Abstract
Prior to acquisition of the first host blood meal, the anautogenous mosquito Culex quinquefasciatus requires a period of time in order to prepare for the blood feeding and, later, vitellogenesis. In the current study, we conducted whole transcriptome analyses of adult female Culex mosquitoes to identify genes that may be necessary for both taking of the blood meal, and processing of the blood meal in adult female mosquitoes Cx. quinquefasciatus. We examined temporal expression of genes for the periods of post eclosion and prior to the female freely taking a blood meal. We further evaluated the temporal expression of certain genes for the periods after the taking of a blood meal to identify genes that may be necessary for both the taking of the blood meal, and the processing of the blood meal. We found that adult females required a minimum of 48 h post-eclosion before they freely took their first blood meal. We hypothesized that gene expression signatures were altered in the mosquitoes before blood feeding in preparation for the acquisition of the blood meal through changes in multiple gene expression. To identify the genes involved in the acquisition of blood feeding, we quantified the gene expression levels of adult female Cx. quinquefasciatus using RNA Seq throughout a pre-blooding period from 2 to 72 h post eclosion at 12 h intervals. A total of 325 genes were determined to be differentially-expressed throughout the pre-blooding period, with the majority of differentially-expressed genes occurring between the 2 h and 12 h post-eclosion time points. Among the up-regulated genes were salivary proteins, cytochrome P450s, odorant-binding proteins, and proteases, while the majority of the down-regulated genes were hypothetical or cuticular genes. In addition, Trypsin was found to be up-regulated immediately following blood feeding, while trypsin and chymotrypsin were up-regulated at 48h and 60h post blood-feeding, respectively, suggesting that these proteases are likely involved in the digestion of the blood meal. Overall, this study reviewed multiple genes that might be involved in the adult female competency for blood meal acquisition in mosquitoes.
Keywords: Vitellogenesis, blood feeding, Culex quinquefasciatus
Introduction
The Southern house mosquito, Culex quinquefasciatus vectors the disease causing agents of multiple diseases, including West Nile Fever, St. Louis Encephalitis, Japanese Encephalitis, and Lymphatic Filariasis. The pathogens vectored by Cx. quinquefasciatus are acquired during the blood meal acquisition, which must be taken by the adult female prior to the formation of each egg raft 1. Newly eclosed females require a period of time before they are capable of taking the blood meal during which, the female mates and continues with the necessary development to be competent for the acquisition of the blood meal itself 2. Many studies have shown that genes and gene up-regulation are involved in the processing of the blood meal and subsequently, vitellogenesis 3-5. Studies that have characterized the transcriptome expression patterns of adult Aedes aegypti 6 and Anopheles gambiae 7 have shown that multiple genes are involved in blood feeding and that there are different expression profiles of these genes both prior to and immediately following the blood meal. Characterizing the temporal characterization of the gene expression profiles of the Cx. quinquefasciatus transcriptome prior to the blood meal would provide valuable insight into the genes possibly needed to prepare the female for the taking of the blood meal. The recent advances in next generation sequencing, including RNA Seq, allows for the characterization of the gene expression profiles without requiring a priori knowledge of which genes to investigate. In the current study, we used RNA Seq sequencing to conduct whole transcriptome analyses of adult female mosquitoes during the post-eclosion and pre-vitellogenic stages of development to identify genes that are up- or down-regulated prior to the female freely taking a blood meal. We further investigated the temporal expression patterns of selected genes following the taking of a blood meal by female mosquitoes to identify genes that may be necessary for both the taking of and the processing of the blood meal in Cx. quinquefasciatus. Overall, our study found that the genes identified in the post-eclosion, but pre-blood-meal-taking time period, represent genes that may be necessary for the female to freely take a blood meal.
Materials and Methods
Mosquito strains
Culex quinquefasciatus strain HAmCqG8, originates from Huntsville, Alabama, and has been continuously reared under laboratory conditions since 2002 8. All mosquito rearing and experimentation was conducted at 25±2oC under a photoperiod of 12:12 (L:D) h and larvae were fed on a diet of Brewer's yeast (Fleishmann, Chesterfield, MO), while adults were fed ad libitum on 10% sucrose. Blood feeding of adult females was performed using warmed horse blood (College of Veterinary Medicine, Auburn University) through a stretched Parafilm membrane covering a 37oC heated water jacketed glass holder.
Pre-determination of time period for mosquitoes to take their first blood meal
The adult mosquitoes of 12 h post eclosion from pupae were divided into 11 groups (collection time points) with a minimum of ~60 mosquitoes each in both sexes (~1:1 ratio). The mosquito groups 1-11 were then independently offered the pre-warmed (37oC) horse blood meal with an ascending order of every 12 h starting from 24 h after eclosion; i.e., 24, 36, 48, 60, and 72 84 96 108 120 132 and 144 h, respectively. Each group of mosquitoes was fed for a single blood meal for 2 h and the number of the blood fed female mosquitoes from each group were checked after blood-feeding. All blood-feeding time points were repeated three times.
