ABSTRACT
Although one pathway for the post-translational targeting of tail-anchored proteins to the endoplasmic reticulum (ER) has been well defined, it is unclear whether additional pathways exist. Here, we provide evidence that a subset of mRNAs encoding tail-anchored proteins, including Sec61β and nesprin-2, is partially localized to the surface of the ER in mammalian cells. In particular, Sec61b mRNA can be targeted to, and later maintained on, the ER using both translation-dependent and -independent mechanisms. Our data suggests that this process is independent of p180 (also known as RRBP1), a known mRNA receptor on the ER, and the transmembrane domain recognition complex (TRC) pathway components, TRC40 (also known as ASNA1) and BAT3 (also known as BAG6). In addition, our data indicates that Sec61b mRNA might access translocon-bound ribosomes. Our results show that certain tail-anchored proteins are likely to be synthesized directly on the ER, and this facilitates their membrane insertion. Thus, it is clear that mammalian cells utilize multiple mechanisms to ensure efficient targeting of tail-anchored proteins to the surface of the ER.
KEY WORDS: Endoplasmic reticulum, mRNA localization, Secretion, Tail-anchored protein
Highlighted Article: The mRNA encoding certain tail-anchored proteins is directly localized to the surface of the endoplasmic reticulum, facilitating the insertion of newly synthesized proteins into the membrane.
INTRODUCTION
One major mechanism that directs proteins to their correct subcellular destination is localization of their mRNA (Holt and Bullock, 2009; Martin and Ephrussi, 2009). Likely the most widespread example is the localization of mRNAs encoding membrane and secreted proteins to the surface of the ER in eukaryotic cells. This localization facilitates the targeting of the encoded proteins to the secretory pathway (Cui and Palazzo, 2014).
Previously it was thought that these mRNAs are exclusively targeted to the ER by their encoded proteins. During their translation, newly synthesized hydrophobic signal sequences or transmembrane domains (TMDs) are recognized as they emerge from the ribosome by the signal recognition particle (SRP), which then redirects the mRNA–ribosome–nascent-chain complex to the ER surface. However, recent studies by our laboratory and other groups demonstrate that a substantial fraction of these mRNAs can be targeted to the ER independently of their translation and the SRP system (Pyhtila et al., 2008; Chen et al., 2011; Cui et al., 2012). This is due in part to the activity of mRNA receptors, such as p180 (also known as RRBP1) (Cui et al., 2012, 2013).
ER localization of mRNAs encoding secretory and membrane-bound proteins might not be universal. Some of these mRNAs appear to be translated by free (i.e. non-ER associated) ribosomes, and their encoded polypeptides are then targeted to the ER post-translationally. One group of membrane proteins thought to be exclusively inserted into membranes post-translationally are tail-anchored proteins (Rabu et al., 2009; Borgese and Fasana, 2011; Hegde and Keenan, 2011). These proteins have a single TMD within the last 50 amino acids from the C-terminus and display their functional N-terminal domain towards the cytosol (Kutay et al., 1993). In mammalian cells, tail-anchored proteins are found on most membranes, including the plasma membrane, ER, Golgi, mitochondria and peroxisomes. In the majority of cases, tail-anchored proteins are first inserted into the ER and then are transported to their proper final destination (Kutay et al., 1995). In addition, it appears that all mitochondrial-targeted and most peroxisome-targeted tail-anchored proteins use specialized pathways. Tail-anchored proteins are involved in many essential cellular processes, such as apoptosis, vesicular transport and protein translocation. Therefore, their correct localization is crucial for cell viability.
For ER-targeted tail-anchored proteins, their targeting is thought to be mediated by the transmembrane domain recognition complex (TRC) pathway. Upon completion of their synthesis, the TMD exits the translating ribosome and is recognized by a series of chaperone proteins which are thought to sort the protein to its proper final destination. These chaperones include SGTA, TRC40 (also known as ASNA1) and BAT3 (also known as BAG6) (Stefanovic and Hegde, 2007; Schuldiner et al., 2008; Jonikas et al., 2009; Leznicki et al., 2010, 2011; Mariappan et al., 2010; Wang et al., 2010). TRC40 then delivers the protein to the ER membrane receptors, WRB and CAML (also known as CAMLG) (Vilardi et al., 2011; Yamamoto and Sakisaka, 2012). Functional orthologs of these proteins in yeast, Get1 and Get2, can mediate membrane insertion (Wang et al., 2014), and the expression of WRB and CAML can complement Get1/2Δ strains (Vilardi et al., 2014). Importantly, this pathway was largely derived from studying the homologous pathway in yeast (the GET pathway) and using mammalian in vitro reconstitution assays. However, it remains unclear whether the GET/TRC system is the sole mechanism responsible for targeting tail-anchored proteins to the ER in vivo. The idea that there are other pathways is supported by the fact that GET or TRC pathway components can be deleted in yeast (Schuldiner et al., 2005) and mammalian cells (Sasaki et al., 2007) with minimal effects on cell viability, despite the fact that some tail-anchored proteins are necessary for cell homeostasis.
