellner H. Expression and glycoengineering of functionally active heteromultimeric IgM in plants. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6263–6268. doi: 10.1073/pnas.1320544111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tchoudakova A., Hensel F., Murillo A., Eng B., Foley M., Smith L., Schoenen F., Hildebrand A., Kelter A.R., Ilag L.L., Vollmers H.P., Brandlein S., McIninch J., Chon J., Lee G., Cacciuttolo M. High level expression of functional human IgMs in human PER.C6 cells. MAbs. 2009;1:163–171. doi: 10.4161/mabs.1.2.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J.P., Katinger H. Human mononoclonal antibody 2G12 defines a distinct neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1994;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolbank S., Kunert R., Stiegler G., Katinger H. Characterization of human class-switched polymeric (immunoglobulin M [IgM] and IgA) anti-human immunodeficiency virus type 1 antibodies 2F5 and 2G12. J. Virol. 2003;77:4095–4103. doi: 10.1128/JVI.77.7.4095-4103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorauer-Uhl K., Wallner J., Lhota G., Katinger H., Kunert R. IgM characterization directly performed in crude culture supernatants by a new simple electrophoretic method. J. Immunol. Methods. 2010;359:21–27. doi: 10.1016/j.jim.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Chromikova V., Mader A., Steinfellner W., Kunert R. Evaluating the bottlenecks of recombinant IgM production in mammalian cells. Cytotechnology. 2014 doi: 10.1007/s10616-014-9693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mader A., Kunert R. Humanization strategies for an anti-idiotypic antibody mimicking HIV-1 gp41. Protein Eng. Des. Sel. 2010;23:947–954. doi: 10.1093/protein/gzq092. [DOI] [PubMed] [Google Scholar]
- 18.Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giudicelli V., Brochet X., Lefranc M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 20.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 21.Hansma H.G., Weisenhorn A.L., Edmundson A.B., Gaub H.E., Hansma P.K. Atomic force microscopy: seeing molecules of lipid and immunoglobulin. Clin. Chem. 1991;37:1497–1501. [PubMed] [Google Scholar]
- 22.Switzer R.C., III, Merril C.R., Shifrin S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal. Biochem. 1979;98:231–237. doi: 10.1016/0003-2697(79)90732-2. [DOI] [PubMed] [Google Scholar]
- 23.Schriebl K., Trummer E., Weik R., Lattenmayer C., Muller D., Kunert R., Katinger H., Vorauer-Uhl K. Applicability of different fluorescent dyes for isoform quantification on linear IPG gels. Electrophoresis. 2007;28:2100–2107. doi: 10.1002/elps.200600695. [DOI] [PubMed] [Google Scholar]
- 24.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 25.Bergmann A., Fritz G., Glatter O. Solving the generalized indirect Fourier transformation (GIFT) by Boltzmann simplex simulated annealing (BSSA) J. Appl. Crystallogr. 2000;33:1212–1216. [Google Scholar]
- 26.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J., Golabek M., de Los Reyes K., Ketas T.J., van Gils M.J., King C.R., Wilson I.A., Ward A.B., Klasse P.J., Moore J.P. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gach J.S., Quendler H., Tong T., Narayan K.M., Du S.X., Whalen R.G., Binley J.M., Forthal D.N., Poignard P., Zwick M.B. A human antibody to the CD4 binding site of gp120 capable of highly potent but sporadic cross clade neutralization of primary HIV-1. PLoS One. 2013;8:e72054. doi: 10.1371/journal.pone.0072054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gach J.S., Achenbach C.J., Chromikova V., Berzins B., Lambert N., Landucci G., Forthal D.N., Katlama C., Jung B.H., Murphy R.L. HIV-1 specific antibody titers and neutralization among chronically infected patients on long-term suppressive antiretroviral therapy (ART): a cross-sectional study. PLoS One. 2014;9:e85371. doi: 10.1371/journal.pone.0085371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelat T., Bedouelle H., Rees A.R., Crennell S.J., Lefranc M.P., Thullier P. Germline humanization of a non-human primate antibody that neutralizes the anthrax toxin, by in vitro and in silico engineering. J. Mol. Biol. 2008;384:1400–1407. doi: 10.1016/j.jmb.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 32.Vermeer A.W., Norde W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys. J. 2000;78:394–404. doi: 10.1016/S0006-3495(00)76602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins S.J., Nealis A.S., Sutton B.J., Feinstein A. Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J. Mol. Biol. 1991;221:1345–1366. doi: 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- 34.Rosenes Z., Mulhern T.D., Hatters D.M., Ilag L.L., Power B.E., Hosking C., Hensel F., Howlett G.J., Mok Y.F. The anti-cancer IgM monoclonal antibody PAT-SM6 binds with high avidity to the unfolded protein response regulator GRP78. PLoS One. 2012;7:e44927. doi: 10.1371/journal.pone.0044927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calarese D.A., Scanlan C.N., Zwick M.B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M.R., Stanfield R.L., Roux K.H., Kelly J.W., Rudd P.M., Dwek R.A., Katinger H., Burton D.R., Wilson I.A. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 36.Huber M., Le K.M., Doores K.J., Fulton Z., Stanfield R.L., Wilson I.A., Burton D.R. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J. Virol. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calarese D.A., Lee H.K., Huang C.Y., Best M.D., Astronomo R.D., Stanfield R.L., Katinger H., Burton D.R., Wong C.H., Wilson I.A. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gach J.S., Furtmüller P.G., Quendler H., Messner P., Wagner R., Katinger H., Kunert R. Proline is not uniquely capable of providing the pivot point for domain swapping in 2G12, a broadly neutralizing antibody against HIV-1. J. Biol. Chem. 2010;285:1122–1127. doi: 10.1074/jbc.M109.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almagro J.C., Fransson J. Humanization of antibodies. Front. Biosci. 2008;13:1619–1633. doi: 10.2741/2786. [DOI] [PubMed] [Google Scholar]
- 40.Anelli T., Ceppi S., Bergamelli L., Cortini M., Masciarelli S., Valetti C., Sitia R. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26:4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandberg H., Lütkemeyer D., Kuprin S., Wrangel M., Almstedt A., Persson P., Ek V., Mikaelsson M. Mapping and partial characterization of proteases expressed by a CHO production cell line. Biotechnol. Bioeng. 2006;95:961–971. doi: 10.1002/bit.21057. [DOI] [PubMed] [Google Scholar]
- 42.Gao S.X., Zhang Y., Stansberry-Perkins K., Buko A., Bai S., Nguyen V., Brader M.L. Fragmentation of a highly purified monoclonal antibody attributed to residual CHO cell protease activity. Biotechnol. Bioeng. 2011;108:977–982. doi: 10.1002/bit.22982. [DOI] [PubMed] [Google Scholar]
- 43.Roux K.H., Zhu P., Seavy M., Kunert R., Seamon V. Electron microscopic and immunochemical analysis of the broadly neutralizing HIV-1-specific, anti-carbohydrate antibody, 2G12. Mol. Immunol. 2004;41:1001–1011. doi: 10.1016/j.molimm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Wang F., Sen S., Zhang Y., Ahmad I., Zhu X., Wilson I.A., Smider V.V., Magliery T.J., Schultz P.G. Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Sun S.B., Sen S., Kim N.J., Magliery T.J., Schultz P.G., Wang F. Mutational analysis of 48G7 reveals that somatic hypermutation affects both antibody stability and binding affinity. J. Am. Chem. Soc. 2013;135:9980–9983. doi: 10.1021/ja402927u. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.