Abstract
Notch signaling has well-defined roles in the assembly of arterial walls and in the development of the endothelium and smooth muscle of the vasculature. Hippo signaling regulates cellular growth in many tissues, and contributes to regulation of organ size, in addition to other functions. Here, we show that the Notch and Hippo pathways converge to regulate smooth muscle differentiation of the neural crest, which is crucial for normal development of the aortic arch arteries and cranial vasculature during embryonic development. Neural crest-specific deletion of the Hippo effectors Yap and Taz produces neural crest precursors that migrate normally, but fail to produce vascular smooth muscle, and Notch target genes such as Jagged1 fail to activate normally. We show that Yap is normally recruited to a tissue-specific Jagged1 enhancer by directly interacting with the Notch intracellular domain (NICD). The Yap-NICD complex is recruited to chromatin by the DNA-binding protein Rbp-J in a Tead-independent fashion. Thus, Hippo signaling can modulate Notch signaling outputs, and components of the Hippo and Notch pathways physically interact. Convergence of Hippo and Notch pathways by the mechanisms described here might be relevant for the function of these signaling cascades in many tissues and in diseases such as cancer.
KEY WORDS: Notch signaling, Hippo signaling, Yap, Taz, Jagged1, Vascular development, Neural crest, Mouse
Highlighted article: Direct interaction between the Hippo effector Yap and the Notch intracellular domain regulates Notch target gene expression during vascular smooth muscle differentiation from neural crest.
INTRODUCTION
The neural crest is a transient, migratory, multipotent cell population that contributes to diverse cell lineages in the developing embryo, including vascular smooth muscle. The differentiation of neural crest into vascular smooth muscle is dependent upon Notch signaling (High and Epstein, 2008). Normally, expression of the Notch ligand Jagged1 by vascular endothelium induces Notch activation in adjacent mesenchyme, resulting in smooth muscle differentiation and transcription of Notch target genes, including Jagged1 itself. Jagged1 can then activate successive layers of neural crest to differentiate into smooth muscle, producing a multi-layered vascular wall (Manderfield et al., 2012). Interruption of this Notch-mediated lateral induction pathway, either by genetic deletion of Jagged1 in endothelial cells or neural crest, or by inhibition of Notch signaling in neural crest, results in an array of aortic arch artery and smooth muscle defects (High et al., 2008, 2007).
Hippo signaling is a highly conserved kinase cascade, classically thought to regulate organ size, although its role in development and disease is rapidly expanding. The upstream Hippo kinases Mst1 and Mst2 phosphorylate kinases Lats1 and Lats2. Lats1 and Lats2 then phosphorylate the downstream effector molecules of Hippo signaling, Yap (Yap1 – Mouse Genome Informatics) and Taz. When phosphorylated, Yap and Taz are removed from the nucleus, thereby terminating transcription of Hippo target genes. Yap and Taz have no known intrinsic DNA-binding capabilities and therefore require interaction with a DNA-binding molecule to mediate their functions as co-activators. In canonical mammalian Hippo signaling, the DNA-binding moiety is one of four homologous Tead factors, but Yap and Taz can associate with numerous other transcription factors, including Pax proteins, Tbx5 and p63/p73 (Tcp1/Iap1 – Mouse Genome Informatics) (Manderfield et al., 2014; Murakami et al., 2005; Strano et al., 2001). The Hippo signaling cascade is therefore poised to integrate and modulate multiple developmental and homeostatic regulatory cascades.
Hippo signaling is crucial for vascular smooth muscle repair and development. For example, Yap expression is significantly increased in smooth muscle cells following carotid artery injury, where it promotes smooth muscle proliferation and migration (Wang et al., 2012). Although Yap and Taz are thought to mediate largely redundant functions in many tissues, deletion of Yap in developing smooth muscle is sufficient to disrupt smooth muscle formation, producing thin arterial walls and enlarged vessel lumens in the left carotid and thoracic arteries (Wang et al., 2014). Yap might function with Tead factors in addition to other transcriptional regulators, including myocardin, to regulate smooth muscle gene expression (Wang et al., 2014; Xie et al., 2012).
Given the diversity and breadth of cellular contexts in which Notch and Hippo signaling function in development and disease, it is not surprising that these two signaling pathways frequently intersect (High and Epstein, 2008; Heallen et al., 2011; von Gise et al., 2012; Afelik and Jensen, 2013; Gao et al., 2013; Li et al., 2009; Makita et al., 2008). For example, hepatocyte-specific Yap overexpression leads to an upregulation of Notch1, Notch2, Jagged1 (Jag1) and the Notch target gene Hes1 (Yimlamai et al., 2014). Evidence suggests that Notch2 is a direct, Tead-dependent Hippo target (Yimlamai et al., 2014; Tschaharganeh et al., 2013). Cdx2, a transcriptional regulator of blastocyst lineage restriction, is regulated by the convergence of both Notch and Hippo signaling on a single enhancer element (Rayon et al., 2014). Genome-wide ChIP-seq for Rbp-J, the DNA-binding mediator of Notch signaling, from neuronal stem cells demonstrates Rbp-J binding throughout the Yap and Tead2 loci, and that transgenic overexpression of the Notch intracellular domain (NICD) leads to increased Yap and Tead2 expression (Li et al., 2012). In Drosophila wing imaginal disks, a complex interaction between Notch and Hippo signaling has been elucidated; Notch can inhibit Yorkie (Yap)- Scalloped (Tead) complexes in a fashion dependent upon the specific ratios of Notch and Hippo signaling components (Djiane et al., 2014).
In this manuscript, we present evidence to support an additional layer of complexity in the convergence of Notch and Hippo signaling. We show that loss of Yap and Taz in neural crest abrogates Notch signaling and smooth muscle differentiation. Notch directly activates expression of Jagged1 in neural crest (Manderfield et al., 2012). Here, we show that the Hes1 promoter and a conserved Jagged1 enhancer are co-activated by NICD/Rbp-J and Yap in a Tead-independent fashion. Further, we demonstrate that Yap and NICD physically interact. The convergence of Notch and Hippo signaling on a common transcriptional complex might have relevance to the understanding of how these pathways co-regulate many aspects of organogenesis, regeneration and cancer.
RESULTS
Using Wnt1-Cre, we have previously shown that genetic deletion of Yap and Taz in premigratory neural crest results in embryonic lethality associated with craniofacial defects and vascular hemorrhages (Manderfield et al., 2014). This is a more dramatic phenotype than the previously reported Yap deletion in smooth muscle, which resulted in only arterial dilation (Wang et al., 2014). Further examination of E10.5 embryos in which Yap and Taz have both been deleted in neural crest, and that also express a Td/tomato reporter allele, demonstrates normal migration and patterning of cardiac neural crest (Fig. 1; supplementary material Fig. S1). Neural crest derivatives surround sections of the brachial arch arteries and the outflow tract of the heart in both control and mutant embryos. However, smooth muscle differentiation of neural crest, as determined by expression of SM22α (Tagln – Mouse Genome Informatics) (Fig. 1), SMA (Acta2 – Mouse Genome Informatics), desmin (Des) or smooth muscle myosin (Fig. 2), is absent in mutant embryos. This defect is in striking contrast to the ability of non-neural crest-derived mesenchyme, which forms some of the vascular smooth muscle of the third aortic arch artery, to differentiate normally (Figs 1 and 2). Both neural crest and non-neural crest mesenchyme surrounding the third aortic arch artery express nuclear Yap, with low levels of phosphorylated Yap (pYap) largely confined to the inner endothelial layer (Fig. 3).
