Abstract
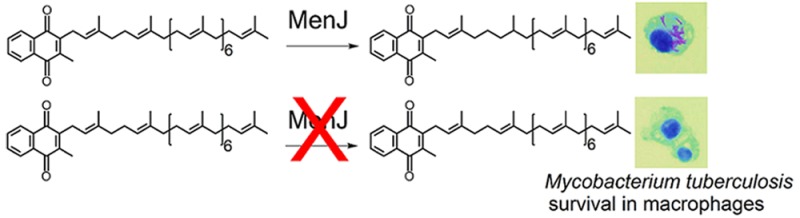
Menaquinone (MK) with partially saturated isoprenyl moieties is found in a wide range of eubacteria and Archaea. In many Gram-positive organisms, including mycobacteria, it is the double bond found in the β-isoprene unit that is reduced. Mass spectral characterization of menaquinone from mycobacterial knockout strains and heterologous expression hosts demonstrates that Rv0561c (designated menJ) encodes an enzyme which reduces the β-isoprene unit of menaquinone in Mycobacterium tuberculosis, forming the predominant form of menaquinone found in mycobacteria. MenJ is highly conserved in mycobacteria species but is not required for growth in culture. Disruption of menJ reduces mycobacterial electron transport efficiency by 3-fold, but mycobacteria are able to maintain ATP levels by increasing the levels of the total menaquinone in the membrane; however, MenJ is required for M. tuberculosis survival in host macrophages. Thus, MK with partially hydrogenated isoprenyl moieties represents a novel virulence factor and MenJ is a contextually essential enzyme and a potential drug target in pathogenic mycobacteria and other Gram-positive pathogens.
Short abstract
Rv0561c (MenJ) reduces the β-isoprene unit of menaquinone in Mycobacterium tuberculosis, which increases electron transport efficiency and is required for bacterial survival in macrophages.
Introduction
Quinones play key roles in the respiratory electron transport systems (ETS) of both prokaryotes and eukaryotes. These lipids shuttle electrons between the membrane-bound protein complexes acting as electron acceptors and donors (Figure 1).1,2 Quinones can be divided into two major structural groups: ubiquinones (or benzoquinones, UQ) and menaquinones (or naphthoquinones, MK).3 Each of these molecules contains an isoprenyl side chain of varying length and degree of regiospecific saturation (reduction or hydrogenation of double bonds). Indeed, the structures of quinones are often species specific and have been used for taxonomic differentiation of bacteria for more than five decades.4,5 The capacity to utilize lipoquinone structure for taxonomy clearly suggests conservation of function, in addition to the conservation of the structural elements of these molecules.
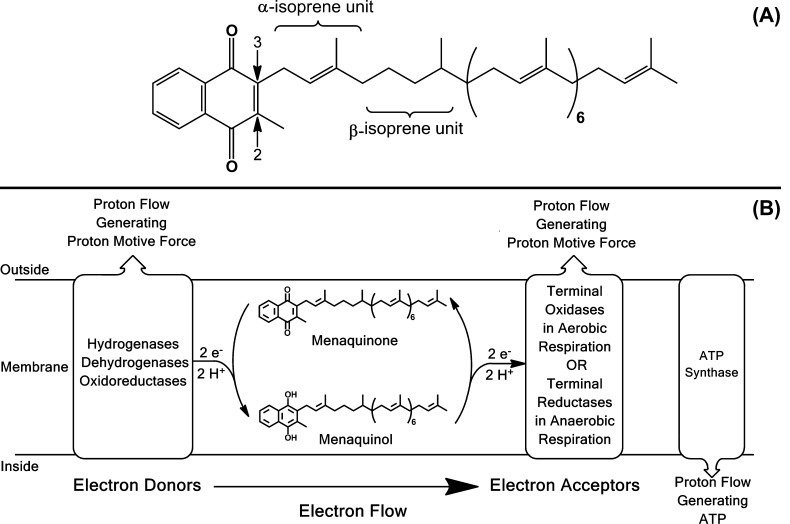
Figure 1.
Structure and role of mycobacterial menaquinone. (A) Mycobacterial menaquinone has 9 isoprene units with the one in the β-position hydrogenated as previously determined7 and is designated MK-9(II-H2) following the IUPAC-IUB recommendations for nomenclature of quinones with isoprenoid side chains (http://www.chem.qmul.ac.uk/iupac/misc/quinone.html). The arrows indicate carbons 2 and 3 of the naphthoquinone ring. (B) The role of menaquinone in mycobacterial respiration and oxidative phosphorylation. In respiration, menaquinone accepts electrons from a variety of electron donors and transfers them to terminal oxidases or reductases. Subsets of the electron donors and acceptors pump protons to the outside of the cell generating proton motive force that can be utilized to generate ATP via oxidative phosphorylation. The energetics of mycobacteria respiration and oxidative phosphorylation have been reviewed recently.56
The specific structural elements of MK play significant roles in altering oxidative phosphorylation.6 Modifications of the naphthoquinone moieties (Figure 1) have been most widely studied. MK molecules typically have a methyl group in the C-2 position of the naphthoquinone and an unsaturated isoprenyl side chain of at least 5 carbon atoms in the C-3 position as seen in Figure 1. Compounds substituted with a hydroxyl group in the C-2 position, or compounds with a non-isoprenoid substituent in the C-3 position, such as the methyl group in dimethylmenadione, allow oxidation but not phosphorylation.6 MK with partially saturated isoprenyl moieties are found in a wide range of bacterial species. Unambiguous localization of the saturated isoprene unit in the second (β) position and the assignment of the absolute configuration of the asymmetric center created in the side chain by the saturation were determined using mass spectrometry (MS) coupled with chemical degradation in studies of authentic samples isolated from mycobacteria and synthetic standards.7 In Gram-positive organisms the reduced double bond is often in the β-isoprene unit, as in mycobacteria, or more distal from the naphthoquinone moiety as in some Gram-negative organisms.4 The role and importance of this modification of quinones has remained enigmatic since its first description 50 years ago;8 however, some reports suggest that small structural changes can cause dramatic changes in the properties of the molecules.9,10
Mycobacteria, like many Gram-positive bacteria, use only MK in their electron transport systems. In mycobacteria, this molecule is predominantly MK-9(II-H2),7,11,12 which contains an isoprenyl side chain of nine isoprene units with the double bond of the one in the β-position hydrogenated (saturated) (Figure 1).11 It has also been reported that the composition of the MK pool in Mycobacterium tuberculosis provides a mechanistic link between the respiratory state of the bacilli and its response to hypoxia.13 Experiments with partially saturated and unsaturated analogues of MK-9(II-H2) suggested that these MK derivatives have roles in signaling and bacterial survival. Thus, identification of the gene encoding the enzyme responsible for hydrogenating a double bond in the isoprenyl side chain represents a significant step in understanding MK synthesis and the role of this modification in M. tuberculosis and other bacteria. This manuscript describes members of a family of enzymes that catalyze the reduction of the side chains of MK, the biological significance of the saturation of the β-isoprene unit of mycobacterial MK, and regulation of MK levels in obligate aerobes. Importantly, saturation of the β-isoprene unit of menaquinone appears to be a virulence factor for a pathogen that kills 1,300,000 people per year.14
Results
Identification of a Potential Menaquinone Reductase
Although little was known about the enzymes involved in the modification of the isoprenyl moiety of MK in bacteria, hydrogenation of isoprene units in structural lipids with isoprenyl moieties has been reported in Archaea, bacteria, and plants.15−17 In this case the hydrogenation reaction is catalyzed by multifunctional geranylgeranyl oxidoreductases (GGRs), which generally saturate the entire isoprenyl moieties of bacteriochlorophyll, chlorophyll, tocopherols, phylloquinone, and Archaeal phospholipids in a stepwise fashion.15−17 Thus, it seemed possible that a menaquinone reductase could have similarity to GGRs at the amino acid level. BLAST searches were conducted using the amino acid sequence of the GGR involved in phospholipid synthesis in Archaeoglobus fulgidus (NCBI GI number: 206557944). These searches resulted in the identification of a putative open reading frame in the M. tuberculosis genome designated Rv0561c, encoding a protein with low (28%) identity with the query GGR. Rv0561c is annotated as a possible oxidoreductase and was predicted to be essential for survival in primary murine macrophages.18,19 In addition, the protein is predicted to have 409 amino acids with molecular mass of ∼43 kDa and isoelectric point 9.08. Rv0561c is not predicted to have trans-membrane domains; however, proteolytic fragments were identified in a cell-wall enriched fraction isolated from M. tuberculosis.20 The protein is very similar to a putative FAD-linked oxidoreductase encoded by Mycobacterium leprae, a putative oxidoreductase from Streptomyces coelicolor, and a bacteriochlorophyll synthase from A. fulgidus. A protein with 74% identity, encoded by MSMEG1132, also predicted to be FAD binding domain-containing, was identified in Mycobacterium smegmatis mc2 155, and subsequent protein sequence alignments indicated that the protein is highly conserved throughout the mycobacteria (Figure S1).
