ABSTRACT
The lipid phosphate phosphatase-related proteins (LPPRs), also known as plasticity-related genes (PRGs), are classified as a new brain-enriched subclass of the lipid phosphate phosphatase (LPP) superfamily. They induce membrane protrusions, neurite outgrowth or dendritic spine formation in cell lines and primary neurons. However, the exact roles of LPPRs and the mechanisms underlying their effects are not certain. Here, we present the results of a large-scale proteome analysis to determine LPPR1-interacting proteins using co-immunoprecipitation coupled to mass spectrometry. We identified putative LPPR1-binding proteins involved in various biological processes. Most interestingly, we identified the interaction of LPPR1 with its family member LPPR3, LPPR4 and LPPR5. Their interactions were characterized by co-immunoprecipitation and colocalization analysis using confocal and super-resolution microscopy. Moreover, co-expressing two LPPR members mutually elevated their protein levels, facilitated their plasma membrane localization and resulted in an increased induction of membrane protrusions as well as the phosphorylation of S6 ribosomal protein. Taken together, we revealed a new functional cooperation between LPPR family members and discovered for the first time that LPPRs likely exert their function through forming complex with its family members.
KEY WORDS: Membrane protrusion, Protein-protein interaction, Filopodia, Actin
Highlighted Article: This paper presents the results of a large-scale proteome analysis to determine LPPR1-interacting proteins and identifies a functional cooperation between LPPR family members.
INTRODUCTION
The lipid phosphate phosphatase-related proteins (LPPRs), also known as plasticity-related genes (PRGs), are a family of six-transmembrane proteins that are enriched in brain, with five members (LPPR1–LPPR5) having been identified to date (Brauer and Nitsch, 2008; Brindley, 2004; Sigal et al., 2005; Strauss and Brauer, 2013). LPPRs are homologous to the lipid phosphate phosphatase (LPP) family proteins, which are important enzymes that dephosphorylate bioactive lipids such as lysophosphatidic acid (LPA) (Brindley, 2004; Sigal et al., 2005). Although LPPRs show a high degree of similarity to the LPPs, owing to non-conservative substitutions in several crucial sites in the catalytic domains, they are unlikely to have a level of enzymatic activity equivalent to the typical LPPs. Therefore the LPPR proteins are designated as LPP-related proteins and comprise of a new subfamily of the LPP superfamily (Brauer and Nitsch, 2008; Brindley, 2004; Sigal et al., 2005).
LPPR family members are all highly expressed in the central nervous system (CNS), specifically in neurons, with some differences in their spatial and temporal expression patterns. The first identified member, LPPR4, was originally named plasticity-related gene-1 (PRG-1) to reflect its regulated expression during brain development and in response to hippocampal lesions (Brauer et al., 2003). The expression of LPPR4 is increased both at the developmental stage characterized by active axon outgrowth and in regrowing axons following injury, and therefore LPPR4 is considered as a regulator of neuronal plasticity. The other four members, named PRG-2 to PRG-5 (corresponding to LPPR3, LPPR1, LPPR2 and LPPR5, respectively), were found by in silico analysis (Brauer et al., 2003; Broggini et al., 2010; Savaskan et al., 2004).
LPPR family members show structural and sequence similarity in terms of their six-transmembrane domains, although each member has a unique C-terminus. In particular, LPPR3 and LPPR4 have long (∼400 amino acids) C-termini, whereas LPPR1, LPPR2 and LPPR5 have very short (∼50 amino acids) C-termini (Strauss and Brauer, 2013), implying that they might exert different functions in cells. LPPR4, being the most studied member, was found to be able to protect against LPA-induced neurite retraction (Brauer et al., 2003). LPPR4 also plays an important role in regulating excitatory neurotransmission, as knockout of LPPR4 leads to seizures and a change in excitatory synaptic efficacy (Trimbuch et al., 2009). LPPR4 has also been shown to inhibit LPA-induced vascular smooth muscle cell migration and proliferation (Gaaya et al., 2012).
Of the other four family members, LPPR1 and LPPR5 influence cell morphology but very little is known about LPPR2 and LPPR3. Overexpression of either LPPR1 or LPPR5 induces membrane protrusions in neuronal and non-neuronal cell lines (Broggini et al., 2010; Savaskan et al., 2004; Sigal et al., 2007). LPPR1 has been reported to promote neurite shaft protrusion in primary neurons (Velmans et al., 2013), whereas LPPR5 has been shown to induce neurite outgrowth (Broggini et al., 2010) and, more recently, to promote dendritic spine formation in hippocampal neurons (Coiro et al., 2014). The current findings suggest that LPPRs play putative roles in axonal outgrowth, regeneration and synaptic plasticity. However, whether the LPPR members have a redundant or distinct functions as well as their mechanisms underlying their effects are still being explored.
In a recent phosphoproteomics screen, we demonstrated that the phosphorylation of LPPR1 was altered in response to chondroitin sulfate proteoglycans (CSPGs) (Yu et al., 2013), important regulators of axonal regeneration and neuronal plasticity. In this study, we aimed to gain more insights into the mechanism of action of LPPR1 by identifying the LPPR1-interacting proteins using affinity purification coupled to mass spectrometry (Berggard et al., 2007; Free et al., 2009). Surprisingly, we found that another three LPPR family members, LPPR3, LPPR4 and LPPR5, were associated with LPPR1. Their interactions were further confirmed by co-immunoprecipitation and colocalization analysis using confocal and super-resolution microscopy. More importantly, we revealed a novel functional cooperation between LPPRs, evidenced as enhanced protein levels and increased plasma membrane localization as well as increased induction of membrane protrusions. Our study thus reveals a novel functional interaction between LPPR family members, suggesting that these molecules might act as a complex in regulating cellular functions.
RESULTS
Importance of the C-terminus of LPPR1 in plasma membrane localization and induction of membrane protrusions
Overexpression of LPPR1 produces membrane protrusions in several different cell types (Savaskan et al., 2004; Sigal et al., 2007). We therefore examined whether LPPR1 overexpression has a similar effect in Neuro2A cells. EGFP–LPPR1 was expressed in Neuro2A cells. Consistent with previous findings in other cell types (Sigal et al., 2007), LPPR1 protein localized to both intracellular membrane structures and plasma membranes of Neuro2A cells (Fig. 1A). Overexpression of a small C-terminal HA-epitope-tagged LPPR1 showed an identical distribution pattern to EGFP–LPPR1 (Fig. 1B). Cells expressing LPPR1 extended many thin, fragile membrane protrusions. Because many protrusions are often destroyed during the staining process, to preserve better cell morphology and also for live-cell imaging purposes, we chose EGFP-tagged LPPR1 in most of the experiments, although the LPPR1–HA construct was also used to make sure similar results were obtained.
Fig. 1.
C-terminal deletion of LPPR1 abolishes its plasma membrane localization. (A,B) Representative images of Neuro2A cells expressing EGFP–LPPR1 (A) or LPPR1–HA (B), showing a similar distribution pattern. (C,D) Representative images of Neuro2A cells expressing EGFP–LPPR1 (C) or EGFP–LPPR1ΔC43 (D) together with a plasma-membrane-targeted mCherry (mem–Cherry) for visualization of overall cell morphology. The inset in C is a higher magnification image of the boxed area, showing the discontinuous and punctate distribution pattern of EGFP–LPPR1 along membrane protrusions (arrows). The inset in D is a higher magnification image of the boxed area, showing that EGFP–LPPR1ΔC43 was absent from membrane protrusions (arrows). (E) Quantification of the relative amount of intracellular and cell surface LPPR1. Quantitative results are mean±s.e.m. ***P<0.001 (two-way ANOVA and Bonferroni's multiple comparisons test). (F,G) Representative images of Neuro2A cells expressing EGFP (F) or EGFP–LPPR1CT (G). (H,I) Representative images and quantification of the protrusion numbers in cells overexpressing EGFP, EGFP–LPPR1, EGFP–LPPR1ΔC43 or EGFP–LPPR1CT. Quantitative results are mean±s.e.m. #P<0.05, ***P<0.001 (one-way ANOVA and Tukey's multiple comparisons test).
The C-terminus of each LPPR member is relatively unique, and a previous report has shown that the C-terminus of LPPR1 is important for filopodial induction (Sigal et al., 2007). This is also the location of the change in phosphorylation induced by CSPGs (Yu et al., 2013). Thus, to gain more information about the role of the C-terminus, we expressed full-length EGFP–LPPR1, a C-terminally truncated LPPR1 (EGFP–LPPR1ΔC43) or just the C-terminal portion (EGFP–LPPR1CT) in Neuro2A cells (supplementary material Fig. S1A). Protein expression was confirmed by western blot analysis with an anti-GFP antibody (supplementary material Fig. S1B). The cellular distribution of these fusion proteins was determined by confocal microscopy and a membrane-targeting mCherry construct was co-expressed to visualize the overall cell morphology. A large amount of EGFP–LPPR1 protein was found in intracellular membrane structures, such as endoplasmic reticulum (ER), with some localized on the plasma membrane protrusions (supplementary material Fig. S1C,D; Fig. 1C). The distribution of EGFP–LPPR1 along the protrusions was discontinuous and often showed a punctate pattern (Fig. 1C). In contrast, EGFP–LPPR1ΔC43 was exclusively localized to intracellular membrane structures with very little on the plasma membrane (Fig. 1D,E). This indicates an important role of the C-terminus of LPPR1 in facilitating protein movement from the ER to the plasma membrane. The C-terminus itself (EGFP–LPPR1CT) showed a diffuse cytoplasmic expression pattern similar to EGFP control (Fig. 1F,G).