RNA extraction
All collected mosquito samples were flash frozen on dry ice and held at -80°C prior to RNA extraction. The hot acid phenol extraction method was used to obtain total RNA 9. A total of 30 µg of RNA was treated with DNase I using the DNA-Free kit from Ambion (Austin, TX) and extracted twice over acid phenol:chloroform (1:1) followed by a final chloroform extraction. The RNA was then precipitated with ethanol and dissolved in sterile distilled water. The total RNAs were subsequently used for either RNA Seq analysis (Hudson Alpha Institute of Biotechnology [HAIB], Huntsville, AL) or for the gene expression analysis.
RNA library preparation, RNA Seq sequencing, Data analysis, and gene expression processing
Total RNA quality was assessed by the HAIB using an Agilent 2100 Bioanalyzer and an Invitrogen Qubit to ensure quality for the libraries for each time point. A total of seven libraries were prepared, one for each time point of 2, 12, 24, 36, 48, 60, and 72 h post-eclosion. The Illumina RNA Sample Prep Kits for mRNA Seq was used to prepare the libraries, which were run using the paired-end 50 nt read module (HAIB), which also conducted the base calling, barcode parsing, and removal of low quality reads. Further cleaning of adapter was performed using Trimmomatic 10. The Cx. quinquefasciatus genome from Vectorbase 11 was used to map the cleaned reads using Tophat 12 and gene expression estimations were performed using Cufflinks, and differential gene expression was tested using Cuffdiff as time series data 13. After analysis, only genes with expression values ≥1, as measured in number of fragments mapped for per kilo (every thousand) bases of gene length for every million fragments sequenced (FPKM), were retained for expression comparisons 14. All data have been submitted to the Gene Expression Omnibus at NCBI as accession #GSE51327.
qPCR gene expression
The total RNA from three independent samples of 20 adult female HAmCqG8 mosquitoes was extracted as previously outlined above. The same methodology for obtaining even-aged females immediately following eclosion was used to obtain the non-blood-fed females for RNA extraction, using the same time points as previously indicated: 2, 12, 24, 36, 48, 60, and 72 h post-eclosion. In order to obtain the material for the RNA extraction for the post-blood meal sampling time points, mosquitoes were initially reared to 7 d of age (post-eclosion) prior to the offering of a blood meal. Blood meals were then offered for a 2 h period at the onset of the scotophase, and blooded females were collected at 2 (immediately after blood feeding), 4, 8, 12, 16, 20, 24, 36, 48, 60, and 72 h since initial blood feeding. Females that had not taken a blood meal were removed from the cages immediately following the blood meal and the remaining females that had taken a blood meal were held in the cages along with the males from the initial population. A total of 20 females were selected for each time point and all collections were repeated in triplicate. Total RNA was treated with DNase I using the DNA-Free kit from Ambion (Austin, TX) as previously described to remove any contaminant DNA. First strand cDNA was generated from the template RNA using the First strand cDNA synthesis kit from Roche (Indianapolis, IN) and an oligo dT primer. RT-qPCR was conducted on an ABI 7500 Real Time PCR system (Applied Biosystems) using the ABI SyBr Green mastermix kit (Life Technologies, Carlsbad, CA) and relative gene expression was determined by using the 2(-∆∆Ct) method 15 using a portion of the 18S rRNA gene as the reference gene and primers were designed according to each of specific gene sequences (Table S1).
Results
Determination of the pre-blood meal time period in Cx. quinquefasciatus
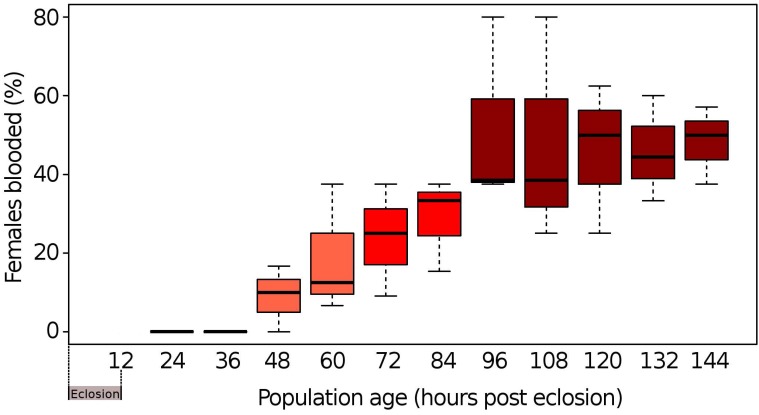
When newly eclosed female Cx. quinquefasciatus were offered a pre-warmed blood meal, our results showed that mosquitoes Cx. quinquefasciatus needed to be a minimum of 48 h old following eclosion to prepare the female for the first blood meal (Fig. 1). When mosquito populations had reached 96 h of post-eclosion, the average number of females taking a blood meal plateaued at ~50% with no observable increase in blood meal taking by females beyond this time point. This suggested that under our experimental conditions, the minimum pre-blood meal competency time period for females was ~48 h, after which females became competent to take a blood meal, reaching maximum blood meal acquisition at ~96 h post-eclosion.