Fig. 1.
Deletion of Yap and Taz in neural crest results in impaired smooth muscle differentiation. (A) Wnt1-Cre; Taz flox/+;Yap flox/+; R26Tom/+ E10.5 embryo imaged in bright-field and fluorescence. (B-H) Transverse sections of E10.5 Wnt1-Cre; Tazflox/+;Yapflox/+; R26Tom/+ embryos stained for tdTomato (RFP) and SM22α. (I) Wnt1-Cre; Tazflox/flox;Yapflox/flox;R26Tom/+ embryo imaged in bright-field and fluorescence. (J-P) Transverse sections of E10.5 Wnt1-Cre; Tazflox/flox;Yapflox/flox;R26Tom/+ embryos stained for tdTomato (RFP) and SM22α. Arrowheads denote sites of decreased smooth muscle differentiation. Branchial arches (ba) are invested with red-fluorescing Wnt1-derived neural crest (A,I). iii, third aortic arch artery; Ao, aortic sac. Images in B,E,H,J,M,P were merged by combining respective red and green channels using Photoshop software (Adobe). Scale bars: 100 μm.
Fig. 2.

Deletion of Yap and Taz in neural crest results in impaired smooth muscle expression. (A-I) Transverse sections of E10.5 Wnt1-Cre; Tazflox/+;Yapflox/+; R26Tom/+ embryos stained for tdTomato (RFP, A), SMA (B), merged RFP/SMA (C), tdTomato (RFP, D), desmin (E) or merged RFP/desmin (F). Yellow arrows denote RFP/desmin double-positive cells. Serial sections stained for tdTomato and Hoechst (RFP, G), smMyosin (H) or smMyosin and Hoechst (I). White arrows denote presumptive RFP/smMyosin double-positive cells. (J-R) Transverse sections of E10.5 Wnt1-Cre; Tazflox/flox;Yapflox/flox;R26Tom/+ embryos stained for tdTomato (RFP, J), SMA (K), merged RFP/SMA (L), tdTomato (RFP, M), desmin (N) or merged RFP/desmin (O). Serial sections stained for tdTomato and Hoechst (RFP, P), smMyosin (Q) or smMyosin and Hoechst (R). White arrows highlight RFP–, smMyosin+ cells. Merged images were generated by combining respective red and green channels using Photoshop (C,F,L,O). iii, third aortic arch artery. Scale bars: 100 μm.
Fig. 3.

Mesenchyme adjacent the third aortic arch artery expresses Yap. (A-E) Transverse sections of wild-type E10.5 embryos stained for Yap (A), Yap, eNOS and Hoechst (B), phospho-Yap (pYap, C), pYap, eNOS and Hoechst (D), pYap and Hoechst (E) or eNOS and Hoechst (F). White arrows denote pYap/eNOS double-positive cells. The boxed area in D is shown at higher magnification with single stains in E,F. iii, third aortic arch artery. Scale bars: 100 μm in A-D; 20 μm in E,F.
Equivalent numbers of migratory RFP+ control or RFP+ Yap/Taz null neural crest cells surround the third arch artery (118.9±12.1 control versus 121.0±13.9 null. (See Materials and Methods for quantification methods.) Both samples exhibit similar proliferation rates (2.6% control versus 1.9% null; supplementary material Fig. S2), supporting the notion that the observed smooth muscle phenotype is a result of impaired differentiation.
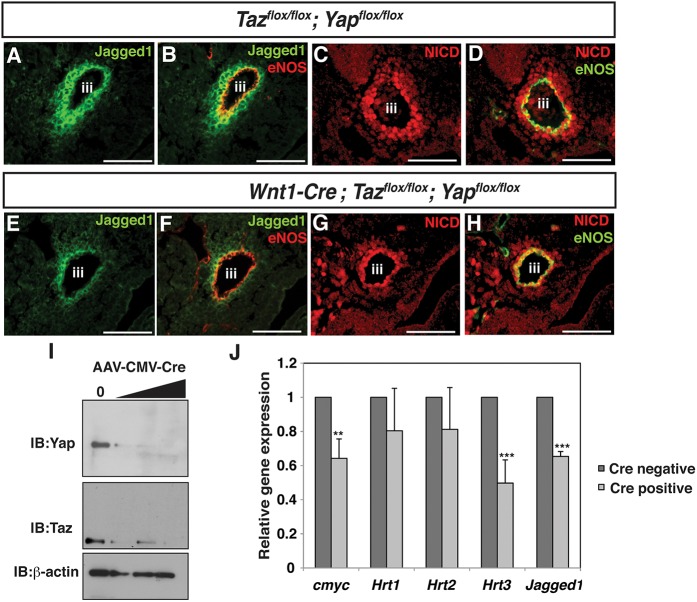
Notch activation and Jagged1 expression are required for proper differentiation of neural crest into smooth muscle and for expression of SM22α and SMA (High et al., 2008, 2007). Loss of Yap/Taz in neural crest results in a dramatic decrease in Jagged1 and NICD expression in mesenchyme surrounding the third aortic arch artery in a region normally populated by neural crest, while endothelial NICD expression is intact (Fig. 4A-D,E-H).
Fig. 4.
Decreased Notch activity following Yap/Taz deletion. (A-D) Transverse sections of E10.5 Tazflox/flox;Yapflox/flox embryos immunostained for Jagged1 (A), Jagged1 with eNOS (B), NICD (C) or NICD with eNOS (D). (E-H) Transverse sections of E10.5 Wnt1-Cre; Tazflox/flox;Yapflox/flox embryos immunostained for Jagged1 (E), Jagged1 with eNOS (F), NICD (G) or NICD with eNOS (H). (I) Anti-Yap and anti-Taz immunoblots from Tazflox/flox;Yapflox/flox MEFs untreated (0) or treated with increasing AAV-CMV-Cre doses [500 genome copies per cell (GC), 1000 GC or 2000 GC]. In parallel, immunoblots of the same protein lysates were probed with anti-actin to demonstrate equivalent protein loading. (J) qRT-PCR of untreated or AAV-CMV-Cre virus-treated (1000 GC) Tazflox/flox;Yapflox/flox MEFs for c-myc, Hrt1, Hrt2, Hrt3 and Jagged1. Data depicted in J are mean+s.e.m. Statistics were completed using Student's t-test. **P<0.01, ***P<0.001. iii, third aortic arch artery. Scale bars: 100 μm.
In order to explore further the effects on Notch signaling resulting from loss of Yap/Taz, we generated murine embryonic fibroblasts (MEFs) from Tazflox/flox;Yapflox/flox embryos. Treatment with adenovirus expressing Cre recombinase results in efficient deletion of Yap and Taz protein (Fig. 4I). In MEFs, deletion of Yap and Taz produces a significant decrease in both Jagged1 and Hrt3 (Heyl – Mouse Genome Informatics) expression. c-myc (Myc), a known Notch and Hippo target (Weng et al., 2006; Neto-Silva et al., 2010), is also significantly decreased (Fig. 4J).