To determine whether deletion of the C-terminus has an effect on the ability of LPPR1 to induce membrane protrusions, we quantified the numbers of F-actin-rich protrusions in Neuro2A cells expressing full-length or C-terminally truncated LPPR1. Compared to cells expressing EGFP, cells expressing EGFP–LPPR1 extended many more protrusions; this effect was significantly attenuated by deletion of the C-terminus (Fig. 1H,I). However, overexpression of LPPR1ΔC43 still caused a slight but significant increase in protrusion formation compared to EGFP alone, which indicates the presence of an additional mechanism promoting protrusion formation independently of its plasma membrane localization. Cells expressing only the C-terminus of LPPR1 (EGFP–LPPR1CT) showed no change in protrusion formation as compared to EGFP control (Fig. 1H,I). Similar results were found in other cell types, including human fibroblasts and Cos-7 cells (supplementary material Fig. S1E; data not shown). LPPR1 overexpression in Cos-7 cells also significantly induced the formation of membrane protrusions, and this effect was attenuated by deletion of its C-terminus (supplementary material Fig. S1E).
Proteome-wide identification of LPPR1-interacting proteins
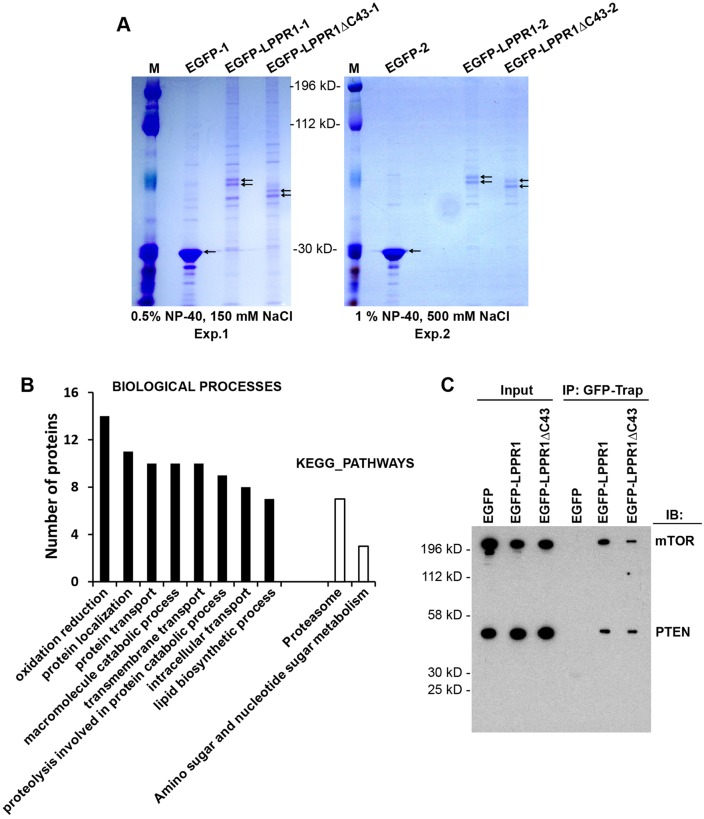
The mechanisms by which LPPR1 elicits protrusions is still an open question. Having characterized the localization pattern of LPPR1, we then sought to identify the interacting proteins of LPPR1 as a way to understand its function. Cell extracts were collected from Neuro2A cells expressing EGFP–LPPR1, EGFP–LPPR1ΔC43 or EGFP. GFP–Trap beads were added to co-immunoprecipitate EGFP-tagged proteins and their binding partners. Because protein–protein interactions can be either weak or strong, considering both the coverage of binding protein identification and the binding specificity, we performed two independent co-immunoprecipitation experiments with varied stringency of the washing step. Consistent with previous reports (Sigal et al., 2007; Velmans et al., 2013), LPPR1 presents as a doublet visualized on PAGE gels after Coomassie Blue staining (Fig. 2A), likely due to post-translational modifications such as glycosylation or phosphorylation. The gel images also highlighted the differences among EGFP, EGFP–LPPR1 and EGFP–LPPR1ΔC43. By increasing both the detergent and the salt concentration in the washing buffer, fewer bands were seen in the second experiment as compared to the first experiment (Fig. 2A).
Fig. 2.
Proteome-wide identification of the LPPR1-interacting proteins. (A) Coomassie Blue staining pattern of the co-immunoprecipitated proteins. Neuro2A cells transfected with pEGFP, pEGFP-LPPR1 or pEGFP-LPPR1ΔC43 constructs were subjected to co-immunoprecipitation using GFP-Trap beads. Two independent co-immunoprecipitation experiments were performed with different detergent and the salt concentrations. M, molecular mass markers. (B) Functional annotation of the putative LPPR1-binding proteins by DAVID. Significantly enriched Gene Ontology (GO) terms for the biological process and KEGG pathways were plotted according to the number of proteins. (C) Validation of the interaction of mTOR and PTEN with LPPR1 using co-immunoprecipitation (IP). IB, immunoblot.
To determine which proteins are present in the complexes, in-gel digestion was performed and the resultant peptides were subjected to liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. Supplementary material Table S1 presents the unique peptides of LPPR1 identified from each sample. It is noteworthy that three peptides (amino acids 281–295, amino acids 296–304 and amino acids 305–325) located at the C-terminus of LPPR1 were only found in full-length EGFP–LPPR1 but not in the C-terminally truncated EGFP–LPPR1ΔC43 sample, and no LPPR1 peptides were present in the EGFP control group.
Overall, the raw data generated by the database search identified 910 proteins from the EGFP–LPPR1 sample and 813 proteins from the EGFP–LPPR1ΔC43 sample in experiment 1, and 531 proteins from the EGFP–LPPR1 sample, and 416 proteins from the EGFP–LPPR1ΔC43 sample in the higher-stringency experiment 2 after subtraction of those also found in the EGFP control samples (supplementary material Table S2). Among them, 496 and 429 proteins from EGFP–LPPR1 and EGFP–LPPR1ΔC43 in experiment 1, and 306 and 236 proteins from EGFP–LPPR1 and EGFP–LPPR1ΔC43 in experiment 2 were assigned based on only a single peptide matching. As proteins identified by one peptide match in proteomics are often found to be false positives (Carr et al., 2004), the data were filtered based on the following dual criteria: proteins identified in both experiments and that had three or more unique peptide matches in at least one of the two experiments. These criteria were met by 107 proteins (supplementary material Table S3). To gain insights into the various functions of the putative LPPR1 binding proteins, these proteins were subjected to Gene Ontology (GO) term annotation using DAVID. The significantly overrepresented GO terms of Biological Process and KEGG Pathways are shown in Fig. 2B, showing that LPPR1 is associated with various biological processes such as protein localization, proteolysis and lipid biosynthesis. A number of proteins involved in lipid biosynthesis, including lysophosphatidylcholine acyltransferase 1, phosphatidylinositol synthase, sphingomyelin phosphodiesterase 4 and phosphatidate cytidylyltransferase 2, were enriched in the protein complexes, which indicated that LPPR1 might play a role in regulating bioactive lipid signaling although LPPR1 itself does not possess lipid phosphatase activity.
Another interesting finding was that both mammalian target of rapamycin (mTOR) and its upstream regulator PTEN were found to be associated with LPPR1. The mTOR peptides were found in both EGFP–LPPR1 and EGFP–LPPR1ΔC43 groups, but not in the EGFP control group. Moreover, it was found in both experiments with four and two unique peptides identified in experiment 1 and the more stringent experiment 2, respectively. For PTEN, only one unique peptide was present in both the EGFP–LPPR1 and EGFP–LPPR1ΔC43 groups and it was identified in experiment 1 but not in the high-stringency experiment 2. To validate the interaction of LPPR1 with mTOR and PTEN, we performed co-immunoprecipitation experiments. Our results showed that both mTOR and PTEN were co-precipitated with LPPR1. This interaction was not dependent on the C-terminus of LPPR1, as both LPPR1 and LPPR1ΔC43 were able to precipitate mTOR and PTEN (Fig. 2C).
Most intriguingly, another three LPPR1 family members: LPPR3, LPPR4 and LPPR5 were identified as binding partners of LPPR1. LPPR3 and LPPR4 seemed to bind both EGFP–LPPR1 and EGFP–LPPR1ΔC43: seven and six unique peptides of LPPR4 were found in the EGFP–LPPR1 and the EGFP–LPPR1ΔC43 group, respectively; nine and four unique peptides of LPPR3 were found in the EGFP–LPPR1 and the EGFP–LPPR1ΔC43 group, respectively; and no peptides were found in the EGFP control group (supplementary material Table S1). This suggests that the binding of LPPR3 and LPPR4 to LPPR1 does not occur through its C-terminus. LPPR5, which shares the highest similarity to LPPR1 among all LPPR members, was also detected by proteomics, although only one unique peptide was present in the EGFP–LPPR1 group.