Figure 1.
Time periods of pre and post blood-feeding of mosquitoes. Box and whisker plot of the percentage of females from even-aged populations of Cx. quinquefasciatus strain HAmCqG8 freely taking an offered blood meal. The black lines within a bar represent the median percentage of females who freely took a blood meal. The upper and lower whiskers represent the highest and lowest observations, respectively, while the bars themselves represent the interquartile range (Q1 - Q3).
Illumina RNA Seq data analysis of the genes during pre-blood meal and mating periods
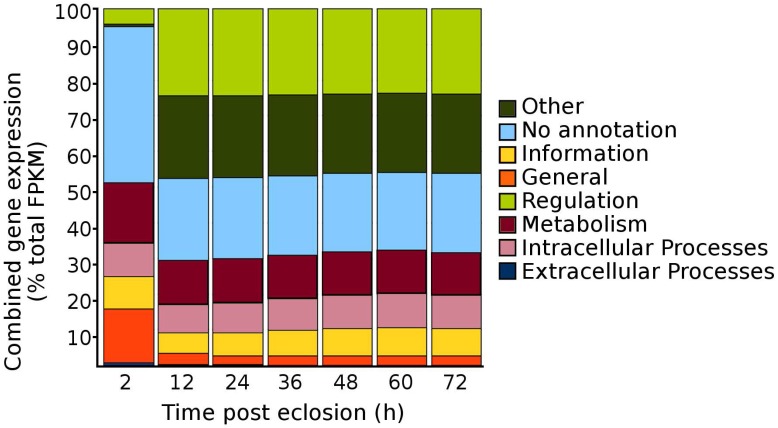
According to the pre-determination of the first blood meal time period, we conducted RNA Seq to characterize the genes that were involved in the taken of blood meal from mosquitoes. Seven time points of mosquitoes post eclosion, i.e., 2, 12, 24, 36 48, 60, and 72 h, were selected for the RNA Seq analysis, covering the time period from eclosion to the first sign of the blood feeding (i.e., by 48 h post-eclosion) and to reaching to the maximum mating period of Culex mosquitoes, i.e., Cx. quinquefasciatus has been shown to begin mating in 24 h after eclosion and reach a maximum by 72 h 2. A total of 200 females collected from each of the time points were pooled for the RNA extraction. Except for the 2 h time point, in which mosquito pupae were allowed to eclose over the 2 h period only, mosquito pupae at all-time points were allowed to eclose over the 12 h period and the females were collected at 12, 24, 36, 48, 60, and 72 h time points after eclosion. Overall, the depths of sequencing for the sample time points ranged from 26 to 51 million paired-end reads (Table 1) and after mapping the reads to the Cx. quinquefasciatus genome, the genes that were identified as expressed (i.e., those genes had an FPKM >1 14), were divided among the Structural Classification Of Proteins (SCOP) general function categories of metabolism, regulation, extra-cellular processes, intra-cellular processes, information, general, other, and no annotation 16-19. When the genes were sorted into their respective SCOP general function categories and their FPKM gene expression values were summed to estimate the total proportion of gene expression within each of the SCOP general function categories for each of the time points, the pattern revealed that there was an overall decrease in total gene expression among the “No annotation” and the “General” SCOP general function categories occurring from 2 to 12 h post-eclosion time points, with a respective increase in total gene expression for the categories of “regulation” and “other” for the 12 h time point (Fig. 2). Beyond the 12 h post-eclosion time point, the total cumulative gene expression profiles within each of the SCOP general function categories were similar up to the 72 h post-eclosion time point that had been tested (Fig. 2). These results suggested that the major global changes in gene expression of adult female Cx. quinquefasciatus during the initial 12 h post-eclosion may be involved in the post eclosion process as well as possibly in the preparation of the female for the taking of a blood meal.
Table 1.
Number of paired end reads from the Illumina HiSeq sequencing and the percentage of reads mapped to the Cx. quinquefasciatus (strain: Johannesburg) predicted transcriptome.
| Sampling time point* | Total paired end reads† | Paired end reads discarded‡ | Paired end reads used for mapping to the Cx. quinquefasciatus genome (JHB v1.2) |
|---|---|---|---|
| 2 h | 39890830 | 2950518 | 36940312 |
| 12 h | 36030651 | 2405121 | 33625530 |
| 24 h | 41173012 | 2979773 | 38193239 |
| 36 h | 35519459 | 1894955 | 33624504 |
| 48 h | 34587710 | 1909928 | 32677782 |
| 60 h | 27128981 | 1276065 | 25852916 |
| 72 h | 54895903 | 3819301 | 51076602 |
*mosquito sampling time point after eclosion
†Total number of FASTQ (DNA sequence with quality scores) reads passing the Illumina quality filter.