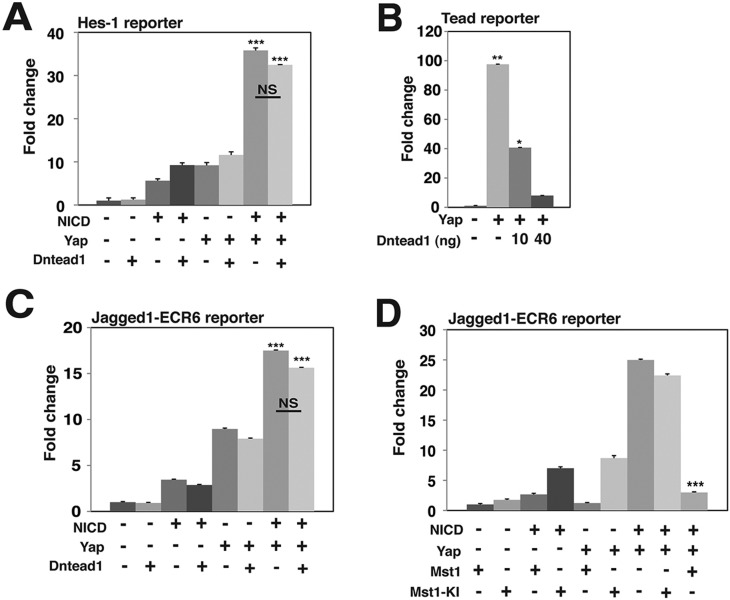
Hes1 is a canonical, direct target of Notch signaling, and an Rbp-J-dependent Notch-responsive Hes1 regulatory element located −194 to +160 relative to the Hes1 transcriptional start site has been previously characterized (Jarriault et al., 1995). We verified that NICD activates a reporter construct containing this Hes1 enhancer (Fig. 5A). Interestingly, NICD activation of this Hes1 reporter is significantly enhanced by the addition of Yap (Fig. 5A). Surprisingly, the activation induced by Yap and NICD is unaffected by co-transfection of a dominant-negative form of Tead (Dntead1) (Fig. 5A), although Dntead1 is able to potently inhibit Yap activation of a control reporter construct containing Tead binding sites (Fig. 5B). Previously, we identified a 617-bp conserved, Notch-responsive intronic enhancer that regulates Jagged1 expression in cardiac neural crest, which we have termed Jagged1-ECR6 (Manderfield et al., 2012). As previously reported, NICD activates a reporter construct containing Jagged1-ECR6 upstream of a synthetic minimal promoter driving luciferase expression. Interestingly, Yap is also able to activate this reporter, and the combination of NICD and Yap generates significantly more activity than either NICD or Yap alone (Fig. 5C). As with Hes1, ECR6 activation induced by Yap, or Yap with NICD, is unaffected by Dntead1 (Fig. 5C). This result suggests that Yap-mediated activation of ECR6 is Tead-independent. Yap/Taz activity is regulated, at least in part, by the Hippo kinase cascade (Mst and Lats), which ultimately control Yap/Taz nuclear localization. As expected, co-transfection of a construct encoding Mst1 abrogates the ability of Yap to activate ECR6, either alone or in combination with NICD (Fig. 5D). A kinase-inactive form of Mst1 failed to inhibit Yap-mediated activation.
Fig. 5.
NICD transcriptional activity is increased in the presence of Yap and the activation is Tead1 independent. Results of dual luciferase reporter assays in HEK293T cells are shown. (A) Hes1 reporter assay in the presence (+) or absence (−) of NICD, Yap or Dntead1, n=3. Complete ANOVA results are included in supplementary material Table S1. (B) 8× GTIIC-Tead-reporter luciferase assay in the presence (+) or absence (−) of Yap or Dntead1, n=3. (C) Jagged1 enhancer element (ECR6)-luciferase reporter assay in the presence (+) or absence (−) of NICD, Yap or Dntead1, n=3. Complete ANOVA results are included in supplementary material Table S2. (D) ECR6-luciferase reporter assay in the presence (+) or absence (−) of NICD, Yap, Mst1 or a kinase-inactive form of Mst1, Mst1-KI, n=4. Complete ANOVA results are included in supplementary material Table S3. All experiments were performed in duplicate for a minimum of three individual occasions, with specific replicate numbers shown in each legend. Data depicted are mean+s.e.m. Statistics were completed using ANOVA with a Tukey–Kramer post-hoc comparison test. ***P<0.001, **P<0.01, *P<0.05.
Chromatin immunoprecipitation (ChIP) experiments suggest that both NICD and Yap occupy ECR6 (Fig. 6A). ChIP with antibodies specific for NICD or Yap produce ∼10-fold enrichment of ECR6 compared with control IgG. No enrichment was detected for a distant upstream Jagged1 conserved putative enhancer, previously termed ECR1, which is 456 bp and not activated by Notch (Manderfield et al., 2012). Importantly, both NICD and Yap occupancy are completely abrogated by mutation of the single Rbp-J binding site located within ECR6, denoted ECR6* (Manderfield et al., 2012). This result suggests that Rbp-J binding is required for recruitment of both NICD and Yap to the Jagged1 enhancer. Accordingly, the presence of the Rbp-J mutation prevented NICD-Yap co-activation of ECR6 in a luciferase assay (Fig. 6B). Co-immunoprecipitation (Co-IP) experiments revealed that NICD and Yap can physically interact (Fig. 6C), supporting the idea that Yap can function as a Notch co-activator.
Fig. 6.
NICD and Yap specifically co-occupy Jagged1 ECR6 and physically interact. (A) Chromatin immunoprecipitation (ChIP) for NICD (FLAG) or Yap from cells transfected with a 3× FLAG-tagged NICD construct, a Yap expression construct and either an ECR1 expression plasmid, an ECR6 expression plasmid or a mutant ECR6 expression plasmid (ECR6*). Data are reported as fold enrichment over an IgG ChIP performed in parallel with the same samples. (B) Results of dual luciferase reporter assays in HEK293T cells with a Jagged1 enhancer element (ECR6), using either a wild-type ECR6-luciferase reporter or a mutant ECR6-luciferase reporter (ECR6* reporter) with a mutated Rbp-J binding site in the presence (+) or absence (−) of NICD or Yap, n=3. (C) Western blots demonstrating co-immunoprecipitation of Yap and NICD. Lysates were immunoprecipitated with either a control IgG or FLAG antibody in the presence (+) or absence (−) of Yap and NICD. The Yap construct contains an N-terminal FLAG epitope tag. The NICD construct contains a C-terminal V5 epitope tag. Input immunoblots confirmed NICD (V5) and Yap (FLAG) expression in specified samples. β-actin immunoblot confirmed protein expression in all samples. (D) ChIP for endogenous Yap from non-transfected MDA-MB-231 cells at Jagged1 genomic loci, ECR1 and ECR6, and the Hes1 promoter. Data are reported as fold enrichment over an IgG ChIP performed in parallel with the same samples. Dual luciferase experiments were performed in duplicate for a minimum of three individual occasions. ChIP experiments were also completed in biological triplicate. Data depicted are mean+s.e.m. Statistics were completed using ANOVA with a Tukey–Kramer post-hoc comparison test. ***P<0.001, **P<0.01, *P<0.05.
MDA-MB-231 breast carcinoma cells express NICD and Yap (Yu et al., 2013; Stylianou et al., 2006). ChIP for endogenous Yap in these cells revealed co-occupancy of the endogenous Hes1 promoter and Jagged1 ECR6 enhancer, but again not of the adjacent genomic region denoted by ECR1 that is not regulated by Notch (Fig. 6D).