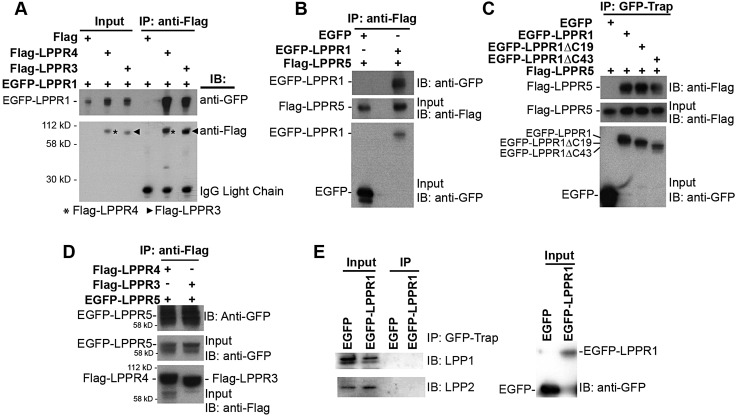
Interactions of LPPR family members
The interaction of LPPR1 with LPPR3 and LPPR4 was validated by reciprocal co-immunoprecipitation. When EGFP–LPPR1 was co-transfected with Flag–LPPR3 or Flag–LPPR4, LPPR1 was co-precipitated with either Flag–LPPR3 or Flag–LPPR4 (Fig. 3A). Many peptides were detected from LPPR3 and LPPR4, but only one peptide from LPPR5 was found by proteomics. However, unlike LPPR3 and LPPR4, which have long C-termini, a large portion of LPPR5 comprises hydrophobic transmembrane regions with very short extramembrane regions, and such proteins are very difficult to be detected by proteomics. We therefore tested the interaction between LPPR1 and LPPR5 by reciprocal co-immunoprecipitation. Flag–LPPR5 was co-expressed with EGFP–LPPR1 or a control EGFP vector in Neuro2A cells; immunoprecipitation was performed using anti-Flag M2 agarose affinity gel followed by immunoblotting with anti-GFP or anti-Flag antibody. We found that EGFP–LPPR1, but not EGFP alone, was co-precipitated with Flag–LPPR5 (Fig. 3B). We also excluded the effect of Flag on the binding as Flag itself was not able to precipitate EGFP–LPPR1 (data not shown). To further identify whether the C-terminal portion of LPPR1 is required for the interaction, the Flag–LPPR5 construct was co-expressed with EGFP, EGFP–LPPR1, EGFP–LPPR1ΔC19 or EGFP–LPPR1ΔC43. GFP–Trap beads were used to immunoprecipitate EGFP-tagged proteins. As shown in Fig. 3C, both full-length LPPR1 (EGFP–LPPR1) and the C-terminally truncated forms of LPPR1 (EGFP–LPPR1ΔC19 and EGFP–LPPR1ΔC43), but not EGFP alone, were able to interact with LPPR5. These results confirmed the interaction between LPPR1 and LPPR5, and that the interaction is not through the C-terminus of LPPR1. Similarly, LPPR5 was co-immunoprecipitated with LPPR3 and LPPR4 (Fig. 3D). We also detected an interaction between EGFP–LPPR1 and LPPR1–HA, as well as interaction between EGFP–LPPR5 and Flag–LPPR5, indicating that besides forming heterodimers, LPPRs can also form homodimers (data not shown). It has been previously demonstrated that LPPs (LPP1–LPP3, also known as PPAP2A, PPAP2C and PPAP2B, respectively) can form both homo- and hetero-oligomers (Long et al., 2008). To determine whether LPPR1, which belongs to a distinct subfamily of the LPP superfamily, can interact with LPPs, we performed co-immunoprecipitation experiments. No interaction was detected between EGFP–LPPR1 and endogenous LPP1 or LPP2 (Fig. 3E).
Fig. 3.
Validation of the interaction between LPPR1 family members. (A) Co-immunoprecipitation (IP) of EGFP–LPPR1 by Flag–LPPR3 or Flag–LPPR4, but not by Flag. (B) Co-immunoprecipitation of EGFP–LPPR1, but not EGFP by the Flag–LPPR5. (C) Co-immunoprecipitation of Flag–LPPR5 by EGFP–LPPR1 or EGFP–LPPR1ΔC, but not by EGFP. The C-terminus of LPPR1 is not required for its interaction with LPPR5. (D) Co-immunoprecipitation of EGFP–LPPR5 by Flag–LPPR3 or Flag–LPPR4. (E) No interaction was detected between EGFP–LPPR1 and LPP1 or LPP2. IB, immunoblot.
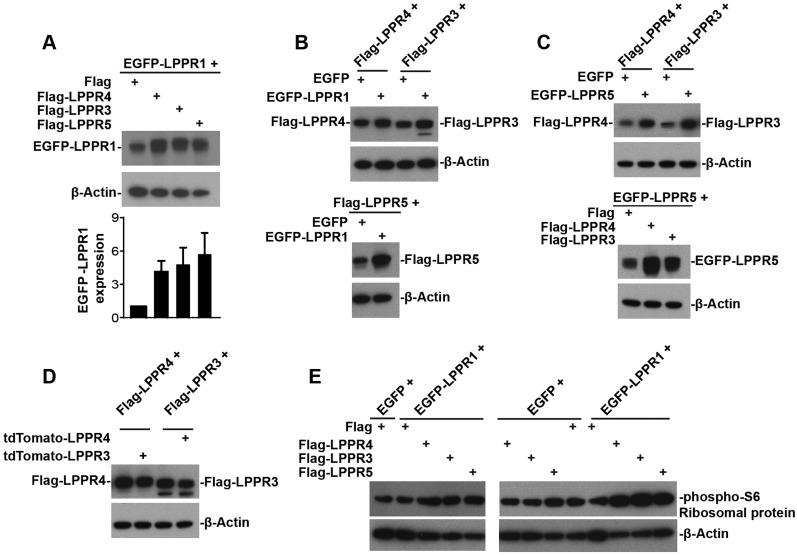
Co-expression of LPPR members increases their protein level
One interesting phenomena we observed in the co-immunoprecipitate results was that, although we loaded the same amount of protein in the input, there was always a higher level of LPPRs in the samples obtained after co-expression of two LPPR members, as compared to when cells were transfected with only one LPPR member together with an empty vector. To confirm this, we expressed various combinations of two LPPR members and detected the protein amount using western blotting. Indeed, we found that, when EGFP–LPPR1 was co-transfected with Flag– LPPR3, Flag–LPPR4 or Flag–LPPR5, the protein levels of EGFP–LPPR1, Flag–LPPR3, Flag–LPPR4 and Flag–LPPR5 were dramatically increased in the corresponding LPPR member co-expression groups, as compared to those expressing only a single LPPR member together with a control vector (Fig. 4A,B). Similarly, a concomitant elevation in the protein levels of LPPR5 and LPPR3, or LPPR5 and LPPR4 was also detected when LPPR5 was co-expressed with LPPR3 or LPPR4 (Fig. 4C). Interestingly, this was not seen when co-expressing LPPR3 with LPPR4 (Fig. 4D), where the protein levels were not altered by each other. Co-expression of EGFP–LPPR1 with Flag–LPPR3, Flag–LPPR4 or Flag–LPPR5 also increased the protein phosphorylation of S6 ribosomal protein, which correlates with an increase in protein synthesis (Fig. 4E). These results together indicate the LPPR members might be co-assembled and function as a complex in cells, which further supports the presence of functional cooperation between LPPR members.
Fig. 4.
Co-transfection two of the LPPR members, except for co-transfection of LPPR3 and LPPR4, increases their protein levels. (A) Western blot and quantification (mean±s.e.m., n=4) showing increase in the EGFP–LPPR1 protein amount when co-transfecting EGFP–LPPR1 with either Flag–LPPR3, Flag–LPPR4 or Flag–LPPR5, compared to co-transfection with Flag control. (B) Western blot showing increase in the protein amount of Flag–LPPR3, Flag–LPPR4 or Flag–LPPR5 when co-transfected with EGFP–LPPR1, compared to co-transfection with EGFP control. (C) Western blot showing increase in the Flag–LPPR3 or the Flag–LPPR4 protein amount upon co-transfection of EGFP–LPPR5 with Flag–LPPR3 or with Flag–LPPR4, coinciding with an increase in EGFP–LPPR5. (D) Western blot showing no change in their protein level upon co-transfection of LPPR3 with LPPR4, compared to transfection with LPPR3 alone or LPPR4 alone. (E) Western blot showing that co-expression of EGFP–LPPR1 with Flag–LPPR3, Flag–LPPR4 or Flag–LPPR5 increased the phosphorylation of S6 (phospho-S6) ribosomal protein.