Mass Spectral Analysis of Menaquinone Isolated from Escherichia coli Cells Expressing MSMEG1132
PCR amplified MSMEG1132 from M. smegmatis mc2 155 was cloned in the pET28a(+) expression vector. E. coli BL21 (DE3) pLysS strains (transformed with empty vector or vector containing MSMEG1132) were grown in LB medium at 37 °C, with shaking, to an OD600 of 0.8. Protein expression was induced with 1 mM IPTG overnight, the cells were harvested by centrifugation, and lipids were extracted. Neutral lipids (containing MK) were analyzed by HPLC coupled to mass spectrometry (LCMS) or tandem MS (LCMS/MS). To perform a detailed structural characterization, a Q-TOF MS was used to study the naphthoquinone ring structures of the MK molecules and an LCQ ion-trap MS was used for analysis of the isoprenyl side chains.21 Identification of MK was accomplished by chemical ionization mass spectrometry with observation of protonated [M + H]+ ions and verification by LCMS/MS. The monoisotopic masses of the quinones (Table S1) were calculated using ChemDraw Ultra 12.0 software (PerkinElmer Informatics).
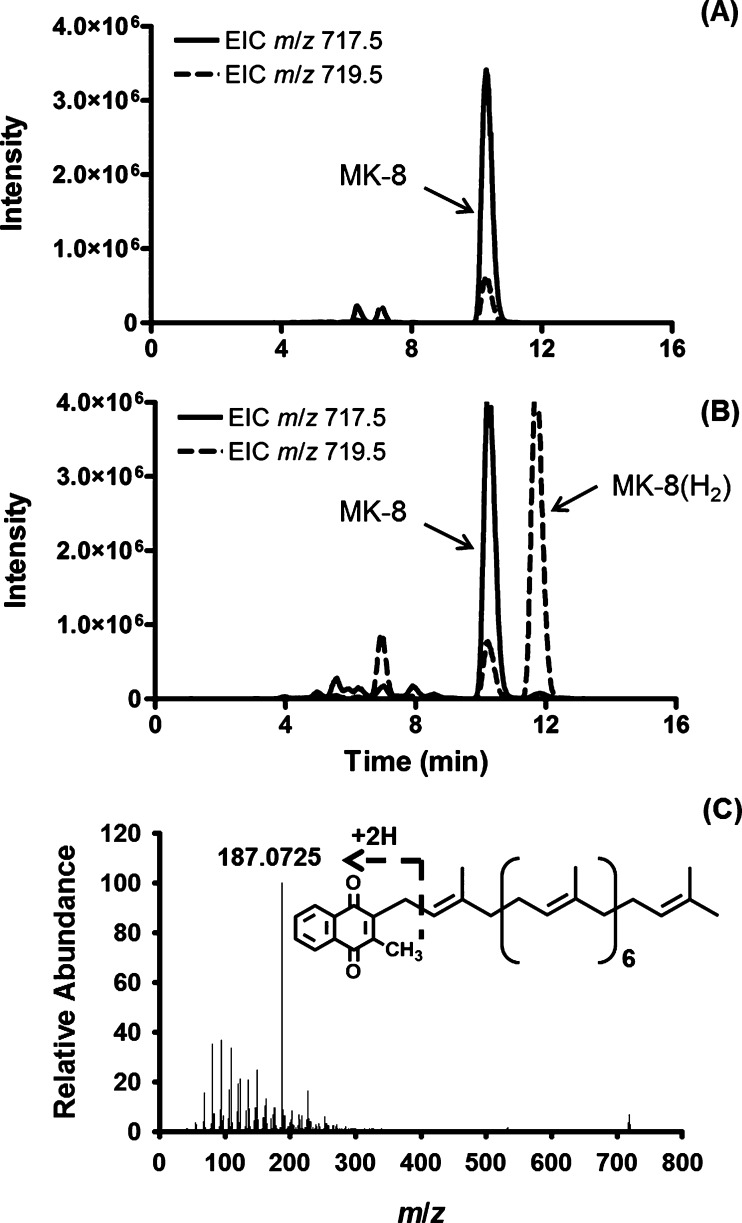
The wild type (WT) facultative anaerobe E. coli synthesizes mainly UQ-8 (ubiquinone with an isoprenyl side chain containing eight isoprene units) under aerobic conditions but produces increased amounts of MK-8 (Table S1) under anaerobic conditions.22 MK-8 from E. coli has 8 isoprene units that all contain a double bond; thus, the monoisotopic mass of this molecule is calculated at 716.5532 Da and the expected m/z value for [M + H]+ would be 717.5605 Da, or 719.5761 Da if a single isoprene unit was reduced. Extracted ion chromatograms (EIC) were generated from total ion chromatograms (TIC) by extracting collected data for ions with [M + H]+ at m/z 717.5 ± 0.5 or [M + H]+ at m/z 719.5 ± 0.5 for MK-8 or MK-8(H2), respectively (Figure 2). The lipids from the E. coli strain transformed with empty vector contained a strong peak with an observed m/z of 717.5512 and a retention time of 10.5 min confirming the presence of MK-8, as previously reported, but did not contain any lipids with m/z values that could be attributed to MK-8(H2) by LCMS/MS. The observed m/z 719.5664 peak coincident with m/z 717.5512 at a retention time of 10.5 min (Figure 2A) represents an isotope peak. However, lipids extracted from E. coli expressing MSMEG1132 contained a new peak, with a retention time of about 11.8 min and an observed m/z value of 719.5662 in addition to the peaks at 10.5 min indicating a gain of function and synthesis of compounds with [M + H]+ ions consistent with the presence of MK-8(H2) (Figure 2B).
Figure 2.
MSMEG1132 catalyzes the reduction of MK-8 when expressed in E. coli. Extracted ion chromatograms derived from Q-TOF LC/MS analysis of partially purified lipids from recombinant E. coli containing pET28a empty vector (A) or vector containing MSMEG1132 (B). Extracted ion chromatograms (EIC) were generated from total ion chromatograms (TIC) by extracting collected data for ions with [M + H]+ at m/z 717.5 ± 0.5 or m/z 719.5 ± 0.5 for MK-8 or MK-8(H2), respectively. (C) Representative tandem MS of ions having m/z 719.5664 identified as MK-8(H2) in panel B; the inset shows the inferred fragmentation generating the fragment ion at m/z of 187.0712.
This increase of ∼2 Da (717.5512 to 719.5662 Da) could, potentially, be attributed to reduction of the ketone functions in the naphthoquinone (as seen in MK’s role in the electron transport system), a double bond in the naphthoquinone moiety, or one of the double bonds in the isoprenyl side chain, which would be consistent with the action of an enzyme synthesizing MK-9(II-H2). Tandem MS of the ions having m/z values of 717.5509 and 719.5664 generated major fragment ions with an expected m/z of 187.0753 [m/z 187.0725 observed (Figure 2C)], which are diagnostic for the naphthoquinone moiety of MK.23 Thus, analysis by Q-TOF MS suggested that the reduction had occurred in the isoprenoid tail, not in the aromatic rings. This conclusion was supported by LCMS/MS analysis performed on the LCQ ion-trap instrument, which generated fragment ions with m/z values of 531.4 (in the vector control samples) and 533.4 (from E. coli expressing MSMEG1132) consistent with ions derived from the isoprenyl side chain of MK-8 or MK-8(H2), respectively (Figures S2A, S2B).
The data presented above strongly suggest that the protein encoded by MSMEG1132 is a reductase that saturates (reduces) a single double bond in the menaquinone of the heterologous host E. coli. A similar series of experiments were attempted in E. coli strains transformed with expression vectors harboring Rv0561c from M. tuberculosis; however, despite the sequence similarity with MSMEG1132, Rv0561c did not reduce the host menaquinone when expressed in E. coli.
Mass Spectral Analysis of Menaquinones from Mycobacteria
In mycobacteria, menaquinone is predominantly MK-9(II-H2),7,11,12 containing an isoprenyl side chain of nine isoprene units with the one in the β-position being reduced (saturated) (Figure 1). Thus, the calculated monoisotopic mass would be 786. 6315 Da and the expected m/z value for [M + H]+ would be 787.6387 Da. If the β-isoprene unit is unreduced, these values would decrease by ∼2 Da.