We next examined the role of the NICD-Yap complex in MOVAS cells, a mouse aortic smooth muscle cell line. RT-PCR analysis confirmed that this cell line expresses the NICD-Yap target genes Jag1 and Hes1, and smooth muscle genes, including Tagln, Acta2, Des1 (Des – Mouse Genome Informatics) and Cnn1 (Fig. 7A). Treatment with the γ-secretase inhibitor DAPT (100 μM) resulted in a dramatic decrease in Jag1 and Hes1 expression, consistent with both genes being Notch signaling targets (Fig. 7B). Overexpression of the upstream Hippo kinase Lats2 led to a robust increase in Yap phosphorylation (pYap) (Fig. 7C), which will remove Yap from the nucleus. The presence of Lats2 inhibited Jag1 and Hes1 expression as well as expression of smooth muscle genes Acta2, Cnn1 and Des1 (Fig. 7D). Hrt1 and Hrt2, two genes unchanged following Yap/Taz deletion in MEFs (Fig. 4J), were similarly unchanged following Lats2 expression in MOVAS cells (Fig. 7D). Co-IP experiments confirm that NICD and Yap can physically interact in MOVAS smooth muscle cells (Fig. 7E). ChIP with native antibodies specific for endogenous Yap or NICD demonstrate specific recruitment and enhanced occupancy to both the endogenous ECR6 enhancer and the endogenous Hes1 promoter compared with ECR1, a distant upstream Jagged1 conserved region (Fig. 7F).
Fig. 7.
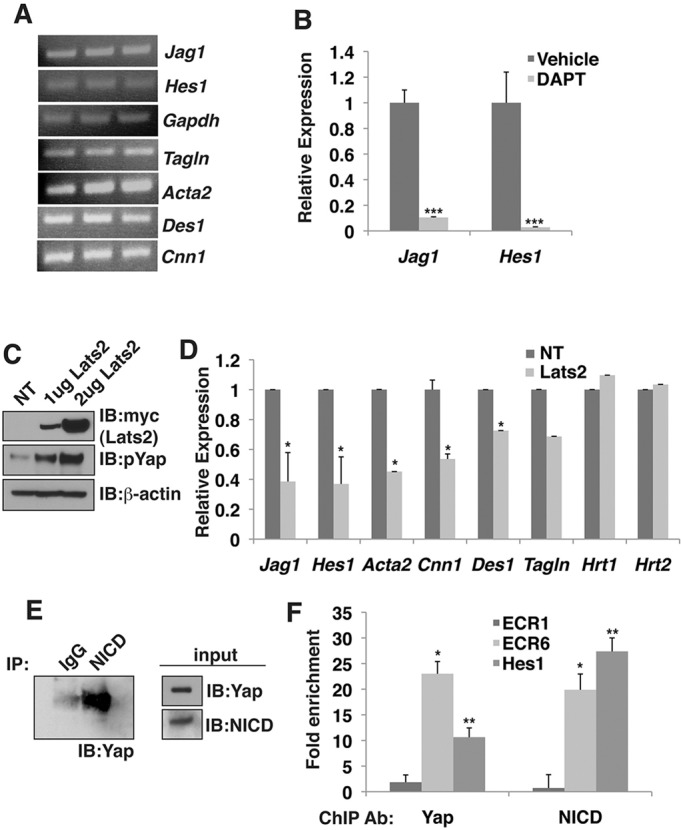
NICD and Yap interact and modulate transcription in smooth muscle cells. (A) RT-PCR from three biological replicates of MOVAS cells. (B) qRT-PCR of vehicle (DMSO)- or DAPT (100 µM)-treated MOVAS cells. Data depicted are mean+s.e.m. and the statistics were completed using Student's t-test; ***P<0.001. (C) Anti-myc (Lats2) and anti-pYap immunoblots from MOVAS cells transfected with increasing amounts of myc-tagged Lats2. In parallel, immunoblots of the same protein lysates were probed with anti-actin to demonstrate equivalent protein loading. (D) qRT-PCR of non-transfected (NT) or Lats2 (2 µg)-transfected MOVAS cells. Data depicted are mean+s.e.m. and the statistics were completed using Student's t-test; *P<0.05. (E) Western blot demonstrating co-immunoprecipitation of Yap and NICD in MOVAS cells. Lysates were immunoprecipitated with either a control IgG or NICD antibody. Input immunoblots confirmed NICD and Yap expression. (F) ChIP for endogenous Yap and NICD in non-transfected MOVAS cells at Jagged1 genomic loci, ECR1 and ECR6, and the Hes1 promoter. Data are reported as fold enrichment over an IgG ChIP performed in parallel with the same samples. Data depicted are mean+s.e.m. The statistics were completed using Student's t-test; **P<0.01, *P<0.05. qRT-PCR and ChIP experiments were completed in biological triplicate.
The Yap protein contains well-defined structural domains, including an N-terminal Tead-binding domain, a C-terminal transactivation domain and two WW domains (Varelas, 2014). WW domains mediate protein-protein interactions with proline-rich or proline-containing motifs. High-affinity binding occurs with proteins containing a PPxY motif, and lower-affinity interactions have been identified with proteins containing PPLP or PPR motifs (where P is a proline residue, Y is a tryptophan residue, L is an leucine residue, R is an arginine residue and x stands for any amino acid) (Varelas, 2014; Russ et al., 2005). Yap interaction with Tead factors is mediated by the N-terminal Tead interaction domain, but Yap can interact with other DNA-binding proteins in a manner dependent upon the WW domains (Zhao et al., 2009). Murine NICD1 has amino acid sequences similar to two of the lower-affinity proline-containing motifs, PPLLP and PPPPR. Mutation of the first Yap WW domain, but not the second WW domain, prevented Yap from augmenting NICD induction of Jagged1-ECR6 (Fig. 8A). As previously reported, none of the Yap WW domain mutants prevented activation of a Tead-dependent reporter (Fig. 8B) (Zhao et al., 2009), and immunoblot analysis confirmed that wild-type and mutant Yap constructs were expressed at similar protein levels (Fig. 8C). We confirmed that NICD could interact with Yap by using the Duolink proximity ligation assay to monitor protein-protein interactions in situ and found that mutation of the first Yap WW domain, but not of the second WW domain, prevented Yap interaction with NICD (Fig. 8D). Taken together, these results suggest that Yap can interact with NICD by utilizing the first WW domain, and that Yap and NICD can be recruited to the Jagged1 enhancer by the DNA-binding protein Rbp-J.
Fig. 8.
NICD-Yap interaction requires the first Yap WW domain. (A) Results of dual luciferase reporter assays in HEK293T cells with an ECR6-luciferase reporter in the presence (+) or absence (−) of NICD, Yap or Yap mutants, Yap-WW1, Yap-WW2 or Yap-WW1WW2, n=4. Complete ANOVA results are included in supplementary material Table S4. (B) Results of dual luciferase reporter assays in HEK293T cells with 8× GTIIC-Tead luciferase reporter in the presence (+) or absence (−) of Yap or Yap mutants, Yap-WW1, Yap-WW2 or Yap-WW1WW2, n=3. (C) Anti-FLAG and anti-Yap immunoblots from HEK293T cells transfected with FLAG-tagged Yap or FLAG-tagged Yap mutants. (D) Examination of NICD-Yap interaction using Duolink reagents. Interaction is documented as red fluorescence. Samples were co-transfected in the presence (+) or absence (−) of the specified plasmids. NICD-V5+Rbp-J served as positive control. The samples in the bottom row were not transfected with NICD-V5, but did include the V5 antibody to serve as negative controls. Scale bars: 100 µm. Dual luciferase experiments were performed in duplicate for a minimum of three individual occasions. Data depicted are mean+s.e.m. Statistics were completed using ANOVA with a Tukey–Kramer post-hoc comparison test. ***P<0.001, **P<0.01.