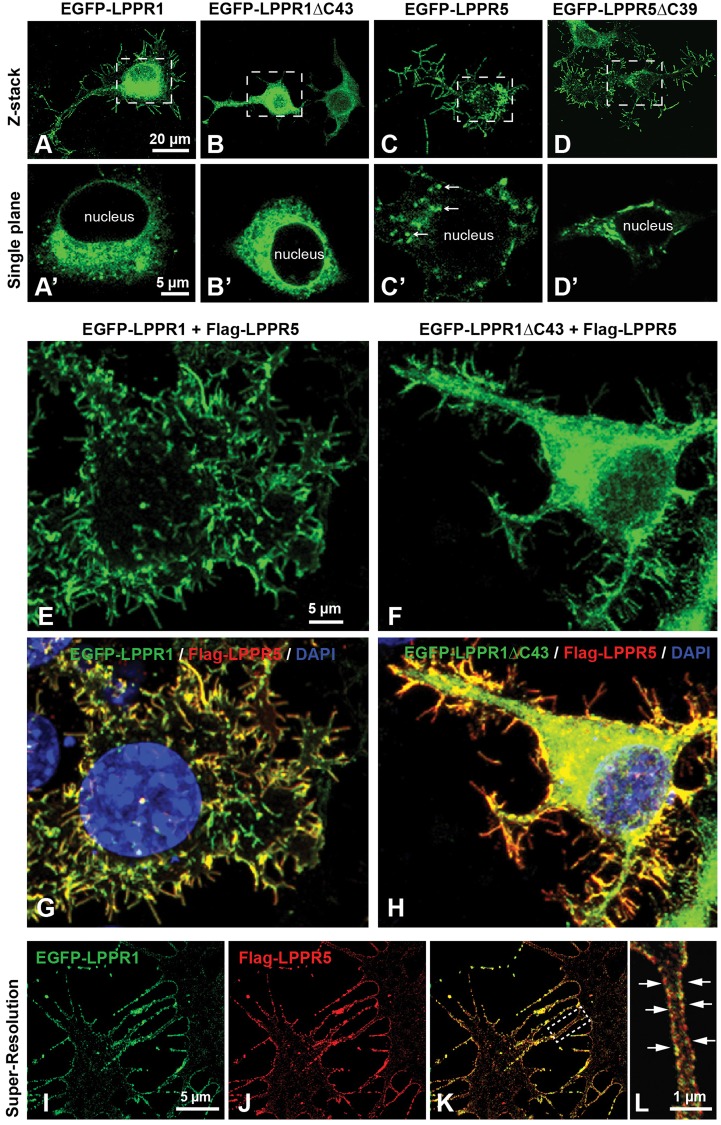
Colocalization of LPPR members at plasma membrane protrusions
To further characterize the interaction between LPPR members, LPPR1 or LPPR1ΔC43 was co-expressed with LPPR5 and confocal microscopy was used to analyze the 3D distribution as well as colocalization. As demonstrated in Fig. 1, when LPPR1 alone was overexpressed in Neuro2A cells, it localized to both intracellular membranes and plasma membrane protrusions (Fig. 5A,A′). Deletion of the C-terminus of LPPR1 abolished its membrane localization: LPPR1ΔC43 protein was mainly found in intracellular membrane compartments (Fig. 5B,B′). In contrast to LPPR1, LPPR5 protein showed a more pronounced localization in plasma membrane protrusions (Fig. 5C). LPPR5 was also seen in some vesicle-like structures inside the cells (Fig. 5C′), but, compared to LPPR1, LPPR5 showed a more exclusive plasma membrane distribution pattern. In addition, unlike LPPR1, deletion of the C-terminal portion of LPPR5 showed little effect on its protein localization. Similar to full-length LPPR5, the LPPR5ΔC39 protein was also present predominantly in plasma membrane protrusions (Fig. 5D) with some located intracellularly (Fig. 5D′).
Fig. 5.
Colocalization of LPPR5 and LPPR1 on plasma membrane protrusions. (A–D) Neuro2A cells were transfected with EGFP–LPPR1, EGFP–LPPR1ΔC43, EGFP–LPPR5 or EGFP–LPPR 5ΔC39 separately. Representative z-stack confocal images collected throughout the depth of cells showing the protein distribution of EGFP–LPPR1 (A), EGFP–LPPRΔC43 (B), EGFP–LPPR5 (C) or EGFP–LPPR5ΔC39 (D). LPPR1 was expressed in endomembrane systems as well as plasma membrane protrusions, and deletion of C-terminus of LPPR1 (LPPR 1ΔC43) abolished its localization towards plasma membrane protrusions. LPPR5 was expressed mainly in plasma membrane protrusions and in some vesicle-like structures inside the cells (arrows, in C′), C-terminal truncated LPPR5 (LPPR5ΔC39) displayed similar distribution pattern as full-length LPPR5. A′–D′ are high magnification single plane images of cell body areas boxed in A–D, respectively. (E–H) Neuro2A cells were co-transfected with EGFP–LPPR1 and Flag–LPPR5, or EGFP–LPPR1ΔC43 and Flag–LPPR5. Cells were stained with anti-Flag antibody and DAPI. Co-expression of LPPR1 with LPPR5, showing LPPR1 was colocalized with LPPR5 at plasma membrane protrusions (E,G). Co-expression of LPPR1ΔC43 with LPPR5, showing colocalization of LPPR1ΔC43 with LPPR5 and that some LPPR1ΔC43 can be targeted to plasma membrane protrusions (F,H). (I–L) STED super-resolution images showing the colocalization of LPPR1 with LPPR5 on plasma membrane protrusions. The boxed area in K is highlighted in L, showing a strong colocalization observed along the two opposed membranes of the filopodia (arrows).
Surprisingly, when LPPR5 was co-expressed with LPPR1, the distribution of LPPR1 became very similar to LPPR5 (i.e. a predominant localization to plasma membrane protrusions; Fig. 5E,G). This phenomenon was consistently observed after co-expressing EGFP–LPPR1 and Flag–LPPR5, LPPR1–HA and EGFP–LPPR5, or LPPR1–HA and Flag–LPPR5 in Neuro-2A cells (data not shown): all displayed a strong colocalization of LPPR1 with LPPR5 and a remarkable increase in membrane protrusions. Co-transfection of EGFP, HA or Flag control vector did not alter the overall distribution pattern of LPPR1 (data not shown). We next performed live-cell imaging of EGFP–LPPR1 and Flag–LPPR5 co-transfected cells stained with CellMask Deep Red, and again the images showed an obvious overlap of EGF–LPPR1 with CellMask plasma membrane staining (supplementary material Fig. S2A–C). The same result was also obtained in LPPR1–HA and EGFP–LPPR5 co-transfected cells (data not shown). Confocal image analysis further demonstrated that the protrusions in Neuro2A cells co-expressing EGFP–LPPR1 and Flag–LPPR5 mainly projected from the ventral cell surface downwards onto the culture plate (supplementary material Fig. S2). Supplementary material Fig. S2D,E shows the top view (x-y) and side views (x-z; y-z) of a representative z-stack of LPPR1 and LPPR5 co-transfected cells. Colocalization analysis using Imaris revealed that 88% of EGFP–LPPR1 colocalized with Flag–LPPR5 and 81% of Flag–LPPR5 colocalized with EGFP–LPPR1. From the 2D-fluorogram shown in supplementary material Fig. S2F, the distribution of pairs of pixel intensities (corresponding to EGFP–LPPR1 and Flag–LPPR5) aligned to a diagonal with a Pearson's colocalization coefficient of 0.81. A z-projection of serial z-stack images also revealed that the majority of the protrusions were located at the bottom of the cells towards the culture surface (supplementary material Fig. S2G,H). A shift of localization was also found for LPPR1ΔC43 when co-expressed with LPPR5. When alone, LPPR1ΔC43 showed an intracellular retention pattern (Fig. 1G; Fig. 5B); however, when co-expressed with LPPR5, a significant amount of LPPR1ΔC43 was found to be targeted to plasma membrane protrusions, although some still remained intracellularly (Fig. 5F,H; supplementary material Fig. S2I–L). Colocalization analysis showed that LPPR1ΔC43 was highly colocalized with LPPR5 with a Pearson's correlation coefficient of 0.85 (supplementary material Fig. S2M,N). However, different from co-expression of full-length LPPR1 and LPPR5, the distribution of which displayed a single cloud with diagonal alignment in the 2D-fluorogram (supplementary material Fig. S2F), the distribution of pairs of pixel intensities for LPPR1ΔC43 and LPPR5 was separated into two clouds in the 2D-fluorogram (supplementary material Fig. S2M). Interestingly, these two populations in the 2D-fluorogram correspond to the signals derived from plasma membrane protrusions and from the intracellular portion, respectively (supplementary material Fig. S2M,N). This indicates that LPPR5 could promote the trafficking of LPPR1ΔC43 from the endomembrane system to plasma membrane protrusions. However, as a result of the C-terminal deletion of LPPR1, a small portion of LPPR5 was also retained intracellularly together with LPPR1ΔC43.