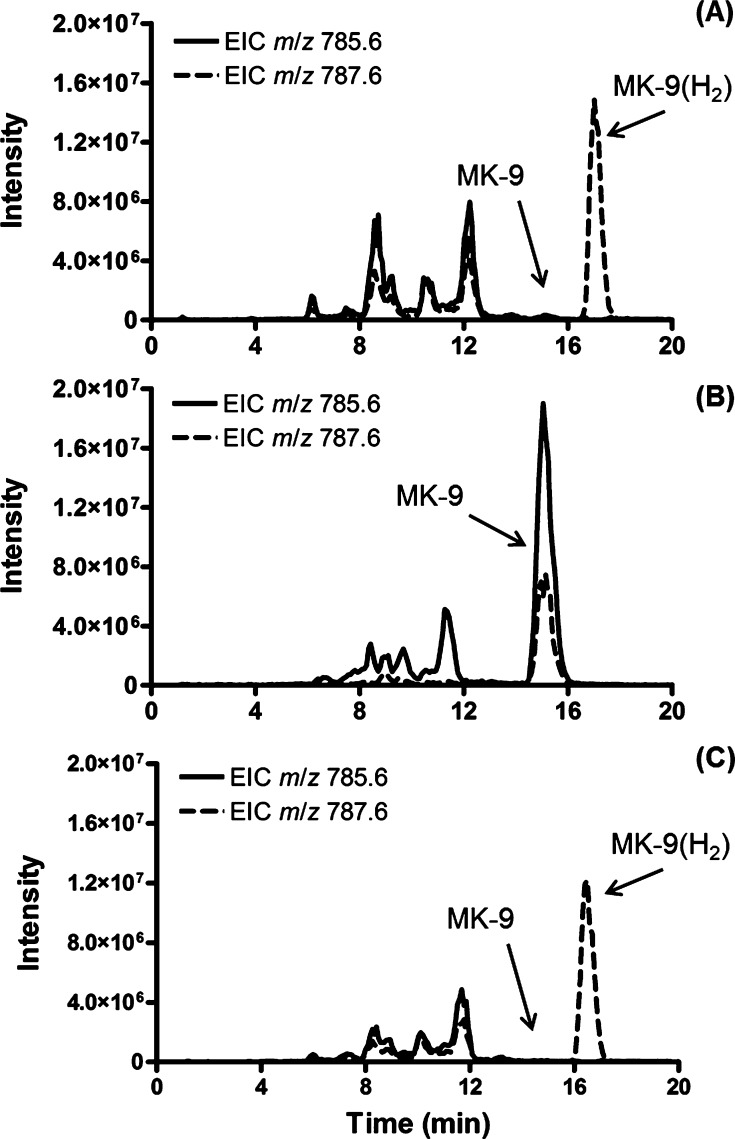
Extraction of TICs generated by LCMS analysis of lipids from M. smegmatis mc2 155 or M. tuberculosis H37Rv for ions with [M + H]+ at m/z 787.6 ± 0.5 or 785.6 ± 0.5 identified features corresponding to both MK-9(H2) (observed m/z 787.6356 at ∼17.5 min) and MK-9 (observed m/z 785.6263 at ∼15.0 min) (Figure 3A). In both cases, MK-9(H2) was dominant and much smaller amounts of MK-9 were evident. The identification of these molecules as MK-9(H2) and MK-9 was verified by the generation of diagnostic fragment ions23 at an observed m/z of 187.0766 (187.0753 calculated) by tandem MS on the Q-TOF instrument as described above. This conclusion was further verified by LCMS/MS analysis of MK-9(H2) ([M + H]+ at m/z 787.6) on an LCQ ion-trap instrument. The resulting spectra were complex, consisting of several series of ions (Figure S3). However, identification of an ion series that retains the naphthoquinone structure and differs by 68 Da (designated n2–n9) and an ion that retains only the isoprenoid moiety (designated i1) serves to identify and locate the reduction in the MK tail. Thus, the molecular ion (m/z 787.6), [M + H – H2O]+ (m/z 769.6) and the n-series ions (m/z 323.2, 391.3, 459.3, 527.3, 595.5, 663.5 and 731.6, respectively) all indicate that the molecule has a single reduced double bond near the naphthoquinone ring. The fragments i1 (m/z 601.5) and n2 (m/z 253.1) would identify the location of that reduction in the β-isoprene unit; however, the spectra are very complex and weak in the n2/n3 region, supporting previous reports that it is not possible to unambiguously assign the position of the reduced double bond by mass spectrometry or NMR analysis.7 Unambiguous localization of this double bond requires large amounts of highly purified material for chemical degradation studies.7,11
Figure 3.
Rv0561c catalyzes the reduction of MK-9 in M. tuberculosis. Extracted ion chromatograms derived from Q-TOF LC/MS analysis of partially purified lipids from WT M. tuberculosis (A), M. tuberculosis H37RvΔRv0561c (B), and M. tuberculosis H37RvΔRv0561c complemented with pNIP40bRv0561c (C). Extracted ion chromatograms (EIC) were generated from total ion chromatograms (TIC) by extracting collected data for ions with [M + H]+ at m/z 785.6 ± 0.5 or m/z 787.6 ± 0.5 for MK-9 or MK-9(H2), respectively.
Deletion of Rv0561c or MSMEG1132 Abrogates Menaquinone Saturation
To further establish the function of Rv0561c and orthologues, MSMEG1132 and Rv0561c knockout mutants were constructed in M. smegmatis mc2 155 and M. tuberculosis H37Rv, respectively. These mutants were generated through homologous recombination after transformation of WT M. smegmatis mc2 155 with pPR27KOMSMEG1132::kan and WT M. tuberculosis H37Rv with pPR27KORv0561c::kan (Table S2, Figure S4), resulting in strains designated as M. smegmatis ΔMSMEG1132 and M. tuberculosis H37RvΔRv0561c, respectively. Analysis of the lipids from both knockout (KO) strains showed complete abolition of the synthesis of MK-9(II-H2) accompanied by accumulation of MK-9 (Figure 3A, B). Complementing either strain with an integrative plasmid allowing expression of the WT Rv0561c gene [pNIP40bRv0561c (Table S3)] restored synthesis of MK-9(H2) and depleted the pool of MK-9 (Figure 3C), clearly demonstrating that MSMEG1132 and Rv0561c encode proteins with analogous function.
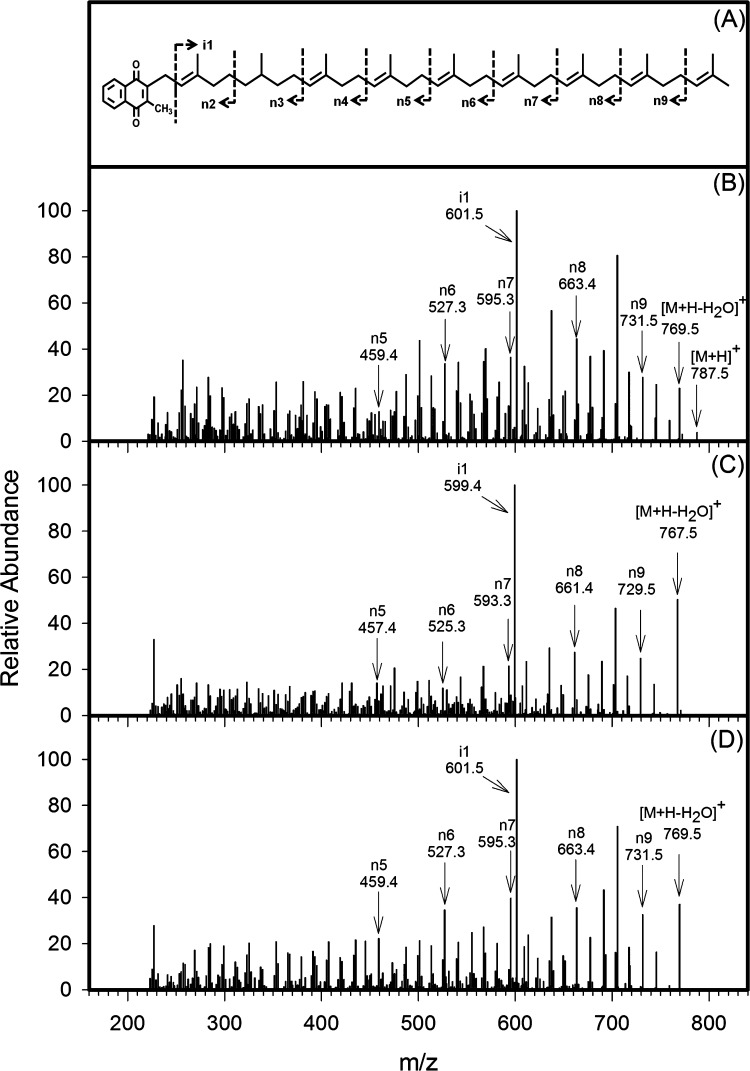
LCMS/MS analysis of the MK-9(H2) peak at 17.5 min and MK-9 peak at 15 min identified in Figure 3 generated diagnostic fragment ions at m/z 187.0766 (187.0754 calculated) by tandem MS on the Q-TOF instrument, again indicating that the modification was in the isoprenoid tail. Further analysis using the ion-trap instrument supported this conclusion. As shown in Figure 4A, MK-9(II-H2) from WT M. tuberculosis H37Rv generated fragment ions n5–n9 and i1 with the predicted m/z values. The MK-9 present in M. tuberculosis H37RvΔRv0561c, however, had strong fragment ions in the n-series with m/z values of 457.4, 525.3, 593.3, 661.4, and 729.5 (Figure 4B). In addition, the i1 fragment had an m/z of 599.4. All of these values indicate that the isoprenoid tail of the MK-9 contained nine double bonds (was fully unsaturated). Analysis of the fragment ions of the complemented strain demonstrated fragment ions in the n-series with m/z values of 459.4, 527.3, 595.3, 663.4, and 731.5 (an increase of ∼2 Da in fragments n5–n9). In addition, the i1 fragment had an m/z of 601.5 (Figure 4C). Thus, the complemented strain contained MK-9(H2) with 8 double bonds in the isoprenoid tail, and the presence of fragments i1 (m/z 601.5) and n5 (m/z 459.4) strongly suggests that the reduced double bond is located near the naphthoquinone ring and the molecule is likely MK-9(II-H2) as previously determined.11
Figure 4.