DISCUSSION
In this study, we demonstrate a crucial role for Hippo signaling in the differentiation of neural crest-derived smooth muscle. We show that deletion of the Hippo effector molecules Yap and Taz in neural crest disrupts Notch signaling, and we provide evidence that Yap can physically and functionally interact with NICD to augment transcription in a manner that is independent of Tead factors.
We have focused on the ability of a Yap-NICD-Rbp-J complex to regulate Jagged1. However, it is unlikely that all NICD/Rbp-J targets require Yap as a co-factor, and we note that Yap/Taz deletion did not alter expression of several Notch target genes, including Hrt1 and Hrt2 in MEFs (Fig. 4J). Moreover, many aspects of Hippo signaling and Yap function are almost certainly Notch independent. Consistent with this conclusion, loss of Yap and Taz in neural crest is embryonic lethal at E10.5, whereas loss of Notch signaling (via expression of a dominant negative mastermind protein or via deletion of Rbp-J) causes a less-severe phenotype, although all exhibit defective smooth muscle differentiation (High et al., 2007; Mead and Yutzey, 2012). Particularly striking is the similarity of the smooth muscle defect in animals in which either Yap/Taz or Rbp-J are deleted in neural crest (Mead and Yutzey, 2012). In both models, neural crest cells migrate to the aortic arch arteries, yet are unable to differentiate to smooth muscle, in contrast to the non-neural crest-derived cells surrounding each arch (Mead and Yutzey, 2012). The likeness of these models supports our findings that Yap could function as a necessary co-factor for the NICD-Rbp-J complex. In the future, it would be interesting to compare ChIP-seq analyses of Yap, Tead, Rbp-J and NICD in neural crest, although the limited material available from microdissected or sorted embryonic tissue will make this approach highly technically challenging at present.
The identification of a Yap-NICD-Rbp-J complex might lead to the re-interpretation of many Hippo signaling phenotypes. Modulation of Hippo signaling has frequently been reported to result in alterations of Notch target gene expression. For example, genetic deletion of Mst1/2 in pancreatic epithelium resulted in increased Yap expression and a robust increase in Hes1 expression (Gao et al., 2013). Yap overexpression in hepatocytes caused upregulation of Notch1/2, Jagged1, Hes1 and the Notch target Sox9 (Yimlamai et al., 2014). Furthermore, recent studies demonstrated that an interleukin-6 co-receptor, gp130 (Il6st – Mouse Genome Informatics), can activate both Hippo and Notch signaling (Taniguchi et al., 2015). This work was interpreted in terms of parallel Hippo and Notch signaling pathways. In light of our results, it would be interesting to revisit these and other prior studies to determine whether a Yap-NICD-Rbp-J complex is contributory.
Hippo signaling has been studied extensively in the context of its ability to regulate organ size and, more recently, with a focus on epithelial-mesenchymal transformation and cancer. However, relatively little work has implicated Hippo in regulation of cell fate or differentiation. Recently, Hippo has been implicated in maintenance of the differentiated hepatocyte fate, and disruption of Hippo resulted in hepatocyte dedifferentiation to a multi-potent progenitor state. Intriguingly, these studies also suggested that Notch signaling was functional downstream of Yap in this setting, although the potential role of a Yap-NICD complex was not examined (Yimlamai et al., 2014).
The identification of a Yap-NICD complex might be particularly important in the understanding of tumor biology. Reactivation of either Notch or Hippo signaling has been implicated in various cancers (Mo et al., 2014; Andersson and Lendahl, 2014). In hepatocellular carcinomas, studies have identified a robust increase in Yap expression, which can increase proliferation and tumor progression (Zender et al., 2006). Subsequently, in a human hepatocellular carcinoma cell line, Yap overexpression has been shown to increase Jagged1 expression through an upstream Tead-dependent enhancer (Tschaharganeh et al., 2013). Interestingly, this study also examined a region of the Jagged1 locus, which includes Jagged1-ECR6, and observed no Tead-Yap-dependent activation, supporting our results that Jagged1-ECR6 activation is Tead independent (Tschaharganeh et al., 2013). Transgenic overexpression of NICD in hepatoblasts also resulted in hepatocellular carcinoma, and further analysis of human hepatocellular carcinoma samples demonstrated that a reactivation of the Notch pathway occurs frequently in these tumors (Villanueva et al., 2012). A similar reactivation of Notch and Hippo signaling has also been observed in breast cancers (Li et al., 2015; Robinson et al., 2011). It will be of interest to determine whether Hippo and Notch functionally interact to promote cancer progression at least in part by activating downstream genes that are regulated by a Yap-NICD-Rbp-J complex.
MATERIALS AND METHODS
Mice
All mice were maintained on a mixed genetic background. Wnt1-Cre (Jiang et al., 2000), Yapflox/+ (Xin et al., 2011) and Tazflox/+ (Xin et al., 2013) alleles were genotyped as previously described. R26tdTomato mice [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J] were obtained from Jackson Labs (strain number 007914) and genotyped as previously described (Madisen et al., 2010). All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Plasmids
The firefly-luciferase reporter construct Jagged1-ECR6 (chr2:136933719-136934335, mm10) was generated previously (Manderfield et al., 2012) but subcloned into pGL4.27 (Promega). The Jagged1-ECR6-mutant construct, abbreviated ECR6*, which contains an altered Rbp-J binding site from 5′-TTTCCCACAGT-3′ to 5′-TGCAGCACAGT-3′, was previous described (Manderfield et al., 2012). The Hes1 firefly-luciferase reporter construct was previously described (High et al., 2007). Murine cleaved notch intracellular domain (NICD) with a C-terminal 3× FLAG epitope tag, and murine Rbp-J with an N-terminal 6× c-myc epitope tag were described previously (Manderfield et al., 2012). We PCR-generated and sequence-verified a murine NICD with a C-terminal V5 epitope tag for biochemical experiments. The Tead reporter, 8× GTIIC, murine Yap, Dntead1, murine Hippo kinases Mst1 and Mst1-KI, and human Hippo kinase LATS2 were all described previously (Manderfield et al., 2014). Wild-type human YAP2, as well as human YAP2 mutants, YAP2-WW1 (W199A, P202A), YAP2-WW2 (W258A, P261A) and YAP2-WW1WW2 (W199A, P202A, W258A, P261A), were previously described and contain two N-terminal FLAG epitope tags [Oka et al. (2008); Addgene plasmids 19045, 19046, 19047 and 19048].