‡Number of reads discarded after adapter clipping.
Figure 2.
Total proportions of cumulative gene expression levels within the Structural Classification of Proteins (SCOP) general function categories for adult sugar-fed female Culex quinquefasciatus, strain HAmCqG8, for the initial 72 h post-eclosion. Gene expression values expressed are summed within each SCOP category to provide an overall profile of the complete distribution of all gene expression within the mosquitoes.
Transcriptome profile: differential gene expression
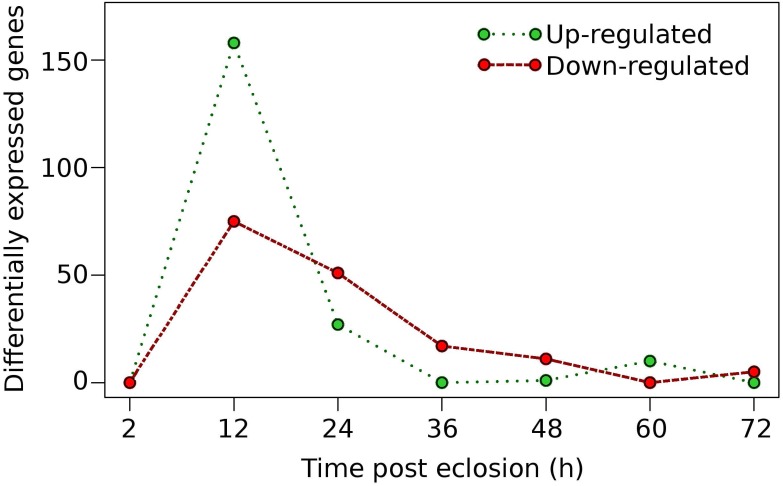
To further identify candidate genes that may be involved in preparing the female for the taking of a blood meal, we investigated the genes that were differentially-expressed throughout the time course investigated, i.e., 12, 24, 36, 48, 60, and 72 h time points after eclosion. Differential gene expression was determined as a time series, where gene differential expression was defined by comparing the expression levels of genes at any one time point with the expression at the prior time point; i.e., the expression at 24 h was compared with the expression at 12 h and so on. Overall, the majority of the genes that were identified as differentially up- and/or down-regulated occurred after 2 h post-eclosion and reached a maximum peak at 12 h, after which, a decrease in the number of differentially expressed genes was observed in each of the successive time points (Fig. 3, Table S2). A total of 325 genes were found to be differentially expressed, both up- and down-regulated genes, during 72 h post-eclosion. Roughly one-third of the differentially-expressed genes identified (101 genes) had no functional annotation in Vectorbase, while the remaining genes had predicted functions (Table S2). At 12 h post-eclosion, the greatest numbers of differentially-expressed genes was observed with 159 genes being up-regulated, and 74 down-regulated (Fig. 2). Among the up-regulated genes at 12 h post-eclosion were 32 salivary proteins and 2 apyrases, which may be involved in the prevention of platelet clotting during blood feeding 20, 10 cytochrome P450s (CPIJ005952, CPIJ011837, CPIJ010225, CPIJ010227, CPIJ000294, CPIJ019586, CPIJ019587, CPIJ020018, CPIJ012470, CPIJ010546 ), which were distributed among families CYP4, CYP6, CYP9, and CYP325, 20 proteases, and also genes involved in embryogenesis including wnt inhibitors and oskar 21, 22. Among the up-regulated genes at 24 h post-eclosion were 9 proteases and 1 olfactory receptor (CPIJ008023), which may be involved in preparation for the blood meal and host seeking, since both proteases and olfactory receptors have been linked to blood meal digestion and host seeking, respectively in mosquitoes 23-25. Among the genes down-regulated at 12 h post-eclosion were 27 hypothetical proteins, 11 cuticle proteins and 5 cytochrome P450 genes (CPIJ011841, CPIJ011840, CPIJ015954, CPIJ015961, and CPIJ015960), which were all in family CYP325. At 24 h post-eclosion, 32 hypothetical genes and 6 cuticle genes, which may be involved in the transition from the pharate to the adult, were down regulated.
Figure 3.
Distribution of differentially-expressed genes in adult sugar-fed female Culex quinquefasciatus for the initial 72h post-eclosion period.