Localization of the site of action of Rv0561c by tandem MS. LCQ ion-trap tandem mass spectra of ions identified as MK-9(H2) in Figure 3A (B); MK-9 in Figure 3B (C); and MK-9(H2) in Figure 3C (D). Diagnostic ions are indicated with the arrows, and the inferred fragmentation pattern is shown in panel A and Figure S3.
Phenotypic Analysis of M. tuberculosis H37ΔRv0561c and M. smegmatis ΔMSMEG1132
M. tuberculosis H37Rv, M. tuberculosis H37ΔRv0561c, and M. tuberculosis H37ΔRv0561c complemented with pNIP40bRv0561c had similar growth rates in aerated 7H9 medium as judged by OD600 or by enumeration of colony forming units (CFU). Similar results were observed for the M. smegmatis ΔMSMEG1132 strain when compared to WT M. smegmatis. Growth rates in hypoxic conditions using a modified Wayne model24 were also established. In this case, the strains were grown in sealed glass tubes containing 7H9 medium ± 1.5 μg/mL methylene blue to verify the conversion from an oxidizing to a reducing environment due to the consumption of oxygen. Again there was no difference in the growth rates of the bacterial strains as assessed by either OD600 or CFU. However, the rate of decolorization of methylene blue was 40% slower in the M. tuberculosis H37ΔRv0561c than in the M. tuberculosis H37Rv culture (0.001 ± 0.0006 OD670/min vs 0.0006 ± 0.00004 OD670/min) and 50% slower in the M. smegmatis ΔMSMEG1132 culture than in the WT M. smegmatis culture (0.01 ± 0.003 OD670/min vs 0.02 ± 0.003 OD670/min), suggesting possible changes in electron transport activity and/or coupling of oxidative phosphorylation.
Loss of Electron Transport Activity in Mycobacteria Is Compensated for by Increased Menaquinone Production
A tetrazolium salt, 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride (INT), was utilized to evaluate electron transport efficiency (AETS). This dye is reduced by electron transport through ETS components prior to and including the quinones in bacteria.25 Cell homogenates were prepared from both WT and ΔMSMEG1132 strains of M. smegmatis and assayed for ability to reduce INT in the presence of succinate, NADPH, and NADH. As designed, the assay measures electron flow from both the succinate and NAD(P)H dehydrogenases through MK and into the artificial acceptor, INT. Results indicate that the M. smegmatis ΔMSMEG1132 homogenate was approximately 3-fold less efficient at reducing INT/ng of MK (Table 1). However, total cellular ATP concentrations and the rate of ATP synthesis in membrane preparations from both the strains were unchanged. Further analysis demonstrated that the cellular content of MK had increased by a factor of 2 in the M. smegmatis ΔMSMEG1132 cells relative to the WT M. smegmatis cells.
Table 1. Comparison of Electron Transport, ATP Synthesis, and Menaquinone Content of M. smegmatisΔMSMEG1132 with those of M. smegmatis mc2 155a.
| traits | M. smegmatis (WT) | M. smegmatis ΔMSMEG1132 | fold change |
|---|---|---|---|
| rate of electron transport (O2 equiv) | 0.12 ± 0.01 μmol of O2/h/ng of menaquinone | 0.04 ± 0.01 μmol of O2/h/ng of menaquinone | 3-fold reduction |
| total cellular ATP | 6.4 ± 0.01 nmol/108 CFU | 6.2 ± 0.08 nmol/108 CFU | no change |
| rate of ATP biosynthesis | 1.3 ± 0.01 nmol/min/108 CFU | 1.4 ± 0.01 nmol/min/108 CFU | no change |
| total menaquinone (MK) | 4.3 ± 0.06 ng/108 CFU | 9.0 ± 1.6 ng/108 CFU | 2-fold increase |
Results are means of replicate observations ± standard deviation.
Saturation of Menaquinone Does Not Affect Proton Motive Force (PMF) or Oxidative Phosphorylation in Mycobacteria
The F0F1 ATP biosynthesis complex utilizes proton motive force (PMF) to drive ATP production. The two components contributing to PMF are membrane potential, ΔΨ, and transmembrane proton concentration gradient, ΔpH; PMF = ΔΨ – ZΔpH, where Z is a constant.26 Thus, the membrane electrical potential (ΔΨ) is a measure of both the integrity of the plasma membrane and the state of energy metabolism in bacteria.27 ΔΨ was investigated qualitatively in intact WT28,29 and ΔMSMEG1132 strains of M. smegmatis. In agreement with the observation that cellular levels of ATP and ATP synthesis were unchanged (Table 1), there were no obvious qualitative changes in ΔΨ in intact bacteria as indicated by a ΔΨ dissipation curve generated by exposure to varying concentrations of valinomycin (Figure S5). Both WT M. smegmatis and the ΔMSMEG1132 strains required exposure to valinomycin at approximately 0.5 to 0.3 μM, respectively, to dissipate 50% of ΔΨ (EC50).
To determine ΔpH across the membranes of intact bacilli, 31P NMR spectroscopy was utilized. Results indicated that the MSMEG1132 deletion had no impact on ΔpH (Table S4) although the control compound, nigericin, clearly dissipated the proton gradient. Taken together with the ΔΨ data it is clear that the knockout strain maintained WT ΔΨ and ΔpH levels. Thus, the mutant maintains cellular integrity in the presence of significantly increased amounts of total MK, a neutral lipid that could disrupt membrane integrity and potentially oxidative phosphorylation if accumulated in large concentrations.
Deletion of Rv0561c Reduces M. tuberculosis H37Rv Virulence
It is well established that M. tuberculosis infects macrophages and is capable of surviving within these cells for long periods of time;30,31 however, a previous report predicted that Rv0561c in M. tuberculosis is essential for survival in primary murine macrophages.18 Thus, a deletion mutant of M. tuberculosis ΔRv0561c and the complemented strain were used to infect J774A.1 cells, a mouse macrophage-like cell line, at an MOI of 5 (bacilli/J774A.1 cell). After incubation of J774A.1 cells with M. tuberculosis H37Rv, H37ΔRv0561c, and H37ΔRv0561c complemented with pNIP40bRv0561c, the capacity of each of these strains to infect and survive in the J774A.1 cells was analyzed. The association of bacteria with J774A.1 cells (either surface adherent or intracellular) was examined via acid-fast staining and fluorescence microscopy (Figures 5A, 5B). All three strains of bacilli were associated with corresponding J774A.1 nuclei, not all J774A.1 cells contain bacilli, and the bacilli are not present in large clusters. The percentage of J774A.1 cells infected with wild type, H37ΔRv0561c, or the complemented bacteria strains and the number of bacilli per J774A.1 cell are shown in Figure 5C. Overall, the results show that adhesion to and infection of J774A.1 cells with H37Rv wild type, H37ΔRv0561c, or complemented H37ΔRv0561c strains were similar for all three strains of bacteria.
Figure 5.
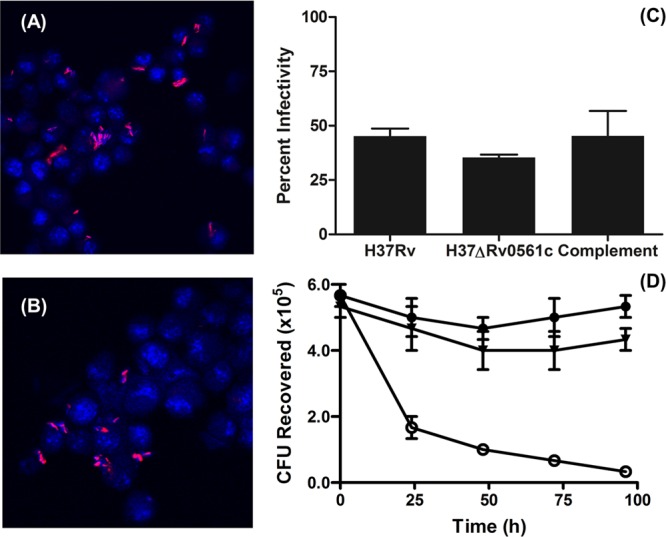
MenJ is required for survival in murine macrophages. Representative fluorescence micrographs of J774A.1 cells infected with M. tuberculosis H37Rv (A) or M. tuberculosis H37RvΔRv0561c (B) at an MOI of 5:1 (bacteria/J774A.1 cell) prior to incubation. Cells were stained with Auramine–Rhodamine and counterstained with DAPI. Micrographs were taken at 60× magnification. Panel C shows the percent infection of J774A.1 cells by M. tuberculosis H37Rv, M. tuberculosis H37RvΔRv0561c, or M. tuberculosis H37RvΔRv0561c complemented with pNIP40bRv0561c. Error bars represent standard deviation of the mean of counts of 5 randomly selected fields. J774A.1 cells infected with M. tuberculosis H37Rv (●), M. tuberculosis H37RvΔRv0561c (○), or M. tuberculosis H37RvΔRv0561c complemented with pNIP40bRv0561c (▼) were aliquoted into culture flasks, incubated at 37 °C under 5% CO2 for the indicated times, lysed, and plated on 7H10 agar plates with appropriate antibiotics to determine CFU present (panel D). Error bars indicate the standard deviation of the mean of three independent experiments.