Cell culture and luciferase assay
HEK293T cells were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% fetal bovine serum, penicillin and streptomycin. Human breast adenocarcinoma MDA-MB-231 cells, used for endogenous ChIP in Fig. 6D, were maintained at 37°C with 5% CO2 in Leibovitz's L-15 media supplemented with 15% fetal bovine serum, penicillin and streptomycin. MOVAS cells, used in the entirety of Fig. 7, were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% fetal bovine serum, penicillin and streptomycin and were given a dose of 0.2 mg/ml G-418 every 10 passages, per supplier's instructions (American Type Culture Collection, #CRL-2797). MOVAS cells were treated with either vehicle (DMSO) or 100 µM DAPT (Sigma-Aldrich, # D5942) for 48 h. All transfections in all cell types were prepared using FuGene6 (Promega). Experiments included 300 ng of the specified firefly-luciferase reporter constructs, 80 ng Yap, 300 ng NICD expression vector and 75 ng pGL2-Basic-renilla luciferase (Promega). In experiments in which the Hippo kinases Mst1 or Mst1-KI were co-expressed, 120 ng of the designated expression vector was included. All transfections maintained an equal concentration of total DNA with the inclusion of the pCMV-Sport6 empty vector (Invitrogen). Cellular extracts were collected 48 h post-transfection and measured in a dual-luciferase assay (Promega) in which the cellular extract was used to assess firefly and renilla luciferase activities. All luciferase activity measurements were normalized to the renilla activity of each sample. All experiments were performed in duplicate on at least three separate occasions. Statistical differences between conditions were analyzed using ANOVA, with a Tukey–Kramer post-hoc comparison test.
Histology, immunofluorescence and in situ hybridization
Samples were harvested, fixed overnight in 4% paraformaldehyde and dehydrated through an ethanol series. All samples were paraffin-embedded and sectioned. All primary antibodies were incubated overnight at 4°C at the specified dilutions. Antibodies used for immunofluorescence were anti-SM22α (1:100; Abcam, #10135), anti-RFP (1:50; Rockland Immunochemicals, #600-401-379), anti-SMA (1:200; Sigma-Aldrich, #A2547), anti-desmin (1:20; Dako, #M0760, Clone D33), anti-smooth muscle myosin heavy chain (smMyosin, 1:100; Thomas Scientific, #BT-562), anti-Yap (1:200; Cell Signaling, #4912S), anti-phospho Yap (1:200; Cell Signaling, MA, #4911), anti-Jagged1 (1:25; Santa Cruz Biotechnology, sc-8303), anti-eNOS (1:250; BD Biosciences, #610296) and anti-NICD (1:25; Cell Signaling, #2421). Hoechst nuclear counterstaining was completed following a standard protocol.
Mouse embryo fibroblast preparation
Mouse embryo fibroblasts (MEFs) were isolated from E14.5 Tazflox/flox;Yapflox/flox embryos as described (Connor, 2001). To delete the floxed Taz and Yap alleles, MEFs were treated for 48 h with an adeno-associated virus expressing a constitutively active Cre-recombinase (AAV1-CMV-Cre, Penn Vector Core, University of Pennsylvania, Philadelphia, PA, USA). Viral treatment is reported as genome copies (GC) per cell.
RNA isolation, complementary DNA synthesis and quantitative real-time PCR
RNA was harvested from one 100-mm dish of untreated or AAV1-CMV-Cre-treated Tazflox/flox;Yapflox/flox MEFs or one well of a 6-well dish of MOVAS cells either vehicle/DAPT-treated or transfected with LATS2 using the Qiagen RNeasy kit following manufacturer's instructions (Qiagen). Complementary DNA (cDNA) was synthesized with the Superscript III system (Invitrogen). Quantitative RT-PCR was performed in triplicate with SYBR Green reagents (Applied Biosystems). Relative gene expression was normalized to Gapdh. Quantitative RT-PCR primer sequences are shown in supplementary material Table S5.
Neural crest migration and proliferation quantitation
Transverse sections from Wnt1-Cre;Tazflox/+;Yapflox/+;R26Tom/+ and Wnt1-Cre;Taz flox/flox;Yap flox/flox;R26Tom/+ E10.5 embryos were co-stained for RFP and phospho-histone H3 (anti-pHH3, 1:200; Cell Signaling, 9706L). ImageJ was used to calculate a total area of red cells 50 μm from the endothelial layer of the third arch artery (247,681.7 µm±25,218.7 µm control versus 245,484.3 µm±28,203.9 µm null). Three sections from three independent embryos of each genotype were included in the analysis. ImageJ was then utilized to calculate the size of a single RFP+ neural crest cell in both genotypes (n=34 cells, 2082.5 µm±65 µm control versus 2029.5 µm±81.7 µm null). To calculate the number of migratory RFP+ neural crest cells in both genotypes, the total red area was divided by the cell size. The number of pHH3+ cells were counted by hand and the percentage of proliferative cells was determined by divided the number of pHH3+ cells by the total number of RFP+ cells. Data are reported as an average of each genotype ±s.e.m. Statistical differences between conditions were analyzed using Student's t-test.
Co-IP
HEK293T cells were grown to 70-90% confluency and transfected with Lipofectamine 2000 (Life Technologies, #11668019). At 48 h post-transfection, cells were collected in IP lysis buffer [50 mM Tris HCl pH 7.5, 150 mM NaCl, 1% Igepal, Calbiochem Protease Inhibitor Set I (EMD Millipore)] and sonicated on ice. Lysates were pre-cleared for 1 h at 4°C with Protein G Dynabeads (Life Technologies, #10003D). 10% of the lysates were separated for input, and equal amounts were incubated with either 5 μg of M2 FLAG mouse monoclonal antibody (Sigma-Aldrich, #F3165) or 5 μg of mouse IgG (Santa Cruz Biotechnology, sc-2025) for 2 h at 4°C. 20 μl of Protein G Dynabeads were subsequently added and incubated overnight at 4°C. The beads were washed three times in 1 ml wash buffer (25 mM Tris HCl pH 7.5, 150 mM NaCl), boiled for 10 min in sample buffer [Nupage LDS sample buffer (Life Technologies), 2-mercaptoethanol, dithiothreitol] and then run on SDS-PAGE.
MOVAS cells were grown to 70-90% confluency, collected in IP lysis buffer and pre-cleared as described above. 10% of the lysates were separated for input and equal amounts were incubated with either 17 mg/ml of anti-NOTCH1 (cleaved N-terminal) rabbit polyclonal sera (Rockland Immunochemicals, #100-401-407) or equal concentration of rabbit IgG (Santa Cruz Biotechnology, sc-2027) for 2 h at 4°C. Samples were then processed as described above.
Western blot
Protein lysates were isolated from MEFs 48 h after viral treatment. MEFs were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-base, pH 7.5, 1% Igepal, 0.5% sodium deoxycholate, 0.1% SDS) plus protease inhibitors (Complete Mini, Roche) and were quantitated with a BCA Assay (Promega). Equal quantity of protein from each condition was run on a 4-12% gradient gel at 120 V for 2 h. Blots were transferred overnight at 4°C, at 20 V onto PVDF membrane (Invitrogen). Membranes were blocked in 10% non-fat dry milk/TBS-T and incubated in primary antibody overnight: anti-Yap rabbit polyclonal (Cell Signaling, #4912S; 1:1000), anti-Taz (V386) rabbit polyclonal (Cell Signaling, #4883; 1:1000) and anti-β-actin rabbit polyclonal (Cell Signaling, #4970; 1:1000). Blots were probed with anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling) for 1 h in 5% non-fat dry milk/TBS-T and then developed using ECL Prime (Amersham).
Protein lysates were also collected from two 100-mm dishes of HEK293T cells transfected with 2 µg of either wild-type YAP2 or YAP2 mutants, YAP2-WW1, YAP2-WW2 and YAP2-WW1WW2. All YAP2 constructs contained two N-terminal FLAG epitope tags to allow for biochemical detection. The samples were lysed in the RIPA buffer and processed exactly as described above. The primary antibodies for detection included anti-Yap rabbit polyclonal (Cell Signaling, #4912S; 1:1000), and anti-M2-FLAG mouse monoclonal (Sigma-Aldrich, #F3165; 1:2500). Blots were probed with anti-rabbit or anti-mouse HRP-conjugated secondary antibodies (Cell Signaling).