While the majority of differential gene expression was observed in the earlier time points, there were a few genes that were up-regulated after 36 h post-eclosion (Fig. 3) with a total of 18, 12, 10, and 5 genes up-regulated for the 36 h, 48 h, 60 h, and 72 h time points, respectively (Table S2). Notably, at 60 h post-eclosion, multiple proteases were identified as up-regulated (CPIJ000990, CPIJ002595, CPIJ003539, and CPIJ007077) as well as ficolin-3, which has been linked to the humoral lectin immune defense response in mosquitoes 26. In addition, our results revealed that some genes were not differentially up- or down-regulated between time points, however the expression of some genes gradually increased in expression over time, notably the lipophorin receptor (CPIJ018375), the 60S protein rpL32 (CPIJ001220), the pro-epidermal growth factor gene (putative vitellogenin receptor) (CPIJ020278), and heavy-chain clathrin (CPIJ014882) (Table 2). Interestingly, work by Clifton and Noriega 27 revealed that the expression of some vitellogenesis-related transcripts, the ribosomal 60S protein rpL32, the lipophorin receptor AaLpRov, the vitellogenin receptor AaVgR, and heavy-chain clathrin (AaCHC) increased during the pre-blooding time period in female Ae. aegypti and that this was significantly influenced by the nutritional status of the female Ae. aegypti prior to the taking of a blood meal. Whether the constantly increased expression of the genes identified in our study following the time points identified was related to the nutrient supply (such as sugar water in our case) or involved in the regulation of development post eclosion or pre-blood feeding needs further investigation.
Table 2.
Expression levels of genes in Culex quinquefasciatus, strain HAmCqG8 for genes previously identified as up-regulated in non-blood-fed female Aedes aegypti and linked to nutritional status with regard to blood-feeding competency.
| Expression level (FPKM† at time point post-eclosion (h) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene number | Predicted function‡ | 2 | 12 | 24 | 36 | 48 | 60 | 72 |
| CPIJ001220 | 60S ribosomal protein L32 | 1100 | 1400 | 2200 | 3330 | 3600 | 4600 | 2900 |
| CPIJ014882 | clathrin heavy chain | 100 | 126 | 154 | 160 | 200 | 150 | 210 |
| CPIJ018375 | lipophorin receptor | 7 | 20 | 25 | 35 | 37 | 33 | 30 |
| CPIJ020278* | pro-epidermal growth factor | 3 | 12 | 93 | 130 | 190 | 200 | 240 |
†Fragments mapped Per Kilo bases of reference sequence for every Million fragments sequenced
‡Predicted function from Vectorbase, v. 1.2. https://www.vectorbase.org/organisms/culex-quinquefasciatus
*Putative vitellogenin receptor based on closest blastx match to Anopheles gambiae
qPCR validation of differential gene expression
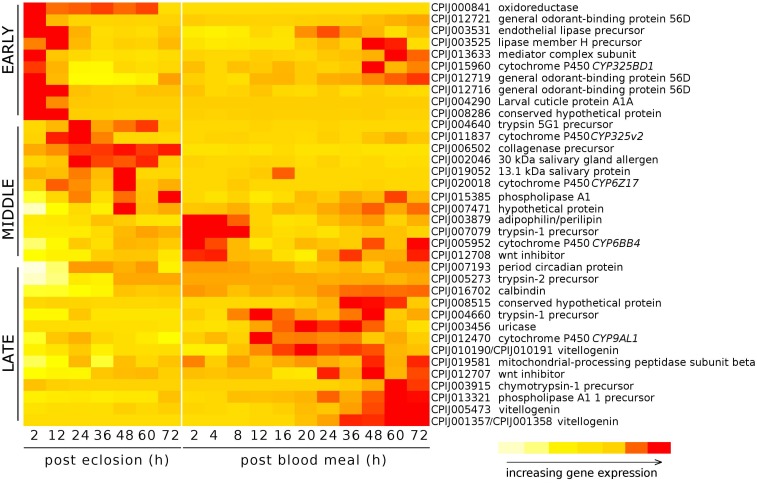
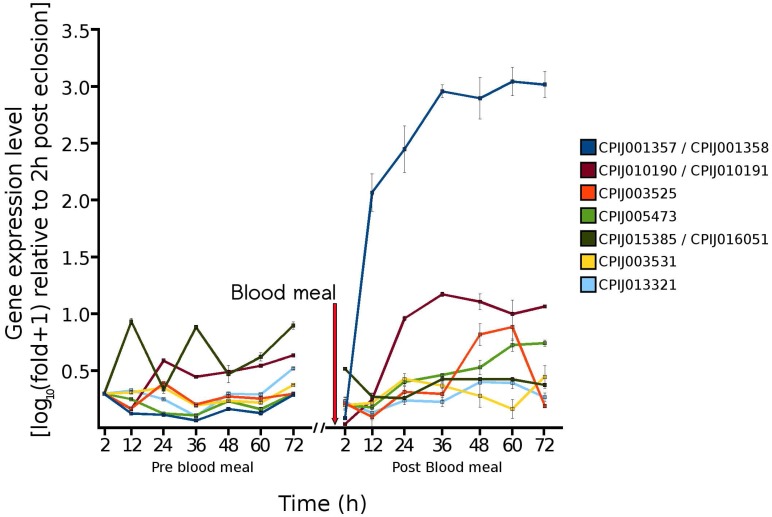
In order to estimate the accuracy of our RNA Seq results, and to determine the expression of selected genes throughout the pre blood-feeding and into the post blood-feeding time periods of Cx. quinquefasciatus, we selected a total of 36 genes from the RNA Seq data that showed the different levels of expression during 72 h post-eclosion time periods. The genes selected included genes that were involved in: host finding or feeding behavior (eg/ odorant-binding proteins) 23, 24, the maturation of the pharate female to the adult (cuticular proteins), the taking of the blood meal (salivary proteins) 28, the digestion of the blood meal (trypsins, collagenase, lipases, uricase) 29, 30, the provisioning of nutrients to the egg (vitellogenins, adipophilin/perilipin) 31, embryogenesis (wnt inhibitor, oskar) 32 as well as proteins with other functions involved in metabolism and development, including cytochrome P450s, calbindins, and oxidoreductases. The expression of these 36 genes was investigated from 2 to 72 h post-eclosion to be consistent with/validate our RNA Seq data. In addition, we further examined the expression of these 36 genes from 2 to 72 h post blood-feeding to evaluate the relationship of their expression changes following the blood feeding. Our qRT-PCR results showed that the expression of these genes was mostly consistent with the RNA Seq data - with similar traces of up- down-, or no change patterns during the different time points of 72 h after eclosion (Fig. 4). Our study showed that, according to the temporal expression, the genes can be grouped into early, middle, and late expression after eclosion and blood feeding. Our results also showed that all three general odorant-binding proteins (CPIJ012716, CPIJ012719, CPIJ012721) had their highest expression at 2 h post-eclosion except CPIJ012716 which also showed high expression at 12 h, indicating that females initially expressed the odorant-binding