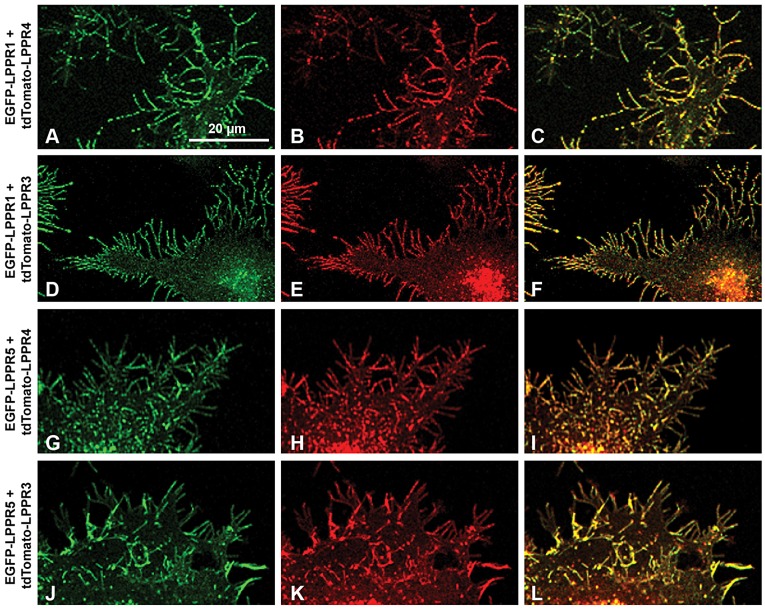
The colocalization of LPPR1 and LPPR5 was further assessed by stimulated emission depletion (STED) microscopy. A strong colocalization of EGFP–LPPR1 and Flag–LPPR5 was observed along the two opposed membranes of the filopodia (Fig. 5I–L). These data further suggest that LPPR1 and LPPR5 form complexes along the plasma membrane protrusions. The colocalization of LPPR1 with LPPR5 at membrane protrusions was also observed in human fibroblasts (supplementary material Fig. S3A–D). In addition, this colocalization was not affected by C-terminal deletion of LPPR5. When LPPR1–HA was co-expressed with EGFP–LPPR5ΔC39, they were still colocalized and present predominantly on membrane protrusions (supplementary material Fig. S3E–H). Likewise, a similar distribution shift was also found upon co-expression of LPPR5 with LPPR3 or LPPR4, and co-expression of LPPR1 with LPPR3 or LPPR4. Like LPPR1, LPPR3 or LPPR4 alone showed a more diffuse distribution pattern localized on both intracellular membranes and cell surface (supplementary material Fig. S3I,K). However, when co-expressed with any two of above LPPR members, they all showed a strong colocalization on plasma membrane protrusions (Fig. 6; supplementary material Fig. S3J,L).
Fig. 6.
Colocalization of LPPR1 family members exclusively at plasma membrane protrusions. Representative images of Neuro2A cells co-transfected with: (A–C) EGFP–LPPR1 and tdTomato–LPPR4; (D–F) EGFP–LPPR1 and tdTtomato–LPPR3; (G–I) EGFP–LPPR5 and tdTomato–LPPR4; and (J–L) EGFP–LPPR5 and tdTomato–LPPR3.
LPPR5 facilitates the localization of LPPR1 to plasma membrane protrusions resulting in enhanced induction of membrane protrusions
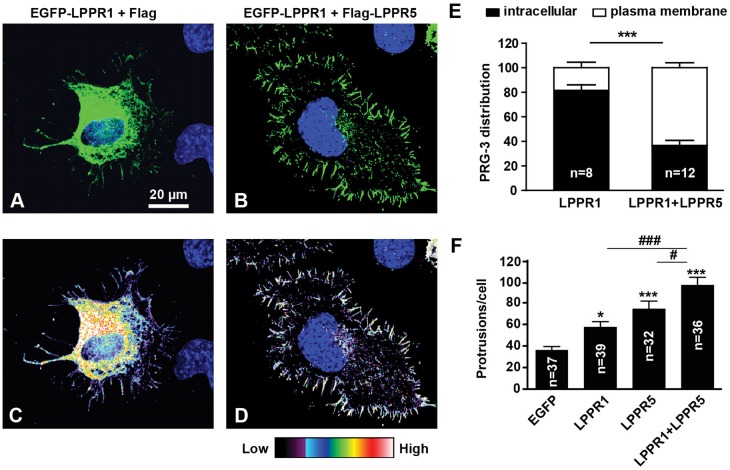
When Neuro2A cells are grown on uncoated surfaces, the protrusions induced by LPPRs extend in random directions, mostly towards the cell periphery and downwards onto the culture plate, which makes them very difficult to count (supplementary material Fig. S4A). In contrast, when plated onto poly-L-lysine (PLL) and laminin-coated surfaces, cells become flat and well spread, and protrusions are now generated predominantly away from the cell periphery (supplementary material Fig. S4B). This enabled us to separate the cell surface signal from the intracellular signal and also to more accurately count protrusions (Fig. 7). The majority of EGFP–LPPR1 was found inside the cells when expressed alone or with control vectors (Fig. 7A,C). However, when co-expressed with LPPR5, there was an increase in LPPR1 protein localized to the cell surface (Fig. 7B,D). The percentage of LPPR1 on the cell surface increased from 20% in control cells to 60% in cells that expressed both LPPR1 and LPPR5 (Fig. 7E).
Fig. 7.
LPPR5 facilitates the localization of LPPR1 to the plasma membrane, which further increases the number of plasma membrane protrusions. (A–E) Representative images and quantification of the distribution shift of LPPR1 by LPPR5. Neuro2A cells plated on laminin were transfected with EGFP–LPPR1 and Flag (A,C), or EGFP–LPPR1 and Flag–LPPR5 (B,D). C and D show the fluorescence intensity of EGFP–LPPR1 in a heat-map format. The percentages of intracellular LPPR1 and cell surface LPPR1 were measured and the quantification (mean±s.e.m.) is shown in E. More EGFP–LPPR1 was localized to plasma membrane when co-expressed with LPPR5. ***P<0.001 (two-way ANOVA and Bonferroni post test). (F) Coexpression of LPPR1 with LPPR5 produced an enhanced induction of plasma membrane protrusions. Results are mean±s.e.m. *P<0.05; ***P<0.001 compared to EGFP control group; #P<0.05 compared to LPPR5 group; ###P<0.001 compared to LPPR1 group (one-way ANOVA and Tukey's multiple comparison test).
Overexpression of either LPPR1 or LPPR5 alone has been reported to induce the formation of plasma membrane protrusions (Broggini et al., 2010; Sigal et al., 2007). We therefore evaluated whether the increased plasma membrane localization of LPPR1 as a result of co-expression with LPPR5 could further increase the formation of plasma membrane protrusions. Cells expressing either LPPR1 or LPPR5 indeed generated more protrusions than control transfected cells, and co-expression of LPPR1 and LPPR5 further increased the induction of protrusions as compared to cells expressing either alone (Fig. 7F). This reinforces the importance of cooperation between LPPR1 and LPPR5 in exerting their biological roles such as promoting cell protrusions.
LPPRs induced actin-dependent plasma membrane protrusion
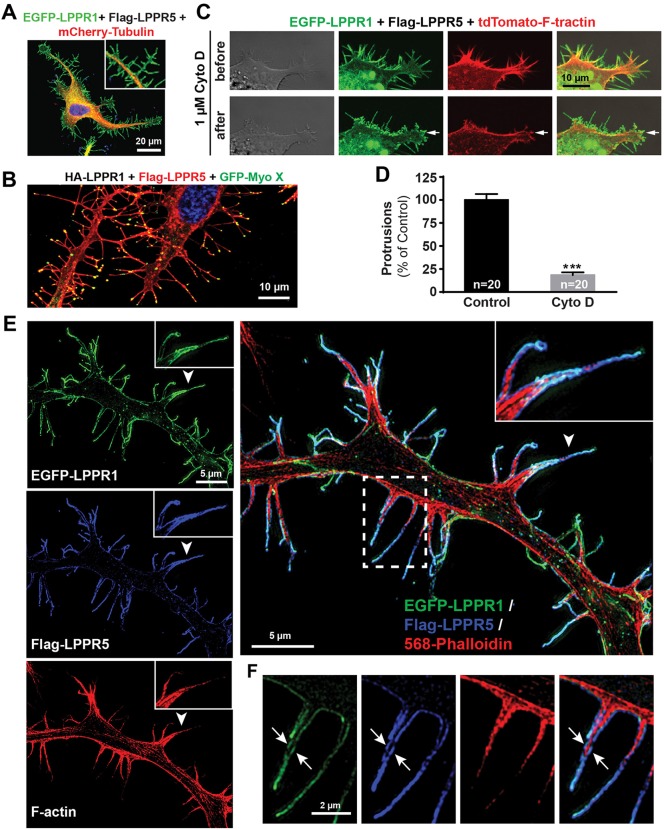
A previous report has shown that the protrusions induced by LPPRs are independent of the classic Cdc42–WASP–Arp2/3 pathway and do not require Ena/Vasp protein (Sigal et al., 2007), which made us wonder whether the protrusions are driven by tubulin. However, we found that tubulin is almost absent in the LPPR-labeled protrusions (Fig. 8A). GFP-tagged EB3 (also known as MAPRE3) was used to visualize microtubule assembly in living cells; we found that EB3 comets rarely moved into the LPPR-labeled protrusions (data not shown). In contrast, the actin-based motor protein myosin X was concentrated at the distal tip of almost every LPPR-labeled protrusion (Fig. 8B). Moreover, treatment of the cells with cytochalasin D resulted in a rapid retraction of the membrane protrusions induced by LPPR1 and LPPR5 (Fig. 8C,D). Structured illumination microscopy (SIM) imaging allowed visualization with a higher resolution of F-actin filaments as well as the LPPR distribution along the protrusions, obtained from cells co-transfected with EGF–LPPR1 and Flag–LPPR5. Fig. 8E,F shows clearly that actin filaments in the protrusions were wrapped by LPPR1 and LPPR5 which were co-distributed along the two opposed membranes of the protrusions. These data indicate that the protrusions induced by LPPRs are dependent on actin.