Protein lysates were collected from one well of a 6-well dish of MOVAS cells transfected with either 1 µg or 2 µg of LATS2. The LATS2 construct contains an N-terminal myc epitope tag for biochemical detection. A non-transfected well of cells was processed in parallel as control. The samples were lysed in the RIPA buffer and processed exactly as described above. The primary antibodies for detection included anti-myc mouse monoclonal (Cell Signaling, #2276; 1:2500), anti-phospho Yap rabbit polyclonal (Cell Signaling, #4911; 1:1000) and anti-β-actin rabbit polyclonal (Cell Signaling, #4967; 1:1000). Blots were probed with anti-rabbit or anti-mouse HRP-conjugated secondary antibodies (Cell Signaling).
Co-IP samples, both from transfected HEK293T and MOVAS cells, and input was processed as described above. The primary antibodies for detection included mouse anti-V5 mouse monoclonal (Life Technologies, #R960; 1:2500), anti-M2-FLAG mouse monoclonal (Sigma-Aldrich, #F3165; 1:2500), anti-β-actin rabbit polyclonal (Cell Signaling, #4967; 1:1000), anti-Yap rabbit polyclonal (Cell Signaling, #4912S; 1:1000) and/or anti-NOTCH1, cleaved N-terminal (NICD, Rockland Immunochemicals, #100-401-407; 1:500). Blots were probed with anti-rabbit or anti-mouse HRP-conjugated secondary antibodies (Cell Signaling).
Proximity ligation assay
The proximity ligation assay was performed using HEK293T cells transfected with 40 ng of the indicated cDNA, including murine NICD with a C-terminal V5 epitope tag, wild-type YAP2, YAP2 mutants, YAP2-WW1 or YAP2-WW2 or murine Rbp-J with an N-terminal 6× c-myc epitope tag. All transfections were completed using FuGene6 (Promega). Primary antibodies were diluted 1:200 and incubated for 1 h at room temperature; they included: anti-V5 mouse monoclonal (Life Technologies, #R960), anti-Yap rabbit polyclonal (Cell Signaling, #4912S) and anti-myc rabbit polyclonal (EMD Millipore, 06-549). Staining was performed using the Duolink in situ Detection Reagents Red Kit (Sigma-Aldrich) following the manufacturer's instructions. DAPI nuclear staining was included in the Duolink mounting media (Sigma-Aldrich).
Chromatin immunoprecipitation (ChIP)
To examine NICD and Yap occupancy at Jagged1-ECR6, chromatin was generated from three 100-mm dishes of HEK293T transfected with 800 ng of the Jagged1-ECR6-pGL4.27, 100 ng of a C-terminal FLAG tagged NICD and 100 ng of a murine Yap expression plasmid. In parallel, chromatin was also isolated from cells transfected with two control conserved regions, either a distant conserved region which was not Notch responsive, Jagged1-ECR1 (chr2:136942017-136942473, mm10), or Jagged1-ECR6-mutant containing a mutated Rbp-J binding site, again both in the presence of FLAG-tagged NICD and Yap (Manderfield et al., 2012). To examine endogenous Yap and/or NICD occupancy at Jagged1-ECR6 and the Hes1 promoter, chromatin was generated from three 100-mm dishes of non-transfected MDA-MB-231 or MOVAS cells. 48 h post-transfection, cells were washed twice in cold PBS and cross-linked with 1% formaldehyde for 10 min at room temperature. Then, the reaction was quenched with 0.14 M glycine for 5 min at room temperature. Cells were collected in PBS, pelleted and resuspended in SDS lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 1% NP-40, 1% SDS, 0.5% deoxycholic acid and protease inhibitors). After 10-min incubation on ice, the chromatin was sonicated using a BioRuptor (Diagenode). 10 µg of chromatin was diluted in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100) and incubated overnight at 4°C with 0.5 µg of specific antibody or control IgG: anti-Yap rabbit polyclonal (Cell Signaling, #4912S), anti-M2-FLAG mouse monoclonal (Sigma-Aldrich, #F3165), anti-NOTCH1, cleaved N-terminal (NICD, Rockland Immunochemicals, #100-401-407), normal-rabbit IgG (Santa Cruz, sc-2027) or normal-mouse IgG (Santa Cruz, sc-2025). Protein-G agarose beads were added to each reaction for 1 h at 4°C. The agarose beads were washed once with each of the following buffers for 5 min at 4°C: low salt buffer, high salt buffer, LiCl buffer and TE buffer. Complexes were eluted in 200 μl elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 15 min. After the reversal of chromatin cross-linking, protein degradation and DNA purification, samples were PCR-amplified using primers for Jagged1 or Hes1 loci:
ECR1-Forward: 5′-TCCCAGCTCATGTATCTTTGCTTGC-3′;
ECR1-Reverse: 5′-GGAGGAATGCAGATCAAAGCGAAGTCT-3′;
ECR6-Forward: 5′-CTACAACCACTAACAGGAGAGC-3′;
ECR6-Reverse: 5′-GCTTCATACTTACAGCAGG-3′;
Hes1-Forward: 5′-TTCCTCCCATTGGCTGAAAG-3′;
Hes1-Reverse: 5′-AGCTCCAGATCCTGTGTGA-3′.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.J.M. designed and performed experiments, analyzed data and wrote the paper. H.A., K.A.E., L.Y.L., F.L. and L.L. designed and performed experiments. R.J. performed experiments and edited the manuscript. E.N.O. provided the Yap and Taz floxed alleles. J.A.E. oversaw the entire project, designed experiments, analyzed data and wrote the paper.