proteins to aid for food searching (sugar or blood). Other genes of interest were the salivary genes (CPIJ002046 and CPIJ019052) which reached maximal gene expression values at 24 and 48 h, respectively, and then remained at low expression levels afterwards (Fig. 4). Trypsin CPIJ007079 was up-regulated immediately following blood feeding, while trypsin CPIJ004660 and chymotrypsin CPIJ003915 were up-regulated at 48 h and 60 h post blood-feeding, respectively. These proteases are likely involved in the digestion of the blood meal, with the first gene representing early trypsin, and the two later two representing late trypsins. Another gene involved in the processing of the blood meal, CPIJ003456 (uricase) 30 showed the highest level of gene expression at 20 h post blood-feeding, which decreased by 60 h post blood-feeding. Among the genes that were predicted to be up-regulated throughout the post-eclosion and pre-blooding time period, the vitellogenin genes CPIJ001357/CPIJ001358 reached maximal gene expression at 60 h post blood-feeding, while the vitellogenin genes CPIJ010190/CPIJ010191 and CPIJ005473 reached maximal expression at 36 h post blood-feeding (Fig. 5). Following the blood meal, two trypsins (CPIJ007079, CPIJ004660) and one chymotrypsin (CPIJ003915) were up-regulated (Fig. 4).
Figure 4.
Heat map displaying the relative increases in gene expression for selected genes for the initial 72h post-eclosion for adult female Cx. quinquefasciatus, strain HAmCqG8, and for 72h post blood meal. Females offered a blood meal were 6 days old at the time of the blood feeding.
Figure 5.
Temporal gene expression of vitellogenin genes in adult female Cx. quinquefasciatus for the 72 h time period immediately following eclosion and for the 72 h time period immediately following the blood meal.
Discussion
Craig 33 indicated that males of Cx. quinquefasciatus needed a minimum of 24 h of post-eclosion development in order to mate and Williams and Patterson 2 also reported that mating reached a maximum by 72 h. Sebastian and DeMeillon 34 found that mating was a pre-requisite for blood feeding in the closely-related mosquito Culex pipiens fatigans and further identified a sex ratio of 2:1 (males:females) that was necessary to maximize insemination, while a sex ratio of 1:1 resulted in an insemination rate of only 83%. Baldini et al. 35 further pointed out that the impact of interactions between male and female factors during mating on the reproductive behavior and physiology of female mosquitoes. Since the sex ratio was held at ~1:1 in our study, this may explain why only 50% of the females took a blood meal 33 and under 1:1 sex ratio conditions tested, some females may have remained un-mated.
Our finding that the 72 h post-eclosion time point resulted in the maximum number of females being competent to take a blood meal suggested that mating may be involved in the preparation of the female for the taking of a blood meal, since mating would have occurred as early as 24 h post-eclosion 2. The predominance of the genes identified as differentially expressed, however, occurred between 2 and 24 h post-eclosion. These genes were likely involved in preparing the female for blood feeding since several of these genes could be attributed to genes that aid in the feeding and digestion of the blood meal, such as trypsins, apyrase, and salivary proteins 36-38, while genes having functions related to continued development after eclosion, such as cuticle structure were down-regulated 39. In addition, 1-day-old females (pre-mating period) have been shown to be capable of provisioning their eggs in the absence of a blood meal if they were exposed to the ecdysone agonist tebufenozide in the immature stages 40. This demonstrates that 24 h old Cx. quinquefasciatus females are capable of vitellogenesis if provided with the appropriate hormonal stimulation 40, thus the genes needed for the female to provision the eggs must be expressed by 24 h post-eclosion. Vitellogenesis and successful completion of egg formation in mosquitoes, however, requires not only ecdysone, but specific amino acids, juvenile hormone, and the activation of various signaling pathways and miRNAs as well 4, 5, 41-43. In addition, in Ae. aegypti, vitellogenesis has been shown to terminate if the nutritional status of the female is not sufficient for complete egg maturation, resulting in the re-absorption of vitellins 27. Therefore since female Cx. quinquefasciatus are capable of vitellogenesis as early as 24 h post-eclosion 40, but do not freely take a blood meal until at least 48 - 96 h post-eclosion, the genes that were identified as up-regulated at 48 and 60 h in our study may also be essential for preparing the female to take and process the blood meal, but not for vitellogenesis.