Fig. 8.
Characterization of the plasma membrane protrusions induced by LPPRs. (A) Representative image of Neuro2A cells co-transfected with EGFP–LPPR1 and Flag–LPPR5 together with Cherry–tubulin, showing the absence of microtubules in the LPPR-induced protrusions. (B) Representative image of Neuro2A cells co-transfected with EGFP–LPPR1 and Flag–LPPR5 together with GFP–myosin-X, showing the localization of myosin X at the tips of the LPPR-induced protrusions. (C) Live-cell imaging showing retraction of LPPR-induced plasma membrane protrusions after treatment with 1 µM cytochalasin D. (D) Quantification of protrusions after cytochalasin D treatment. (E,F) SIM super-resolution images demonstrating the presence of actin filaments labeled by phalloidin (red) inside membrane protrusions labeled for LPPR1 (green) and LPPR5 (blue). The arrowhead indicates the area that is enlarged in the inserts. The dashed boxed area in E is highlighted in F showing actin filaments (red) surrounded by LPPR1 (green) and LPPR5 (blue) distributed along the two opposed membrane layers (arrows).
As we found interaction of mTOR with LPPR1 and moreover an increase in phosphorylation of S6 ribosomal protein by co-expression of LPPR proteins, we tested whether blockade of the mTOR pathway by rapamycin would alter the formation of protrusions induced by LPPR proteins. Treatment with rapamycin of LPPR1 and LPPR5 co-transfected cells had no obvious effect on LPPR protein distribution or protrusion formation, although the phosphorylation of S6 ribosomal protein was significantly reduced (supplementary material Fig. S4C,D), excluding mTOR as a signaling pathway for LPPR-induced protrusions. Thus, the exact mechanism by which LPPRs induce membrane protrusions and the potential roles of mTOR signaling downstream of LPPRs still requires further investigation.
DISCUSSION
In this study, with the aim of elucidating the function of LPPR1, we used affinity purification coupled to mass spectrometry to identify its interacting proteins. Analysis of the putative binding partners revealed that LPPR1 might be involved in multiple biological processes such as protein localization, protein catabolic process, transmembrane transport, lipid biosynthetic process and proteolysis. Although most LPPR members, including LPPR1 and LPPR5, show no phosphatase activity, some previous reports have indicated that LPPRs might modulate LPA-induced signaling (Brauer et al., 2003; Broggini et al., 2010; Savaskan et al., 2004; Sigal et al., 2007). Our results showed that several important enzymes involved in phospholipid metabolism were present in the LPPR1 protein complexes, which suggests that LPPRs are involved in the regulation of phospholipid signaling.
The most salient finding was that we identified interactions between LPPR1 and three other members of the LPPR family, LPPR3, LPPR4 and LPPR5. Co-expression of pairs of LPPR proteins provided further evidence of the strong binding and functional interaction between these LPPR members. The fact that the LPPR protein levels and their plasma membrane targeting are significantly increased when they are expressed in pairs is likely a result of increased protein stability through their interaction. This is reminiscent of the increased maturation and assembly of integrins when they are co-expressed (Heino et al., 1989). Further evidence for a functional consequence of these associations is that co-expression LPPR members increased the generation of membrane protrusions beyond that of any single LPPR family member. This is likely due to the higher level of protein expression as well as the increased protein translocation from intracellular compartments to the cell membrane. Overall, these data strongly support the idea that the LPPR proteins are acting as a complex.
Both LPPs and Drosophila LPP homologs have previously been reported to form oligomers. Although oligomerization seems to occur commonly among LPP superfamily members, some specificity does exist. The Drosophila LPP homologue Wunen forms only homodimers, but not heterodimers, with Wunen-2 or human LPP1 and LPP3 (Burnett et al., 2004), whereas LPP1, LPP2 and LPP3 form both homo- and hetero-oligomers (Long et al., 2008). The dimerization between Wunen monomers was prevented by point mutation of a catalytic site and required the C-terminal portion (Burnett et al., 2004). In case of LPPRs, both homo- and hetero-oligomers exist and their interactions are independent of their C-terminal portion. Moreover, we detected no interaction between LPPR1 and LPPs either by proteomics or co-immunoprecipitation.
Among the five LPPR family members, LPPR3 and LPPR4 have a long C-terminus whereas LPPR1, LPPR2 and LPPR5 have a very short C-terminus. The hydrophilic C-terminus of each LPPR member positioned at the cytoplasmic side is unique and does not show significant sequence or domain similarity to those in the other LPPRs (Brauer and Nitsch, 2008; Strauss and Brauer, 2013). The long C-terminal of LPPR4 is believed to play a role in intracellular interactions and signaling. The C-terminus of LPPR1 or LPPR5 has also been reported to be required for its biological function (Broggini et al., 2010; Sigal et al., 2007). Our results showed that the interactions of LPPR3, LPPR4 and LPPR5 with LPPR1 were independent of its C-terminus, as they bound to both full-length and C-terminally truncated LPPR1. This implied that each LPPR protein, as a subunit of the LPPR complex, might exert distinct roles through its unique C-terminal portion. However, the role each LPPR member plays in the complex still needs further investigation.
The question arises as to whether these interactions take place between the endogenous proteins. Unfortunately, due to the lack of high-quality commercial antibodies for the LPPR family proteins, we could not determine their endogenous localization patterns and interactions. Spurious results can result from overexpression studies, and thus we designed our methods to prevent this possibility. First, we fused LPPR protein to different tags (i.e. EGFP, tdTomato, Flag or HA) in order to exclude the possible interference caused by these tags, and empty vector controls were used in every experiment. Second, we tested the interaction of LPPR members in different cell types such as Neuro-2A cells, Cos-7 cells and human fibroblasts, as well as primary mouse cerebellar neurons (data not shown). Overall, we find very reproducible results using different tags or different cell types; the expression pattern of each LPPR fusion protein was also consistent with previous reports (Broggini et al., 2010; Sigal et al., 2007). Considering the strong interaction we detected in the overexpression system, and the association of endogenous LPPR members with recombinant LPPR1 detected by proteomics, we believe interactions between endogenous LPPR members do also occur within cells.
Our study identified mTOR as binding partner of LPPR1; we also detected an increase in phosphorylation of the S6 ribosomal protein, a downstream target of mTOR, which was induced upon co-expression of LPPR proteins. However, we found that inhibition of the mTOR pathway with rapamycin had no effect on the generation of protrusions in response to LPPR expression, although the amount of phosphorylated S6 ribosomal protein was vastly reduced. Most recently, thin fragile actin-rich membrane protrusions (termed ‘cytonemes’) have been produced in cells in response to the overexpression of the seven-transmembrane domain molecules Lgr4 and Lgr5 (Snyder et al., 2015). Lgr-induced protrusions share many common features with those we describe here, including being dependent upon membrane localization, sensitivity to actin-depolymerization and being terminated by myosin X. Cytonemes have been identified as organizers of signaling molecules (Roy et al., 2014) and it might be that the activation of the mTOR pathway is a result of, rather than a cause of, protrusion formation.
In summary, this study suggested that, instead of playing separate or redundant roles in cells, the LPPR protein family members can also act together as a complex to exert their cellular function, which leads us in a new direct for exploring the exact molecular properties of LPPR family proteins. Although the precise functions of LPPR family members remain to be elucidated, our data suggest that these proteins likely act in concert to carry out their biological effects.
MATERIALS AND METHODS
DNA constructs and cell cultures
LPPR1, LPPR3, LPPR4 and LPPR5 cDNA were amplified from a mouse brain cDNA library. Full-length LPPR1 (amino acids 1–325), LPPR1 lacking the last 43 or 19 amino acids of the C-terminal region (LPPR1ΔC43, amino acids 1–282; LPPR1ΔC19, amino acids 1–306) and the C-terminus of LPPR1 (LPPR1CT; amino acids 283–325) were fused into the pEGFP-C1 vector using the In-Fusion cloning system (Clontech, Mountain View, CA). A construct expressing C-terminal HA-tagged LPPR1 (LPPR1–HA) was kindly provided by Andrew J. Morris (University of Kentucky, Lexington, KY) (Sigal et al., 2007). EGFP-tagged full-length LPPR5 (EGFP–LPPR5), LPPR5 with a 39-amino-acid C-terminal truncation (EGFP–LPPR5ΔC39) and 3×Flag-tagged LPPR5 (Flag–LPPR5) constructs were obtained by cloning LPPR5 or LPPR5ΔC39 cDNA into pEGFP-C1 or pCMV-3×Flag. The Flag–LPPR3, Flag–LPPR4, tdTomato–LPPR3 and tdTomato–LPPR4 were constructed using the Gateway cloning system (Life Technologies, Frederick, MD). The pmCherry-Mem construct which fused mCherry to a membrane localization sequence (Mem–Cherry) was from Clontech. The tdTomato–F-tractin construct was a gift of Michael Schell (Johns Hopkins University, Baltimore, MD); the GFP–Myosin-X construct was a gift of Richard Cheney (University of North Carolina, Chapel Hill, NC); the mCherry–tubulin construct was a gift of Roger Tsien (University of California, San Diego, CA).