Funding
This work was supported by the Spain Fund for Cardiovascular Research; the Cotswold Foundation; the WW Smith Endowed Chair; and the National Institutes of Health (NIH) [U01 HL100405 to J.A.E.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125807/-/DC1
References
- Afelik S. and Jensen J. (2013). Notch signaling in the pancreas: patterning and cell fate specification. Wiley Interdiscip. Rev. Dev. Biol. 2, 531-544. 10.1002/wdev.99 [DOI] [PubMed] [Google Scholar]
- Andersson E. R. and Lendahl U. (2014). Therapeutic modulation of Notch signalling — are we there yet? Nat. Rev. Drug Discov. 13, 357-378. 10.1038/nrd4252 [DOI] [PubMed] [Google Scholar]
- Connor D. A. (2001). Mouse embryo fibroblast (MEF) feeder cell preparation. Curr. Protoc. Mol. Biol. Chapter 23:Unit 23.2 10.1002/0471142727.mb2302s51 [DOI] [PubMed] [Google Scholar]
- Djiane A., Zaessinger S., Babaoğlan A. B. and Bray S. J. (2014). Notch inhibits Yorkie activity in Drosophila wing discs. PLoS ONE 9, e106211 10.1371/journal.pone.0106211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Zhou D., Yang C., Singh T., Penzo-Méndez A., Maddipati R., Tzatsos A., Bardeesy N., Avruch J. and Stanger B. Z. (2013). Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology 144, 1543-1553.e1. 10.1053/j.gastro.2013.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L. and Martin J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- High F. A. and Epstein J. A. (2008). The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9, 49-61. 10.1038/nrg2279 [DOI] [PubMed] [Google Scholar]
- High F. A., Zhang M., Proweller A., Tu L., Parmacek M. S., Pear W. S. and Epstein J. A. (2007). An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 117, 353-363. 10.1172/JCI30070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- High F. A., Lu M. M., Pear W. S., Loomes K. M., Kaestner K. H. and Epstein J. A. (2008). Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. USA 105, 1955-1959. 10.1073/pnas.0709663105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R. and Israel A. (1995). Signalling downstream of activated mammalian Notch. Nature 377, 355-358. 10.1038/377355a0 [DOI] [PubMed] [Google Scholar]
- Jiang X., Rowitch D. H., Soriano P., McMahon A. P. and Sucov H. M. (2000). Fate of the mammalian cardiac neural crest. Development 127, 1607-1616. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang X., Leathers R., Makino A., Huang C., Parsa P., Macias J., Yuan J. X.-J., Jamieson S. W. and Thistlethwaite P. A. (2009). Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat. Med. 15, 1289-1297. 10.1038/nm.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hibbs M. A., Gard A. L., Shylo N. A. and Yun K. (2012). Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells 30, 741-752. 10.1002/stem.1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-W., Shen H., Frangou C., Yang N., Guo J., Xu B., Bshara W., Shepherd L., Zhu Q., Wang J. et al. (2015). Characterization of TAZ domains important for the induction of breast cancer stem cell properties and tumorigenesis. Cell Cycle 14, 146-156. 10.4161/15384101.2014.967106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R., Uchijima Y., Nishiyama K., Amano T., Chen Q., Takeuchi T., Mitani A., Nagase T., Yatomi Y., Aburatani H. et al. (2008). Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am. J. Physiol. Renal Physiol. 294, F542-F553. 10.1152/ajprenal.00201.2007 [DOI] [PubMed] [Google Scholar]
- Manderfield L. J., High F. A., Engleka K. A., Liu F., Li L., Rentschler S. and Epstein J. A. (2012). Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation 125, 314-323. 10.1161/CIRCULATIONAHA.111.047159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield L. J., Engleka K. A., Aghajanian H., Gupta M., Yang S., Li L., Baggs J. E., Hogenesch J. B., Olson E. N. and Epstein J. A. (2014). Pax3 and hippo signaling coordinate melanocyte gene expression in neural crest. Cell Rep. 9, 1885-1895. 10.1016/j.celrep.2014.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead T. J. and Yutzey K. E. (2012). Notch pathway regulation of neural crest cell development in vivo. Dev. Dyn. 241, 376-389. 10.1002/dvdy.23717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J.-S., Park H. W. and Guan K.-L. (2014). The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 15, 642-656. 10.15252/embr.201438638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Nakagawa M., Olson E. N. and Nakagawa O. (2005). A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. USA 102, 18034-18039. 10.1073/pnas.0509109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R. M., de Beco S. and Johnston L. A. (2010). Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell 19, 507-520. 10.1016/j.devcel.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Mazack V. and Sudol M. (2008). Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J. Biol. Chem. 283, 27534-27546. 10.1074/jbc.M804380200 [DOI] [PubMed] [Google Scholar]
- Rayon T., Menchero S., Nieto A., Xenopoulos P., Crespo M., Cockburn K., Cañon S., Sasaki H., Hadjantonakis A.-K., de la Pompa J. L. et al. (2014). Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 30, 410-422. 10.1016/j.devcel.2014.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. R., Kalyana-Sundaram S., Wu Y.-M., Shankar S., Cao X., Ateeq B., Asangani I. A., Iyer M., Maher C. A., Grasso C. S. et al. (2011). Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 17, 1646-1651. 10.1038/nm.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ W. P., Lowery D. M., Mishra P., Yaffe M. B. and Ranganathan R. (2005). Natural-like function in artificial WW domains. Nature 437, 579-583. 10.1038/nature03990 [DOI] [PubMed] [Google Scholar]
- Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., Oren M., Sudol M., Cesareni G. and Blandino G. (2001). Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276, 15164-15173. 10.1074/jbc.M010484200 [DOI] [PubMed] [Google Scholar]
- Stylianou S., Clarke R. B. and Brennan K. (2006). Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517-1525. 10.1158/0008-5472.CAN-05-3054 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Wu L.-W., Grivennikov S. I., de Jong P. R., Lian I., Yu F.-X., Wang K., Ho S. B., Boland B. S., Chang J. T. et al. (2015). A gp130–Src–YAP module links inflammation to epithelial regeneration. Nature 519, 57-62. 10.1038/nature14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh D. F., Chen X., Latzko P., Malz M., Gaida M. M., Felix K., Ladu S., Singer S., Pinna F., Gretz N. et al. (2013). Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology 144, 1530-1542.e12. 10.1053/j.gastro.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. (2014). The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141, 1614-1626. 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Villanueva A., Alsinet C., Yanger K., Hoshida Y., Zong Y., Toffanin S., Rodriguez-Carunchio L., Solé M., Thung S., Stanger B. Z. et al. (2012). Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 143, 1660-1669.e7. 10.1053/j.gastro.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., Lin Z., Schlegelmilch K., Honor L. B., Pan G. M., Buck J. N., Ma Q., Ishiwata T., Zhou B., Camargo F. D. et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 109, 2394-2399. 10.1073/pnas.1116136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hu G., Gao X., Wang Y., Zhang W., Harmon E. Y., Zhi X., Xu Z., Lennartz M. R., Barroso M. et al. (2012). The induction of yes-associated protein expression after arterial injury is crucial for smooth muscle phenotypic modulation and neointima formation. Arterioscler. Thromb. Vasc. Biol. 32, 2662-2669. 10.1161/ATVBAHA.112.254730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hu G., Liu F., Wang X., Wu M., Schwarz J. J. and Zhou J. (2014). Deletion of yes-associated protein (YAP) specifically in cardiac and vascular smooth muscle cells reveals a crucial role for YAP in mouse cardiovascular development. Circ. Res. 114, 957-965. 10.1161/CIRCRESAHA.114.303411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A. P., Millholland J. M., Yashiro-Ohtani Y., Arcangeli M. L., Lau A., Wai C., Del Bianco C., Rodriguez C. G., Sai H., Tobias J. et al. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096-2109. 10.1101/gad.1450406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Guo Y., Zhu T., Zhang J., Ma P. X. and Chen Y. E. (2012). Yap1 protein regulates vascular smooth muscle cell phenotypic switch by interaction with myocardin. J. Biol. Chem. 287, 14598-14605. 10.1074/jbc.M111.329268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Qi X., McAnally J., Schwartz R. J., Richardson J. A., Bassel-Duby R. and Olson E. N. (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70 10.1126/scisignal.2002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Murakami M., Qi X., McAnally J., Porrello E. R., Mahmoud A. I., Tan W., Shelton J. M. et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839-13844. 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D., Christodoulou C., Galli G. G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B. Z. and Camargo F. D. (2014). Hippo pathway activity influences liver cell fate. Cell 157, 1324-1338. 10.1016/j.cell.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., Zhang Y., Park H. W., Jewell J. L., Chen Q., Deng Y., Pan D., Taylor S. S., Lai Z.-C. and Guan K.-L. (2013). Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27, 1223-1232. 10.1101/gad.219402.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.-T., Luk J. M., Wigler M., Hannon G. J. et al. (2006). Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253-1267. 10.1016/j.cell.2006.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Kim J., Ye X., Lai Z.-C. and Guan K.-L. (2009). Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 69, 1089-1098. 10.1158/0008-5472.CAN-08-2997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.