We further identified that several of the genes known to be involved in the processing of the blood meal in mosquitoes were either comparable to those in other mosquito species, or had later expression. For example, the uricase gene CPIJ003456 reached a maximal expression at 20 h post blood-feeding and decreased by 60 h post blood-feeding. A similar result had been identified by Isoe and Scaraffia 30 in Ae. aegypti, and suggested that uricase was primarily involved in ammonia metabolism as related to the processing of the blood meal. Other genes, such as the major vitellogenin genes CPIJ001357 and CPIJ001358 reached maximal expression at 36 h, which was delayed compared to the major vitellogenin gene in Ae. aegypti, which was shown to reach maximal expression at 24 h post blood-feeding, suggesting that vitellogenesis is a slower process in Cx. quinquefasciatus than in Ae. aegypti 31.
While our study suggested that there is likely a complex of factors necessary to prepare the female for the taking of a blood meal that include not only the control of vitellogenesis in Cx. quinquefasciatus, but the processing of the blood meal and preparation of the female for egg development and oviposition as well, further studies are needed to decipher these functions of these genes.
Supplementary Material
Tables S1 and S2.
Acknowledgments
The authors are grateful to Drs. Peter W. Atkinson, Peter Arensburger and the Culex quinquefasciatus genome community for the efforts they have devoted to determining the genome sequence and making the information available in VectorBase. We would also like to thank the Hudson Alpha Institute of Biotechnology for their expertise in conducting the RNA sequencing work and for all of their help and support with this study. This study was supported by an NIH grant (1R21AI076893) to N.L., AAES Hatch/Multistate Grants ALA08-045 to N.L., and ALA015-1-10026 to N.L.
Author Contributions
Performed the experiments: WRR. Analyzed the data: WRR NL LZ. Contributed reagents/materials/analysis tools: NL LZ. Wrote the paper: WRR NL LZ.
References
- 1.Cupp EW, Hassan HK, Yue X. et al. West Nile Virus infection in mosquitoes in the mid-south USA, 2002-2005. J Med Entomol. 2005;44:117–125. doi: 10.1603/0022-2585(2007)44[117:wnviim]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams FM, Patterson RS. Effect of age on mating of Culex pipiens quinquefasciatus. Florida Entomol. 1969;52:259–261. [Google Scholar]
- 3.Chen L, Zhu J, Sun G. et al. The early gene Broad is involved in the ecdysteroid hierarchy governing vitellogenesis of the mosquito Aedes aegypti. J Mol Endocrinol. 2004;33:743–761. doi: 10.1677/jme.1.01531. [DOI] [PubMed] [Google Scholar]
- 4.Hansen IA, Attardo GM, Roy SG. et al. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J Biol Chem. 2005;280:20565–20572. doi: 10.1074/jbc.M500712200. [DOI] [PubMed] [Google Scholar]
- 5.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107:22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price DP, Nagarajan V, Churbanov A. et al. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post-blood meal. PLoS ONE. 2011 doi: 10.1371/journal.pone.0022573. doi:10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marinotti O, Calvo E, Nguyen QK. et al. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Cupp EW, Micher KM. et al. Insecticide resistance and cross-resistance in Alabama and Florida strains of Culex quinquefasciatus (S.) J Med Entomol. 2004;41:408–413. doi: 10.1603/0022-2585-41.3.408. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Lohse M, Bolger AM, Nagel A. et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Megy K, Hammond M, Lawson D. et al. Genomic resources for invertebrate vectors of human pathogens, and the role of VectorBase. Infection Genet Evol. 2009;9:308–313. doi: 10.1016/j.meegid.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts A, Trapnell C, Donaghey J. et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:doi. doi: 10.1186/gb-2011-12-3-r22. 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan Q, Schones DE, Eun SH, Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol; 2010. doi:10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Murzin AG, Brenner SE, Hubbard T. et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 17.Andreeva A, Howorth D, Brenner SE. et al. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel C, Berzuini C, Bashton M. et al. Supra-domains - evolutionary units larger than single protein domains. J Mol Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Vogel C, Teichmann SA, Pereira-Leal JB. The relationship between domain duplication and recombination. J Mol Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 20.Smith TM, Hicks-Berger CA, Kim S. et al. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotides. Arch Biochem Biophys. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- 21.Chang CW, Nashchekin D, Wheatley L. et al. Anterior-posterior axis specification in Drosophila oocytes: identification of novel bicoid and oskar mRNA localization factors. Genetics. 2011;188:883–896. doi: 10.1534/genetics.111.129312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaglarz MK, Kloc M, Jankowska W. et al. Nuage morphogenesis becomes more complex: two translocation pathways and two forms of nuage coexist in Drosophila germline syncytia. Cell Tissue Res. 2011;344:169–181. doi: 10.1007/s00441-011-1145-2. [DOI] [PubMed] [Google Scholar]