The exposure of the J774A.1 cells to the H37ΔRv0561c bacilli resulted in a proportion of infected J774A.1 cells that was similar to that for infections with M. tuberculosis H37Rv or complemented H37ΔRv0561c cultures (Figure 5C). However, despite having similar levels of infection at the earliest time point, the survival of the H37ΔRv0561c bacilli in the macrophages was dramatically reduced (Figure 5D), an effect that was obviated by complementation.
Discussion
Rv0561c and MSMEG1132 encode enzymes which catalyze the reduction of MK-9 in M. tuberculosis and M. smegmatis to form dihydromenaquinone, likely MK-9(II-H2), respectively. It is proposed that the genes Rv0561c and MSMEG1132 be designated menJ as the encoded enzymes catalyze the reduction of MK-9 in the MK biosynthesis pathway (Table 2). Thus, a previously undescribed gene has been added to the known genes, menA through menI20,32−39 encoding enzymes which convert chorismate into MK. This gene is highly conserved in Mycobacterium spp., which encode orthologues with greater than 68% identity at the amino acid level. Conserved genes can also be identified in other Gram-positive species with partially saturated MK side chains of known structure such as Corynebacterium diphtheriae, Nocardia spp., and Streptomyces species,40 with 54, 60–63, and 33–40% identity, respectively. However, other Gram-positive species with partially saturated MK side chains of known structure such as Brevibacterium lipolyticum, Nocardioides albus, Oerskovia turbata, and Actinomadura madurae do not appear to have proteins with significant similarity to MenJ even though they have the appropriate modifications of their MK. The same trend is true of Gram-negative eubacteria that partially saturate the isoprenoid side chain of menaquinone. In this case, Desulfobulbus propionicus, Desulfovibrio salexigens, and Desulfovibrio africanus have proteins with 25–30% identity when compared to MenJ at the amino acid level, while genomes of Desulfobulbus elongatus and Thermoleophilum album apparently do not encode proteins with sequence similarity to MenJ. The few Archaea with characterized partially saturated MK side chains do appear to have orthologues of MenJ, although with less than 30% identity. Thus, while the saturation of the isoprenyl side chain of MK is widespread in bacteria and Archaea, there seem to be other enzymes capable of reducing the double bonds in menaquinone that are unrelated to MenJ.
Table 2. MenJ Catalyzes the Final Step in Mycobacterial MK-9(II-H2) Biosynthesisa.
| biosynthetic pathway | enzyme | M. tuberculosis gene | ref |
|---|---|---|---|
| chorismate | |||
| ↓ | MenF | Rv3215 | |
| isochorismate | |||
| ↓ | MenD | Rv0555 | |
| 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate | |||
| ↓ | MenH | Rv2715 | |
| 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate | |||
| ↓ | MenC | Rv0553 | |
| 2-succinylbenzoic acid | |||
| ↓ | MenE | Rv0166 | (37) |
| 2-succinylbenzoyl-CoA | |||
| ↓ | MenB | Rv0548c | (34) |
| 1,4-dihydroxy-2-naphthoate-CoA | |||
| ↓ | MenI | Rv1847 | |
| 1,4-dihydroxy-2-naphthoate | |||
| ↓ | MenA | Rv0534c | (48) |
| demethylmenaquinone | |||
| ↓ | MenG | Rv0558, Rv0560c | |
| menaquinone (MK-9) | |||
| ↓ | MenJ | Rv0561c | this study |
| saturated menaquinone [MK-9(II-H2)] |
Homologues from mycobacteria that have been empirically identified are indicated with the appropriate reference. Putative homologues in Mycobacterium tuberculosis H37Rv genome are based on BLAST searches and annotations from online databases.
MenJ bears sequence similarity to the geranylgeranyl reductases (GGRs) often found in Archaea, plants, and, likely, other organisms. The enzyme under study has 67% sequence identity with a GGR from Arabidopsis thaliana and 28% similarity to a GGR identified in A. fulgidus. The GGR from A. fulgidus was previously reported to synthesize compounds with properties that suggest that they are quinones with partially saturated isoprenyl side chains when expressed in E. coli.16 Currently, virtually all homologues of MenJ are annotated in databases as GGRs, as the MK reductases were previously unknown.
The MS data indicate that mycobacterial MenJ is a reductase that reduces a single double bond in the isoprenoid tail of MK. A series of fragment ions differing in mass by m/z 68, assigned to cleavages between the sp3 and sp2 carbons within an isoprene unit, was utilized to illustrate the differences in MK from WT and mutant strains of bacilli. Other series of fragments were observed in the spectra, including one also differing in mass by m/z 68, assigned to cleavages between the two sp3 carbons that join isoprene units. These fragments are not indicated in the figures or described in the text as they do not provide additional information regarding the position of the saturation. It was not possible to unambiguously identify the precise location of the isoprene unit reduced by MenJ, but the data presented indicates that the reduction takes place near the naphthoquinone moiety of MK. Partial saturation of the side chain of MK has been reported in many eubacteria and Archaea;4 however, due to analytical complexity the exact position of the saturation has only been determined in a few bacterial species. In Gram-positive eubacteria it is generally internal isoprene units that are reduced, usually the β-isoprene unit as seen in mycobacteria,11 but in some cases other isoprene units as well.7 Although some Actinomycetes do possess MK with saturated terminal isoprene units (more commonly seen in Gram-negative eubacteria), these are invariably accompanied by other internal saturated units close to the ring system.4 All hydrogenated MK isolated from Gram-positive eubacteria, which have only one isoprene unit hydrogenated, have the β-isoprene unit from the naphthoquinone system saturated7,11 as seen in Mycobacterium spp. Thus, the present data coupled with earlier analytical studies that determined that Mycobacterium spp. reduce the β-isoprene unit of MK producing MK-9(II-H2)8−10,21 strongly suggests that MenJ catalyzes the saturation of the β-isoprene unit of MK and that this enzyme is involved in the synthesis of MK-9(II-H2), rather than electron transport where reduction of the ketone residues of the naphthoquinone would be expected.
Detailed structural and mechanistic studies of a digeranylgeranylglycerophospolipid reductase from Thermoplasma acidophilum indicate that the enzyme belongs to the p-hydroxybenzoate hydroxylase (PHBH) SCOP superfamily and shares a common mechanism with other PHBH enzymes in which FAD switches between two conformations corresponding to the reductive and oxidative half-cycles.41 Although, MenJ has only 20% sequence identity with this GGR, it seems likely that the reaction mechanism is similar. Studies designed to evaluate this hypothesis are ongoing.