Here, we demonstrate that some mRNAs encoding the tail-anchored proteins, such as nesprin-2 and Sec61β, associate with the ER. Our data suggests that the ER association of Sec61b mRNA is not dependent on TRC40, BAT3 or p180. Interestingly, overexpression of Sec61b mRNA displaces other mRNAs from the ER, including those that are anchored by translocon-bound ribosomes. This indicates that certain mRNAs encoding tail-anchored proteins can access translocon-bound ribosomes on the surface of the ER and suggests a new alternative pathway for their targeting.
RESULTS
Sec61b mRNA is partially localized on the ER
It is currently believed that mRNAs encoding tail-anchored proteins are first translated by free ribosomes, and that the encoded polypeptide is later post-translationally targeted to the ER through the TRC pathway (Rabu et al., 2009; Borgese and Fasana, 2011; Hegde and Keenan, 2011).
To assess the distribution of endogenous mRNA in human cells, we stained U2OS cells with a panel of fluorescent in situ hybridization (FISH) probes. By simultaneously staining with many probes, one can efficiently visualize individual mRNA molecules (Coassin et al., 2014), as can be seen in Fig. 1. To determine whether these RNAs were tethered to the ER we repeated the experiment in cells that were treated with digitonin, which permeabilizes the plasma membrane and thus extracts the cytosol and removes any molecule that is not associated with the ER (Lerner et al., 2003; Cui et al., 2012; Cui and Palazzo, 2012). By comparing the number of puncta in non-extracted versus extracted cells, we can determine the percentage of mRNAs that are anchored to the ER.
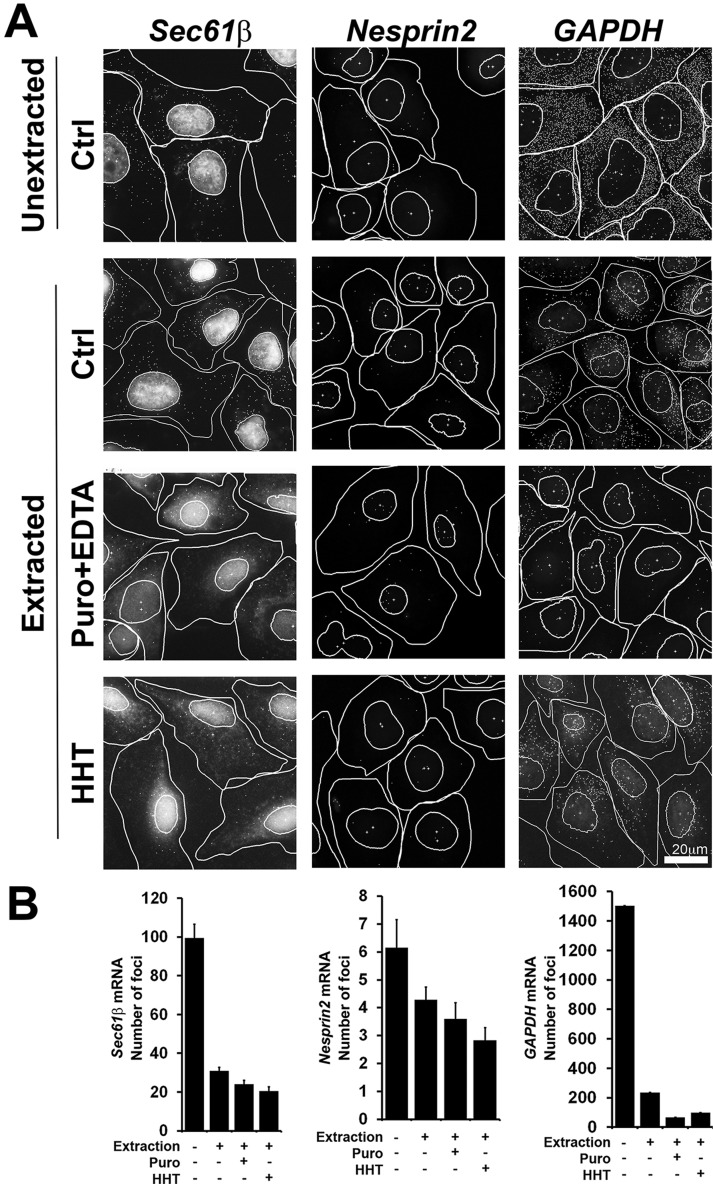
Fig. 1.
Endogenous Sec61b and nesprin-2 mRNA associates with the ER membrane. U2OS cells were either: fixed (Unextracted); first extracted with digitonin and then fixed (Extracted); or pre-treated with puromycin (Puro) or homoharringtonine (HHT) for 30 min, extracted with digitonin in the presence or absence of EDTA and then fixed. Cells were stained with a pool of FISH probes to visualize individual endogenous human Sec61b, nesprin-2 or GAPDH mRNA molecules. Each cell was visualized by phase microscopy to determine the cell contours. mRNA foci were identified using the NIS-element ‘Spot Detection’ function (see Materials and Methods section). (A) mRNA FISH signals overlaid with the contours of the cells and nuclei, and with the detected foci highlighted by the spot detection function. (B) The number of cytoplasmic (i.e. non-nuclear) foci were determined for each condition. Each bar is the mean±s.e.m. of 30 cells. Scale bar: 20 µm.