Neuro2A cells, Cos-7 cells, HEK293 cells or human fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were transfected with the indicated constructs using Lipofectamine LTX reagent (Life Technologies). Cells were fixed for imaging or cell lysates were collected for western blot analysis at 20–24 h after transfection. In some cases, Neuro2A cells were grown on PLL and laminin-coated (5 µg/ml) surfaces.
Affinity purification of LPPR1 and its binding proteins
Neuro2A cells growing on 10-cm dishes were transfected with pEGFP, pEGFP-LPPR1 or pEGFP-LPPR1ΔC43. After 24 h, cells were washed with PBS, followed by extraction with 0.5 ml/dish of lysis buffer [10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% NP-40 and 1× protease inhibitor cocktail (Calbiochem, La Jolla, CA)]. Cells were lysed for 15 min on a rotating device at 4°C and kept on ice for another 15 min. Cell extracts were centrifuged at 12,000 g for 15 min at 4°C to remove insoluble material. Affinity purification of EGFP fusion proteins and their interacting proteins were performed using Chromotek GFP-Trap agarose beads (Allele Biotechnology, San Diego, CA) according to the manufacturer's instructions. Briefly, two experiments were performed from two independent cultures. For the first experiment, the GFP-Trap beads were first washed three times with dilution buffer [10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA and 1× protease inhibitor cocktail] and resuspended in 400 µl of dilution buffer. The same volume of cell lysate was added into the beads and incubated with end-over-end rotation at 4°C for 2 h. Samples were then washed three times with 1 ml of washing buffer (0.5% NP-40 in dilution buffer). Protein complexes were eluted using 20 μl of 2× SDS sample buffer containing 5% β-mercaptoethanol and boiled for 10 min. For the second immunoprecipitation experiment, we increased both the detergent concentration (1% NP-40) and salt concentration (500 mM NaCl) in order to increase the stringency of the affinity purification.
In-gel digestion and LC-MS/MS analysis
The eluted proteins were separated using a 4–12% Bis-Tris gradient gel (Invitrogen) and stained with Coomassie Blue. Independent lanes corresponding to GFP-Trap immunoprecipitated protein complexes from extracts of cells transfected with pEGFP, pEGFP-LPPR1 or pEGFP-LPPR1ΔC43 were cut into 20 gel slices per lane and subjected to in-gel digestion. Briefly, individual gel slices were diced into small cube pieces and placed into 1.5 ml tubes. After destaining Coomassie Blue with 25 mM NH4HCO3 in 50% acetonitrile (ACN), the gel samples were reduced at 56°C for 30 min in 10 mM DTT in 25 nM NH4HCO3 followed by alkylation in the dark for 30 min with 55 mM iodoacetamide in 25 nM NH4HCO3 at room temperature. The gel pieces were dried out using a Speed Vac prior to trypsin digestion. A solution of 12.5 ng/μl trypsin in 25 mM NH4HCO3 was added in a volume just enough to cover the gel pieces. Trypsin digestion was performed overnight at 37°C. Sequential extraction of peptides from the gel was performed twice with 5% formic acid in 50% ACN. The extracts were pooled together and the organic solvent was dried off in a Speed Vac. The dried peptide extract was resuspended in 20 μl of 0.1% formic acid and desalted with μ-C18 ZipTips. The peptides bound to the ZipTip were eluted with 60% ACN in 0.1% formic acid solution. After drying in the Speed Vac, the peptide sample was resuspended in 0.1% formic acid and subjected for LC-MS/MS analysis.
LC-MS/MS was performed using an Eksigent nanoLC-Ultra 2D system (Dublin, CA) coupled to an Orbitrap Elite mass spectrometer (Thermo Scientific, San Jose, CA). The peptide sample was first loaded onto a Zorbax 300SB-C18 trap column (Agilent, Palo Alto, CA), and then separated on a reversed-phase BetaBasic C18 PicoFrit analytical column (New Objective, Woburn, MA) using a linear gradient of 5–35% B (buffer A, 0.1% formic acid in water; buffer B, 0.1% formic acid in acetonitrile). Eluted peptides were sprayed into the Orbitrap Elite equipped with a nano-spray ionization source. Survey mass spectrometry spectra were acquired in the Orbitrap, and data-dependent MS/MS scans were performed in the linear ion trap with dynamic exclusion.
Database search and bioinformatics
Raw data files generated from the Orbitrap Elite were searched by the MASCOT search engine using the Proteome Discoverer software (ver.1.4, Thermo Scientific). Protein identifications were assigned by searching against the mouse UniprotKB/SwissProt database (release date, 2 June 2013; number of entries, 16648). Peptide identifications were restricted to tryptic peptides with no more than two miscleavages. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine, N-acetylation as well as deamidation of asparagine and glutamine were searched as variable modifications. Database searches were performed with a peptide precursor ion tolerance of 25 ppm and a MS/MS fragment mass tolerance of 0.8 Da. Peptide-spectrum matches (PSMs) were filtered to achieve an estimated false discovery rate (FDR) of 1% based on a target-decoy database search strategy.
Co-immunoprecipitation and western blot analysis
To validate the interaction between LPPR1 and other LPPR members, EGFP–LPPR1 or EGFP–LPPR1ΔC43 were co-transfected with Flag-tagged LPPR3, LPPR4 or LPPR5 in Neuro2A cells, the cell lysates were collected and immunoprecipitated either with EZview Red ANTI-FLAG M2 Affinity Gel (Sigma) or with Chromotek GFP-Trap agarose beads (Allele Biotechnology). Empty EGFP and Flag vectors were used as controls. The precipitated products were blotted with chicken anti-GFP antibody (1:2000; Abcam) or anti-Flag M2 antibody (1:1000; Sigma). To determine the interaction of LPPR1 with LPP1 and LPP2, the EGFP- or EGFP–LPPR1-transfected HEK293 cell lysate was subjected to co-immunoprecipitation using GFP-Trap beads and the eluted proteins were blotted with rabbit anti-LPP1 antibody (1:250; Exalpha Biologicals, Shirley, MA) or with rabbit anti-LPP2 antibody (1:500; Exalpha Biologicals). To validate the interaction of LPPR1 with mTOR and PTEN, the EGFP, EGFP–LPPR1 or EGFP-LPPR1ΔC43 transfected cell lysate was subjected to co-immunoprecipitation using GFP-Trap beads and the eluted proteins were blotted with rabbit anti-mTOR antibody (1:1000; Cell Signaling Technology, Beverly, MA) or with rabbit anti-PTEN antibody (1:1000; Cell Signaling Technology). To access the alteration in LPPR protein levels after co-expression of two LPPR members, the co-transfected cell lysates were blotted with chicken anti-GFP antibody (1:2000; Abcam), anti-Flag M2 antibody (1:1000; Sigma) or rabbit monoclonal anti-phospho-S6 ribosomal protein (Ser235/236) antibody (1:1000; Cell Signaling Technology).
Confocal microscopy
Neuro2A cells transfected with the indicated constructs were fixed with 4% PFA and stained with one of the following antibodies: rabbit anti-HA antibody (1:200; Santa Cruz Biotechnology), mouse anti-Flag antibody (1:1000) or rabbit anti-calreticulin (1:100; Millipore). Stacks of images were collected using a Zeiss LSM510 or a Zeiss LSM780 confocal microscope (Carl Zeiss MicroImaging, Jena, Germany) with a 63×1.4 NA oil immersion objective. To address the relationship between LPPR1 and LPPR5, stacks of images collected throughout the depth of cell were analyzed in 3D with Imaris software v7.6.3 (Bitplane, Belfast, UK). Quantification of colocalization was assessed in 3D, and pixel co-distribution was calculated for green and red staining patterns. Colocalized pixels were displayed as a two-color histogram (scattergram-fluorogram). 2D fluorograms represent quantification of the colocalization as a distribution of pairs of pixel intensities (aligned to a diagonal indicates perfect colocalization; randomly scattered, or towards one channel indicates lack of colocalization). In addition Pearson coefficients in the colocalized volume were computed and compared (1, perfect correlation; 0, no correlation; −1, perfect inverse correlation). To quantify the distribution shift of EGFP–LPPR1 after co-expressing with Flag–LPPR5, cells growing on laminin-coated surfaces were co-transfected with EGFP–LPPR1 and Flag, or with EGFP–LPPR5 and Flag–LPPR5. Stacks of images were obtained and the intracellular and total fluorescence intensity of EGFP–LPPR1 for individual cell was measured using ImageJ (Schneider et al., 2012). The percentages of intracellular and cell surface LPPR1 were calculated based on the intracellular EGFP–LPPR1 fluorescence intensity and the total EGFP–LPPR1 fluorescence intensity.