The generation of the deletion mutants described herein have provided the first opportunity to determine the biochemical role of saturation of an isoprene unit in the β-position of the isoprenoid tail of menaquinone. The disruption of menJ in M. tuberculosis and M. smegmatis presented interesting phenotypes. There were no differences in the growth rate of either mutant strain relative to the wild type in aerobic culture, confirming the prediction that this gene was not required for bacterial growth in the culture.18 In addition, M. smegmatis ΔMSMEG1132 grew at the same rate as WT M. smegmatis in a modified Wayne model24 of gradually increasing hypoxia in sealed tubes. However, in control experiments where methylene blue was added to the tubes to verify the conversion from an oxidizing to a reducing environment, it was observed that decolorization of the indicator occurred more slowly in the tubes containing the mutant cultures relative to the tubes containing WT cultures. This observation combined with similar bacteria numbers, doubling times, rates of ATP synthesis, or cellular ATP content suggested that the degree of coupling of oxidative phosphorylation may have been altered. However, subsequent studies clearly indicated that the efficiency of the electron transport (AETS) of the MK-9 was 3-fold less than that of the MK-9(II-H2). Quantitation of total MK, both MK-9 and MK-9(II-H2), per CFU indicated that the levels of total MK in the ΔMSMEG1132 strain was ∼2-fold higher than in the WT indicating that the bacteria were able to compensate for the reduced efficiency of electron transport of the MK-9 relative to the MK-9(II-H2) by increasing the levels of the total menaquinone in the membrane. The ability to maintain ATP synthetic rates despite reduced AETS strongly suggests that the bacilli can sense the electron transport capacity and have the ability to regulate the amounts of MK-9 and/or MK-9(II-H2) in the membrane. Subsequent measurement of ΔΨ and ΔpH indicated that the accumulation of increased levels of MK-9 in M. smegmatis ΔMSMEG1132 did not alter the integrity of the cell membrane and that deletion of menJ had little or no effect on the PMF generated by the bacilli. Thus, the mechanism(s) behind the reduction of electron transfer efficiency in the KO strains is not clear. It is possible that structural changes induced by the reduction of double bond in MK-9(II-H2) result in a kinetic effect due to reduced binding affinity to either the dehydrogenases or the cytochrome complex. Alternatively, it is possible that the reduction of a single double bond alters the reducing potential of the MK. While there is little known about the electrochemical effects of having a single reduced double bond in the β-isoprene unit of MK, it has been reported that vitamin K1 [MK-4(II,III,IV-H6)] has a much more negative Em value than MK-4 (−500 to −700 mV vs −20 to −30 mV, respectively).9,10
The most significant phenotype observed in the M. tuberculosis ΔRv0561c strain was decreased survival in J774A.1 cells. When J774A.1 cells were incubated with mutant or WT M. tuberculosis at five bacilli/J774A.1 cell, it was clear that M. tuberculosis ΔRv0561c was capable of infecting the host cells but was incapable of surviving in the macrophage, whereas complementation restored intracellular survival to WT levels. The mechanism for this loss of virulence is not obvious. The fact that there is increased MK-9 in the membranes of the ΔMSMEG1132 strain that compensates for the loss of electron transfer efficiency suggests that uncoupling of oxidative phosphorylation does not play a dominant role in the loss of virulence. If reduction of the β-isoprene results in a significantly more stable oxidized state of the molecule, it is possible that this is an adaptation to life in hypoxic conditions, which would be consistent with the observation that the wild type bacteria consume reduced amounts of oxygen relative to the mutant bacteria when entering hypoxia. In addition, benefits derived from a more negative Em would come at the cost of being able to utilize some electron donors. However, the results are in agreement with high density transposon mutagenesis experiments that identified Rv0561c as one of 126 genes required for M. tuberculosis survival in both activated and inactivated primary macrophages derived from bone marrow precursors from C57BL/6 mice.18
It has previously been reported that the structural composition of the MK in the MK pool of M. tuberculosis provides a mechanistic link between the respiratory state of the bacilli and response to hypoxia.13 The data presented here indicate that the reduction of a single double bond in the isoprenoid side chain of MK, converting MK-9 to MK-9(II-H2), increases the efficiency of the mycobacterial ETS system. In addition, the fact that the deletion mutants grow as well as the WT bacteria in aerobic and hypoxic conditions but have severely attenuated virulence in macrophages suggests that MK-9(II-H2), or possibly MenJ itself, functions as a virulence factor for M. tuberculosis.
Recently, MK synthesis has been proposed to be an attractive drug target in M. tuberculosis and potentially other Gram-positive pathogens.42−44 MenJ is clearly nonessential for mycobacterial growth in culture and, thus, would not be considered to be a classic target for small molecule inhibitors. However, there is interest in targeting virulence as a new paradigm for antimicrobial therapy that may circumvent, or slow, resistance mechanisms.45−47 Typically, this would mean targeting toxin function or delivery, regulation of virulence expression or cellular adhesion.45 MenJ and MK-9(II-H2) do not fall into any of these categories, but could be included in the proposed category of an in vivo essential gene target.45 That is, MenJ or MK-9(II-H2), while not classically essential for mycobacterial survival in culture, appears to be contextually essential for mycobacterial survival in macrophages, suggesting that MenJ represents a testable system of a contextual drug target in M. tuberculosis.
In conclusion, a family of enzymes that reduce the isoprenyl side chain, rather than the naphthoquinone carbonyls, of menaquinone was identified in mycobacteria, and similar enzymes were found in other Gram-positive and -negative bacteria. In mycobacteria saturation of the β-isoprene unit of mycobacterial menaquinone results in a 3-fold increase in the efficiency of electron transport. Deletion of menJ (reduction/hydrogenation) activity was not found to be essential for mycobacterial growth in culture; however, deletion of menJ reduces M. tuberculosis survival in macrophages. Thus, saturation of the β-isoprene unit of mycobacterial menaquinone represents a novel virulence factor for M. tuberculosis, which may constitute a contextual drug target.
Methods
Materials
The origins of bacterial strains are as described in Table S2. High Fidelity Taq polymerase was from Roche Diagnostics, and BCA Protein Assay Kits were purchased from Thermo Scientific Pierce. The ATPlite luminescence assay system kit was from PerkinElmer Inc. Cell Culture Inserts (3 μm) and Radio-Immunoprecipitation Assay (RIPA) lysis buffer were from Millipore Corp. 3,3′-Dipropylthiadicarbocyanine iodide [DiSC3(5)], valinomycin, nigericin, and DAPI (4′,6-diamidino-2-phenylindole) were supplied by Life Technologies. Auramine–Rhodamine T mycobacterial stain and Prolong Gold Anti-Fade Reagent were from BD Biosciences and Invitrogen, respectively. Tissue culture flasks and cell scrapers were obtained from Corning and BD Falcon, respectively. Hygromycin B was from Calbiochem. Kanamycin, vitamin K2 (MK-4), catechol, 7H9, 7H10, OADC polyvinylpyrrolidone (PVP 40), protease inhibitors, lysozyme, typsin, and DNase I were obtained from Sigma-Aldrich, as were all other chemicals unless otherwise noted.
Identification and Cloning of menJ from M. tuberculosis H37Rv
BLAST searches were conducted on the TubercuList and NCBI Web sites. Multiple alignments of the amino acid sequences were generated using Multalin.48 Genes from M. tuberculosis H37Rv (Rv0561c) and its orthologue in M. smegmatis mc2 155 (MSMEG1132) were from amplified genomic DNA from M. tuberculosis H37Rv and M. smegmatis mc2 155 using the primers listed in Table S3. Taq DNA polymerase was used for PCR amplification. Amplified gene products were cloned into appropriate vectors wherever necessary using standard molecular biology techniques.49 Fidelity of the clones was confirmed by restriction digestion and sequencing.
Construction of Deletion Mutants
The Ts/sacB method50 was used to achieve allelic replacement at the MSMEG1132 and Rv0561c locus of M. smegmatis mc2 155 and M. tuberculosis H37Rv respectively as described in the Supporting Information.
Lipid Extraction, Identification, and Quantification
Lipid extraction from E. coli and Mycobacterium spp. was done essentially as described earlier51 using chloroform/methanol (2:1, by vol). The extracted lipid was partially purified on a silicic acid column eluted with chloroform and subjected to liquid chromatography–mass spectrometry (LCMS). An aliquot was applied to a reverse-phase Hypersil ODS column (Agilent) connected to an Agilent 1200 series high-performance liquid chromatography (HPLC) system. Separation was achieved using a gradient running from water to 90% methanol over 40 min at 0.3 mL/min at 40 °C. The eluate was directly introduced into an Agilent 6250 quadrupole time-of-flight (Q-TOF) mass spectrometer equipped with an Agilent multimode source operated in the simultaneous electrospray ionization and atmospheric pressure chemical ionization mode. Nebulizing gas temperature was 350 °C, and nebulizer pressure was 30 psi. Data obtained were analyzed using Agilent Mass Hunter Workstation software.
Samples were also analyzed using an Agilent Technologies HP1100 series HPLC connected to a Thermo 2000 Finnigan LCQ-Duo ion-trap mass spectrometer. HPLC separation was achieved using a reverse-phase XBridge C18 3.5 μm 2.1 × 150 mm column (Waters) and a gradient running from 100% methanol to methanol/isopropanol (1:1 v/v) over 50 min at 0.4 mL/min and 40 °C. Eluted molecules were subjected to positive ion MS using APCI as the ionization interface. Capillary temperature was 150 °C, and APCI vaporizer temperature was 450 °C. Electrospray needle voltage was 4.5 kV. Sheath gas flow was maintained at 40 units. Data acquisition and analysis were performed using Xcaliber software from Thermo Scientific. In all cases samples were spiked with known amounts of vitamin K2 (MK-4) as an internal standard and peak area52 was used to quantitate MK.
Preparation of Inverted Membrane Vesicles
Preparation of inverted membrane vesicles for estimation of ATP synthetic rates from M. smegmatis cells was done essentially as previously described by Koul et al.53 The membrane vesicles were resuspended in 50 mM MOPS buffer (pH 7.5) containing 2 mM MgCl2, and protein content was estimated using a BCA kit.
ATP Estimation
Total cellular ATP from both wild type and KO mutant was measured from fully grown cultures, and rate of ATP synthesis was estimated from inverted membrane vesicles of M. smegmatis strains using an ATPlite luminescence assay system kit (PerkinElmer) following the manufacturer’s directions.