- 23.Fox AN, Pitts RJ, Robertson HM. et al. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci USA. 2001;98:14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc Natl Acad Sci USA. 2009;106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isoe J, Rascon Jr AA, Kunz S. et al. Molecular genetic analysis of midgut serine proteases in Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2009;39:903. doi: 10.1016/j.ibmb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimopoulos G, Christophides GK, Meister S. et al. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clifton ME, Noriega FG. The fate of follicles after a blood meal is dependent on previtellogenic nutrition and juvenile hormone in Aedes aegypti. J Insect Physiol. 2012;58:1007–1019. doi: 10.1016/j.jinsphys.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvo E, Sanchez-Vargas I, Favreau AJ. et al. An insight into the sialotranscriptome of the West Nile mosquito vector, Culex tarsalis. BMC Genomics. 2010;11:51. doi: 10.1186/1471-2164-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noriega FG, Shah D, Wells MA. Juvenile hormone regulates early trypsin gene expression. Insect Mol Biol. 1997;6:63–66. doi: 10.1046/j.1365-2583.1997.00154.x. [DOI] [PubMed] [Google Scholar]
- 30.Isoe J, Scaraffia PY. Urea synthesis and excretion in Aedes aegypti mosquitoes are regulated by a unique cross-talk mechanism. PLoS ONE. 2013;8:e65393. doi: 10.1371/journal.pone.0065393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isoe J, Hagedorn HH. Mosquito vitellogenin genes: comparative sequence analysis, gene duplication, and the role of rare synonymous codon usage in regulating expression. J Insect Sci. 2007;7:1–49. doi: 10.1673/031.007.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juhn J, James AA. Oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Mol Biol. 2006;15:363–372. doi: 10.1111/j.1365-2583.2006.00655.x. [DOI] [PubMed] [Google Scholar]
- 33.Craig GB. Mosquitoes: females monogamy induced by male accessory gland substance. Science. 1967;156:1499–1501. doi: 10.1126/science.156.3781.1499. [DOI] [PubMed] [Google Scholar]
- 34.Sebastian A, Meillon BD. Experiments on the mating of Culex pipiens fatigans in the laboratory. Bull Org Mond Sante & Bull Wld Hlth Org. 1967;36:47–52. [PMC free article] [PubMed] [Google Scholar]
- 35.Baldini F, Gabrieli P, South A. et al. The interaction between a sexually transferred steroid hormone and a female protein regulates oogenesis in the malaria mosquito Anopheles gambiae. PLoS Biol. 2013 doi: 10.1371/journal.pbio.1001695. doi:10.1371/journal.pbio.1001695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barillas-Mury CV, Noriega FG, Wells MA. Early trypsin activity is part of the signal transduction system that activates transcription of the late trypsin gene in the midgut of the mosquito Aedes aegypti. Insect Biochem Mol Biol. 1995;25:241–246. doi: 10.1016/0965-1748(94)00061-l. [DOI] [PubMed] [Google Scholar]
- 37.Chamopagne DE, Smart CT, Ribeiro JMC. et al. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5'-nucleotidase family. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631.. doi: 10.1371/journal.ppat.1002631. doi:10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riehle MA, Garczynski SF, Crim JW. et al. Neuropeptides and peptide hormones in Anopheles gambiae. Science. 2002;298:172–175. doi: 10.1126/science.1076827. [DOI] [PubMed] [Google Scholar]
- 40.Gelbič C, Rozsypalová P. Autogeny induced by a nonsteroidal ecdysone agonist tebufenozide in the mosquito Culex quinquefasciatus. Physiol Entomol. 2012;37:19–126. [Google Scholar]
- 41.Hansen IA, Attardo GM, Park JH. et al. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc Natl Acad Sci USA. 2004;101:10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiao SH, Hansen IA, Zhu J. et al. Juvenile hormone connects larval nutrition with target of rapamycin signaling in the mosquito Aedes aegypti. J Insect Physiol. 2008;54:231–239. doi: 10.1016/j.jinsphys.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulia-nuss M, Robertson AE, Brown MR. et al. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE. 2011 doi: 10.1371/journal.pone.0020401. doi:10.1371/journal.pone.0020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2.