Super-resolution microscopy
To further assess the colocalization of LPPR family members on the plasma membrane protrusions, super-resolution microscopy was performed using stimulated emission depletion (STED) methodology. Neuro2A cells co-transfected with EGFP–LPPR1 and Flag–LPPR5 were stained with mouse anti-Flag primary antibody followed by a Cy3-conjugated F(ab′)2 fragment donkey anti-mouse-IgG antibody. Two-color STED images were obtained using a Leica SP8 STED 3X system (Leica Microsystems), equipped with a white light laser and a 592 nm and 660 nm STED depletion lasers. A 100×1.4 NA oil immersion objective lens (HCX PL APO STED white, Leica Microsystems, Mannheim, Germany) was used for imaging. To better visualize the actin filaments within the LPPR-induced membrane protrusions, super-resolution microscopy was performed using 3D structured illumination (3D-SIM). Neuro2A cells co-transfected with EGFP–LPPR1 and Flag–LPPR5 were stained with anti-Flag antibody followed by Alexa-Fluor-647-conjugated goat anti-mouse-IgG secondary antibody, and with Alexa-Fluor-568–phalloidin to label the F-actin. 3D-SIM microscopy was performed using the Delta-Vision OMX V4 (GE Healthcare, Issaquah, WA) imaging system. Images were taken with an Olympus PlanApo N 60×1.42 NA oil objective. Stacks of z-sections were taken over a cell thickness at a spacing of every 125 nm. The microscope was calibrated prior to experiments to calculate both the lateral and axial limits of image resolution under our experimental conditions. All raw images were processed and reconstructed in 3D using DeltaVison SoftWoRx software.
Quantification of membrane protrusions
Cos-7 cells growing on plain glass coverslips or Neuro2A cells growing on laminin-coated coverslips were transfected with the indicated control or LPPR constructs. For Cos-7 cells, a membrane-targeting mCherry (mem–mCherry) construct was co-transfected for visualizing the overall cell morphology and to allow counting of protrusions. The density of protrusions along the edge of each cell was calculated. For Neuro2A cells, cells were stained with phalloidin to label F-actin, and the total number of F-actin-rich protrusions for each cell was counted. Data were collected from at least three independent experiments and are presented as mean±s.e.m. Statistical analysis was performed using one-way ANOVA and Tukey's multiple comparison test. The level of significance was set at P<0.05.
Live cell imaging
Neuro2A cells co-transfected with pEGFP-LPPR1 and pCMV-Flag-LPPR5 were stained with a plasma membrane staining dye (CellMask deep red, Invitrogen) in living cells 24 h after transfection and live-cell images were acquired with a 63×1.4 NA oil immersion objective using a Leica SP5 confocal system (Leica Microsystems). In other cases, Neuro2A cells were co-transfected with pEGFP-LPPR1 and pCMV-Flag-LPPR5 together with tdTomato–F-tractin to label the actin filaments. The effect of actin polymerization on the motility of LPPR-induced plasma membrane protrusions was tested by adding 1 µM cytochalasin D. Time-lapse images of the same area were taken before and after adding cytochalasin D using a Zeiss LSM510 confocal microscope.
Supplementary Material
Acknowledgements
We thank Dr Guanghui Wang and Dr Marjan Gucek (NHLBI Proteomics Core Facility) for mass spectrometry assistance and Dr Aviva Symes (USUHS) for valuable advice. We also thank Dr Andrew J. Morris (University of Kentucky) for providing the HA-LPPR1 construct.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
P.Y. was responsible for experimental design, data acquisition, data interpretation and manuscript writing. C.A., D.M. and X.W. were responsible for data acquisition. Y.K. and J.H. were responsible for data interpretation and critical manuscript revision. H.G. was responsible for study conception and design, data interpretation and manuscript writing. All authors read and approved the final manuscript.
Funding
This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health. C.A. was supported by the National Instiutes of Health [grant numbers R01 NS064250-05 to Dr Jeffrey S. Urbach and T32 NS041218 to Georgetown University]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.169789/-/DC1
References
- Berggard T., Linse S. and James P. (2007). Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833-2842. 10.1002/pmic.200700131 [DOI] [PubMed] [Google Scholar]
- Brauer A. U. and Nitsch R. (2008). Plasticity-related genes (PRGs/LRPs): a brain-specific class of lysophospholipid-modifying proteins. Biochim. Biophys. Acta 1781, 595-600. 10.1016/j.bbalip.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Brauer A. U., Savaskan N. E., Kuhn H., Prehn S., Ninnemann O. and Nitsch R. (2003). A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat. Neurosci. 6, 572-578. 10.1038/nn1052 [DOI] [PubMed] [Google Scholar]
- Brindley D. N. (2004). Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J. Cell Biochem. 92, 900-912. 10.1002/jcb.20126 [DOI] [PubMed] [Google Scholar]
- Broggini T., Nitsch R. and Savaskan N. E. (2010). Plasticity-related gene 5 (PRG5) induces filopodia and neurite growth and impedes lysophosphatidic acid- and nogo-A-mediated axonal retraction. Mol. Biol. Cell 21, 521-537. 10.1091/mbc.E09-06-0506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C., Makridou P., Hewlett L. and Howard K. (2004). Lipid phosphate phosphatases dimerise, but this interaction is not required for in vivo activity. BMC Biochem. 5, 2 10.1186/1471-2091-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr S., Aebersold R., Baldwin M., Burlingame A., Clauser K. and Nesvizhskii A. (2004). The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol. Cell Proteomics 3, 531-533. 10.1074/mcp.T400006-MCP200 [DOI] [PubMed] [Google Scholar]
- Coiro P., Stoenica L., Strauss U. and Brauer A. U. (2014). Plasticity-related gene 5 promotes spine formation in murine hippocampal neurons. J. Biol. Chem. 289, 24956-24970. 10.1074/jbc.M114.597880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free R. B., Hazelwood L. A. and Sibley D. R. (2009). Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr. Protoc. Neurosci. Chapter 5, Unit 5 28 10.1002/0471142301.ns0528s46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaaya A., Poirier O., Mougenot N., Hery T., Atassi F., Marchand A., Saulnier-Blache J.-S., Amour J., Vogt J., Lompre A.-M. et al. (2012). Plasticity-related gene-1 inhibits lysophosphatidic acid-induced vascular smooth muscle cell migration and proliferation and prevents neointima formation. Am. J. Physiol. Cell Physiol. 303, C1104-C1114. 10.1152/ajpcell.00051.2012 [DOI] [PubMed] [Google Scholar]
- Heino J., Ignotz R. A., Hemler M. E., Crouse C. and Massague J. (1989). Regulation of cell adhesion receptors by transforming growth factor-beta. Concomitant regulation of integrins that share a common beta 1 subunit. J. Biol. Chem. 264, 380-388. [PubMed] [Google Scholar]
- Long J. S., Pyne N. J. and Pyne S. (2008). Lipid phosphate phosphatases form homo- and hetero-oligomers: catalytic competency, subcellular distribution and function. Biochem. J. 411, 371-377. 10.1042/BJ20071607 [DOI] [PubMed] [Google Scholar]
- Roy S., Huang H., Liu S. and Kornberg T. B. (2014). Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science 343, 1244624 10.1126/science.1244624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaskan N. E., Brauer A. U. and Nitsch R. (2004). Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur. J. Neurosci. 19, 212-220. 10.1046/j.1460-9568.2003.03078.x [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal Y. J., McDermott M. I. and Morris A. J. (2005). Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem. J. 387, 281-293. 10.1042/BJ20041771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal Y. J., Quintero O. A., Cheney R. E. and Morris A. J. (2007). Cdc42 and ARP2/3-independent regulation of filopodia by an integral membrane lipid-phosphatase-related protein. J. Cell Sci. 120, 340-352. 10.1242/jcs.03335 [DOI] [PubMed] [Google Scholar]
- Snyder J. C., Rochelle L. K., Marion S., Lyerly H. K., Barak L. S. and Caron M. G. (2015). Lgr4 and Lgr5 drive the formation of long actin-rich cytoneme-like membrane protrusions. J. Cell Sci. 128, 1230-1240. 10.1242/jcs.166322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss U. and Brauer A. U. (2013). Current views on regulation and function of plasticity-related genes (PRGs/LPPRs) in the brain. Biochim. Biophys. Acta 1831, 133-138. 10.1016/j.bbalip.2012.08.010 [DOI] [PubMed] [Google Scholar]
- Trimbuch T., Beed P., Vogt J., Schuchmann S., Maier N., Kintscher M., Breustedt J., Schuelke M., Streu N., Kieselmann O. et al. (2009). Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell 138, 1222-1235. 10.1016/j.cell.2009.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmans T., Battefeld A., Geist B., Farres A. S., Strauss U. and Brauer A. U. (2013). Plasticity-related gene 3 promotes neurite shaft protrusion. BMC Neurosci. 14, 36 10.1186/1471-2202-14-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Pisitkun T., Wang G., Wang R., Katagiri Y., Gucek M., Knepper M. A. and Geller H. M. (2013). Global analysis of neuronal phosphoproteome regulation by chondroitin sulfate proteoglycans. PLoS ONE 8, e59285 10.1371/journal.pone.0059285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.