Electron Transport Activity (AETS) Measurements in Vitro
AETS was determined as previously54 with minor modifications. Briefly, M. smegmatis cells were harvested and disrupted by sonication in lysis buffer [0.05 M phosphate buffer pH 8.0 containing 0.15% polyvinylpyrrolidone (PVP-40), 100 μM MgSO4, 1.5 mM NaCl, and 0.2% Triton X-100]. Sonication was carried out on ice for 10 min with 90 s on and 30 s off pulse cycles. Protein concentration was determined using the BCA kit. An appropriate amount of protein was transferred to a 96 well plate, and volume was adjusted to 50 μL with lysis buffer. Subsequently, 50 μL of 4 mM 2-(p-idophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT) and 150 μL of substrate solution containing 1.0 mM NADH, 0.2 mM NADPH, 130 mM sodium succinate (hexahydrate), and 0.2% Triton X-100 in 0.05 M phosphate buffer (pH 8.0) were added. OD490 was recorded in kinetic mode using a Biotek Synergy HT spectrophotometer. AETS is expressed as O2 equivalents in μmol of O2/h/ng of menaquinone.
M. tuberculosis Survival in Macrophages
Macrophage-like cells, J774A.1 (ATCC TIB-67), were infected by incubating adherent cells with M. tuberculosis H37Rv, H37ΔRv0561c, or H37ΔRv0561c complemented with Rv0561c. Prior to infection, bacterial stock cultures (∼108 cell/mL in 7H9) were sonicated for 30 s in a bath sonicator and washed twice in RPMI 1640 containing 2% human serum and 0.05% Tween 80. The washed bacilli were resuspended in phosphate buffer saline (PBS) and diluted appropriately. Macrophage J774A.1 cells were grown to near confluence in 75 cm2 tissue culture flasks in RPMI medium containing 2% human serum. Cells were scraped from representative flasks and counted. Five bacilli per J774A.1 cell (MOI of 5:1) were added to the remaining flasks and incubated at 37 °C under 5% CO2 for 2 h. After infection, the adherent macrophage cells were removed from the surface of the flasks with a cell scraper and resuspended in RPMI media with multiple gentle passes in a disposable pipet to disrupt clumps. Subsequently free bacilli were removed from the culture by filtration through 3 μm cell culture inserts (Millipore Corp.) which retained macrophages and allowed free bacilli to pass through. Macrophage cells retained on the filters were resuspended in PBS. Aliquots of the suspensions were concentrated on slides by centrifugation using a Shandon CytoSpin Centrifuge. Slides were heat fixed and stained using Auramine–Rhodamine T mycobacterial stain as recommended by the manufacturer, and slides were then washed with acid–alcohol and counterstained with DAPI followed by aqueous mount on slides with the Prolong Gold Anti-Fade reagent. Photographs were taken using an LSM510 META confocal microscope as described earlier.55 Images were captured and analyzed using Zeiss LSM image analyzer version 4.0 to ensure that all extracellular bacilli had been removed and to quantitate the number of macrophages and proportion of infected macrophages recovered. The remaining cells were added to 6 well plates at 8.3 × 105 cells/well and cultured at 37 °C and 5% CO2 in RPMI 1640 media containing 2% human serum. After 24, 48, 72, and 96 h of culture, the adherent cells were washed with PBS, scraped from the bottom of the wells and lysed using 1× Radio-Immunoprecipitation Assay (RIPA) lysis buffer, and homogenized in a Bullet Blender (Next Advance, Inc.). 10-fold serial dilutions of the resulting solution were spread on 7H10 plates containing kanamycin (for deletion strains) and hygromycin (for complemented strains) and incubated at 37 °C. After 3 weeks, the colony forming units (CFU) were enumerated. All results presented are representative of multiple experiments and replicates. Error bars indicate standard deviation from the mean.
Acknowledgments
This research was funded via NIH/NIAID Grant AI04915.
Glossary
Abbreviations
- AETS
electron transport activity
- BCA
bicinchoninic acid
- CFU
colony forming units
- DAPI
4′,6-diamidino-2-phenylindole
- DiSC3(5)
3,3′-dipropylthiadicarbocyanine iodide
- Em
midpoint potential of electron couple
- ETS
electron transport system
- GGR
geranylgeranyl reductase
- INT
2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride
- KO
knockout
- MOI
multiplicity of infection
- MK
menaquinone
- MK-9(II-H2)
menaquinone with 9 isoprene units in the side chain, with the one in the second (β) position saturated
- PBS
phosphate buffered saline
- PHBH
p-hydroxybenzoate hydroxylase
- PMF
proton motive force
- UQ
ubiquinone
- WT
wild type
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.5b00212.
Details of genetic manipulation, growth curves, ΔΨ and ΔpH methods and data, and supporting MS data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Pandya K. P.; King H. K. Ubiquinone and menaquinone in bacteria: a comparative study of some bacterial respiratory systems. Arch. Biochem. Biophys. 1966, 114, 154–157. 10.1016/0003-9861(66)90316-X. [DOI] [PubMed] [Google Scholar]
- Weinstein E. A.; Yano T.; Li L. S.; Avarbock D.; Avarbock A.; Helm D.; McColm A. A.; Duncan K.; Lonsdale J. T.; Rubin H. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 4548–4553. 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothery R. A.; Chatterjee I.; Kiema G.; McDermott M. T.; Weiner J. H. Hydroxylated naphthoquinones as substrates for Escherichia coli anaerobic reductases. Biochem. J. 1998, 332 (Part 1), 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. D.; Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 1981, 45, 316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz L.; Gradzki Z.; Kalinowski M. Phenotypic properties and virulence factors of Trueperella pyogenes. Med. Weter. 2014, 70, 73–80. [Google Scholar]
- Brodie A. F.; Ballantine J. Oxidative phosphorylation in fractionated bacterial systems. III. Specificity of vitamin K reactivation. J. Biol. Chem. 1960, 235, 232–237. [PubMed] [Google Scholar]
- Azerad R.; Cyrot-Pelletier M. O. Structure and configuration of the polyprenoid side chain of dihydromenaquinones from Myco- and Corynebacteria. Biochimie 1973, 55, 591–603. 10.1016/S0300-9084(73)80421-3. [DOI] [PubMed] [Google Scholar]
- Gale P. H.; Brodie A. F.; Arison B. H.; Folkers K.; Trenner N. R.; Page A. C. Characterization of Vitamin K9(H) from Mycobacterium phlei. Biochemistry 1963, 2, 200–203. 10.1021/bi00901a038. [DOI] [PubMed] [Google Scholar]
- Itoh S.; Iwaki M. Vitamin-K1 (Phylloquinone) Restores the Turnover of Fes Centers in the Ether-Extracted Spinach Ps I Particles. FEBS Lett. 1989, 243, 47–52. 10.1016/0014-5793(89)81215-3. [DOI] [Google Scholar]
- Munge B.; Das S. K.; Ilagan R.; Pendon Z.; Yang J.; Frank H. A.; Rusling J. F. Electron transfer reactions of redox cofactors in spinach photosystem I reaction center protein in lipid films on electrodes. J. Am. Chem. Soc. 2003, 125, 12457–12463. 10.1021/ja036671p. [DOI] [PubMed] [Google Scholar]
- Azerad R.; Cyrot M. O.; Lederer E. Structure of the dihydromenaquinone-9 of Mycobacterium phlei. Biochem. Biophys. Res. Commun. 1967, 27, 249–252. 10.1016/S0006-291X(67)80069-X. [DOI] [PubMed] [Google Scholar]
- Dunphy P. J.; Gutnick D. L.; Phillips P. G.; Brodie A. F. A new natural naphthoquinone in Mycobacterium phlei. Cis-dihydromenaquinone-9, structure and function. J. Biol. Chem. 1968, 243, 398–407. [PubMed] [Google Scholar]
- Honaker R. W.; Dhiman R. K.; Narayanasamy P.; Crick D. C.; Voskuil M. I. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 2010, 192, 6447–6455. 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaziou P.; Sismanidis C.; Floyd K.; Raviglione M. Global epidemiology of tuberculosis. Cold Spring Harbor Perspect. Med. 2015, 5, a017798. 10.1101/cshperspect.a017798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller Y.; Bouvier F.; d’Harlingue A.; Camara B. Metabolic compartmentation of plastid prenyllipid biosynthesis--evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 1998, 251, 413–417. 10.1046/j.1432-1327.1998.2510413.x. [DOI] [PubMed] [Google Scholar]
- Hemmi H.; Takahashi Y.; Shibuya K.; Nakayama T.; Nishino T. Menaquinone-specific prenyl reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Bacteriol. 2005, 187, 1937–1944. 10.1128/JB.187.6.1937-1944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addlesee H. A.; Hunter C. N. Rhodospirillum rubrum possesses a variant of the bchP gene, encoding geranylgeranyl-bacteriopheophytin reductase. J. Bacteriol. 2002, 184, 1578–1586. 10.1128/JB.184.6.1578-1586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J.; Bloom B. R.; Rubin E. J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 8327–8332. 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E.; Gawronski J. D.; Dejesus M. A.; Ioerger T. R.; Akerley B. J.; Sassetti C. M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan C.; Sharma V.; Hudspeth M. E.; Meganathan R. Menaquinone (vitamin K2) biosynthesis: Evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J. Bacteriol. 1992, 174, 8111–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsclaw C. M.; Sogi K. M.; Gilmore S. A.; Schelle M. W.; Leavell M. D.; Bertozzi C. R.; Leary J. A. Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem. Biol. 2008, 3, 619–624. 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. C.; Kuritzkes D. R.. Pathways for anaerobic electron transport. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology; Neidhardt F. C., Ingraham K. B., Magasanik B., Schaechter M., Umbarger H. E., Eds.; American Society for Microbiology: Washington, DC, 1987; pp 201–221. [Google Scholar]
- Geyer R.; Peacock A. D.; White D. C.; Lytle C.; Van Berkel G. J. Atmospheric pressure chemical ionization and atmospheric pressure photoionization for simultaneous mass spectrometric analysis of microbial respiratory ubiquinones and menaquinones. J. Mass Spectrom. 2004, 39, 922–929. 10.1002/jms.670. [DOI] [PubMed] [Google Scholar]
- Abomoelak B.; Hoye E. A.; Chi J.; Marcus S. A.; Laval F.; Bannantine J. P.; Ward S. K.; Daffe M.; Liu H. D.; Talaat A. M. mosR, A novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J. Bacteriol. 2009, 191, 5941–5952. 10.1128/JB.00778-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J.; McFeters G. A. Mechanisms of INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride), and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli K-12. J. Microbiol. Methods 1997, 29, 161–175. 10.1016/S0167-7012(97)00036-5. [DOI] [PubMed] [Google Scholar]
- Breeuwer P.; Abee T.. Assessment of the membrane potential, intracellular pH and respiration of bacteria employing fluorescence techniques. In Molecular Microbial Ecology Manual, 2nd ed.; Kowalchuk G. A., de Bruijn F. J., Head I. M., Akkermans A. D., van Elsas J. D., Eds.; Springer Netherlands: 2004; published electronically. [Google Scholar]
- Shapiro H. M. Membrane potential estimation by flow cytometry. Methods 2000, 21, 271–279. 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- Li K.; Schurig-Briccio L. A.; Feng X.; Upadhyay A.; Pujari V.; Lechartier B.; Fontes F. L.; Yang H.; Rao G.; Zhu W.; Gulati A.; No J. H.; Cintra G.; Bogue S.; Liu Y. L.; Molohon K.; Orlean P.; Mitchell D. A.; Freitas-Junior L.; Ren F.; Sun H.; Jiang T.; Li Y.; Guo R. T.; Cole S. T.; Gennis R. B.; Crick D. C.; Oldfield E. Multitarget drug discovery for tuberculosis and other infectious diseases. J. Med. Chem. 2014, 57, 3126–3139. 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Upadhyay A.; Fontes F. L.; North E. J.; Wang Y. H.; Crans D. C.; Grzegorzewicz A. E.; Jones V.; Franzblau S. G.; Lee R. E.; Crick D. C.; Jackson M. Novel Insights into the Mechanism of Inhibition of MmpL3, a Target of Multiple Pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 6413–6423. 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J. A.; Hart P. D. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 1971, 134, 713–740. 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. G. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001, 2, 569–577. 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- Daruwala R.; Kwon O.; Meganathan R.; Hudspeth M. E. A new isochorismate synthase specifically involved in menaquinone (vitamin K2) biosynthesis encoded by the menF gene. FEMS Microbiol. Lett. 1996, 140, 159–163. 10.1111/j.1574-6968.1996.tb08330.x. [DOI] [PubMed] [Google Scholar]
- Sharma V.; Meganathan R.; Hudspeth M. E. Menaquinone (vitamin K2) biosynthesis: cloning, nucleotide sequence, and expression of the menC gene from Escherichia coli. J. Bacteriol. 1993, 175, 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.; Hudspeth M. E.; Meganathan R. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menE gene from Escherichia coli. Gene 1996, 168, 43–48. 10.1016/0378-1119(95)00721-0. [DOI] [PubMed] [Google Scholar]
- Suvarna K.; Stevenson D.; Meganathan R.; Hudspeth M. E. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J. Bacteriol. 1998, 180, 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio J. J.; Theis K.; Feng Y. G.; Gajda R.; Machutta C.; Tonge P. J.; Kisker C. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K-2 biosynthesis. J. Biol. Chem. 2003, 278, 42352–42360. 10.1074/jbc.M307399200. [DOI] [PubMed] [Google Scholar]
- Chen M.; Ma X.; Chen X.; Jiang M.; Song H.; Guo Z. Identification of a hotdog fold thioesterase involved in the biosynthesis of menaquinone in Escherichia coli. J. Bacteriol. 2013, 195, 2768–2775. 10.1128/JB.00141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M.; Chen X.; Guo Z. F.; Cao Y.; Chen M.; Guo Z. Identification and characterization of (1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase in the menaquinone biosynthesis of Escherichia coli. Biochemistry 2008, 47, 3426–3434. 10.1021/bi7023755. [DOI] [PubMed] [Google Scholar]
- Lu X. Q.; Zhang H. N.; Tonge P. J.; Tan D. S. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 2008, 18, 5963–5966. 10.1016/j.bmcl.2008.07.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. D.; Kroppenstedt R. M. Structures of the partially saturated menaquinones of Glycomyces rutgersensis. FEMS Microbiol. Lett. 1987, 44, 215–219. 10.1111/j.1574-6968.1987.tb02270.x. [DOI] [Google Scholar]
- Xu Q. P.; Eguchi T.; Mathews I. I.; Rife C. L.; Chiu H. J.; Farr C. L.; Feuerhelm J.; Jaroszewski L.; Klock H. E.; Knuth M. W.; Miller M. D.; Weekes D.; Elsliger M. A.; Deacon A. M.; Godzik A.; Lesley S. A.; Wilson I. A. Insights into Substrate Specificity of Geranylgeranyl Reductases Revealed by the Structure of Digeranylgeranylglycerophospholipid Reductase, an Essential Enzyme in the Biosynthesis of Archaeal Membrane Lipids. J. Mol. Biol. 2010, 404, 403–417. 10.1016/j.jmb.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Q.; Zhou R.; Sharma I.; Li X. K.; Kumar G.; Swaminathan S.; Tonge P. J.; Tan D. S. Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. ChemBioChem 2012, 13, 129–136. 10.1002/cbic.201100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. K.; Liu N. N.; Zhang H. N.; Knudson S. E.; Li H. J.; Lai C. T.; Simmerling C.; Slayden R. A.; Tonge P. J. CoA adducts of 4-Oxo-4-phenylbut-2-enoates: Inhibitors of MenB from the M. tuberculosis menaquinone biosynthesis pathway. ACS Med. Chem. Lett. 2011, 2, 818–823. 10.1021/ml200141e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J.; Siricilla S.; Wan B.; Crick D. C.; Lenaerts A. J.; Franzblau S. G.; Kurosu M. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 2012, 55, 3739–3755. 10.1021/jm201608g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A. E.; Pierson E.; Hung D. T. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Allen R. C.; Popat R.; Diggle S. P.; Brown S. P. Targeting virulence: can we make evolution-proof drugs?. Nat. Rev. Microbiol. 2014, 12, 300–308. 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- Rasko D. A.; Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discovery 2010, 9, 117–128. 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E. F.; Sambrook J.. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: New York, 1983. [Google Scholar]
- Pelicic V.; Jackson M.; Reyrat J. M.; Jacobs W. R. Jr.; Gicquel B.; Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 10955–10960. 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman R. K.; Mahapatra S.; Slayden R. A.; Boyne M. E.; Lenaerts A.; Hinshaw J. C.; Angala S. K.; Chatterjee D.; Biswas K.; Narayanasamy P.; Kurosu M.; Crick D. C. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 2009, 72, 85–97. 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasinger V. C.; Zeng M.; Yau Y. Current status and advances in quantitative proteomic mass spectrometry. Int. J. Proteomics 2013, 2013, 1–12. 10.1155/2013/180605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A.; Dendouga N.; Vergauwen K.; Molenberghs B.; Vranckx L.; Willebrords R.; Ristic Z.; Lill H.; Dorange I.; Guillemont J.; Bald D.; Andries K. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 2007, 3, 323–324. 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- Packard T. T.; Williams P. J. L. Rates of respiratory oxygen-consumption and electron-transport in surface seawater from the northwest Atlantic. Oceanol. Acta 1981, 4, 351–358. [Google Scholar]
- Ryan G. J.; Hoff D. R.; Driver E. R.; Voskuil M. I.; Gonzalez-Juarrero M.; Basaraba R. J.; Crick D. C.; Spencer J. S.; Lenaerts A. J. Multiple Mycobacterium tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PLoS One 2010, 5, e11108. 10.1371/journal.pone.0011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G. M.; Hards K.; Vilcheze C.; Hartman T.; Berney M. Energetics of Respiration and Oxidative Phosphorylation in Mycobacteria. Microbiol. Spectrum 2014, 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.