First, we examined the localization of Sec61b mRNA, which encodes a tail-anchored protein. Sec61β is a component of the translocon, the major protein-conducting channel in the ER, and has been widely used as a model TRC pathway substrate (Borgese and Fasana, 2011). Surprisingly, we found that ∼30% of the endogenous Sec61b mRNA was resistant to digitonin extraction (Fig. 1A,B). To test whether the localization of Sec61b mRNA was translation dependent, we examined the mRNA localization in cells treated with either homoharringtonine (HHT), or with puromycin followed by extraction with EDTA (Puro+EDTA), two treatments that effectively dissociate ribosomes from mRNA (Cui et al., 2012). To our surprise, most of the ER-localized mRNA was unaffected by these treatments.
Next, we monitored the localization of nesprin-2 (SYNE2) mRNA, which encodes a giant tail-anchored protein (796 kDa) that is present on the outer nuclear envelope and is involved in nuclear positioning (Luxton et al., 2010). After extraction, about two thirds of the foci remained, indicating that some of this mRNA was anchored to the ER (Fig. 1A,B). To ensure that the FISH signal was specific, we also probed cells that were depleted of the endogenous nesprin-2 mRNA using RNA interference (RNAi). Indeed, small hairpin RNA (shRNA)-treated cells lost 90% of their signal (supplementary material Fig. S1), indicating that our nesprin-2 probes detected the intended target. Like Sec61b, nesprin-2 mRNA largely remained associated with the ER in cells treated with HHT, or puromycin and EDTA. Thus nesprin-2, like Sec61b, can associate with the ER membrane, and this activity is mostly independent of translation.
To determine whether partial ER association was a general phenomenon for all mRNAs, we next investigated the localization of an mRNA encoding a cytosolic protein, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). We could reproducibly find 15% of the GAPDH puncta in digitonin-extracted cells (Fig. 1A,B). However, in contrast to what we had seen for Sec61b and nesprin-2, most of the GAPDH mRNAs were extracted in cells treated with either HHT, or puromycin+EDTA (Fig. 1), suggesting that the small amount of ER association was mediated by translating ribosomes.
Thus, we conclude that at least two endogenous mRNAs that encode tail-anchored proteins are also associated with the ER, and this was mostly mediated by contacts that did not involve the ribosome.
The ORF of Sec61b mRNA is required to anchor to the ER independently of translation
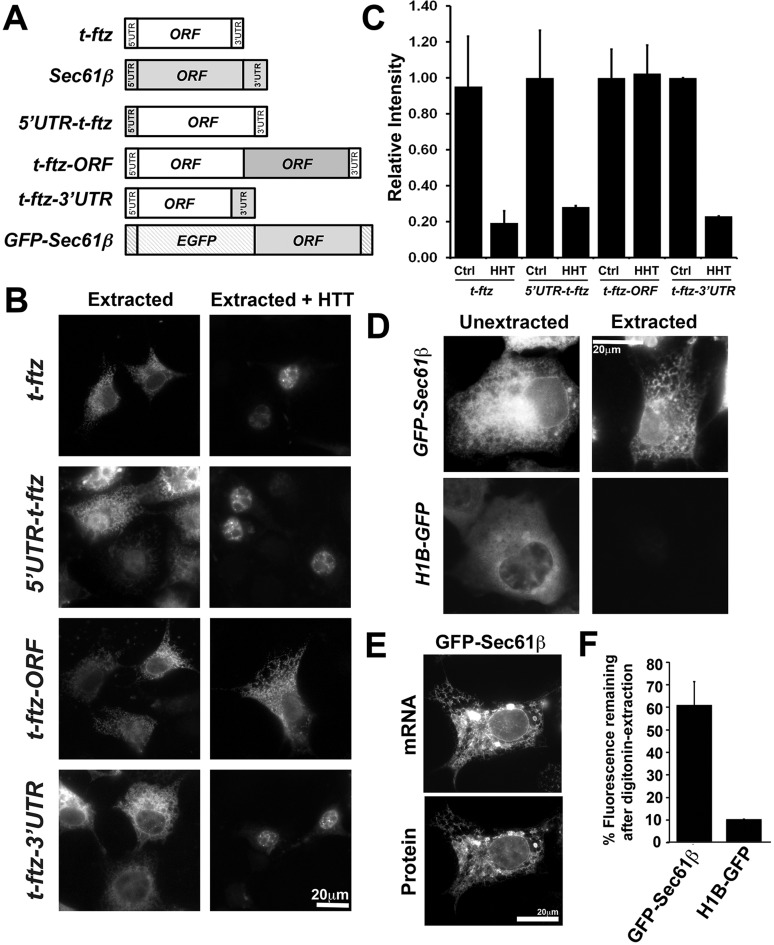
We next wanted to identify the region of Sec61b mRNA responsible for its ER anchorage. We followed a strategy that we had previously used to identify regions in the placental alkaline phosphatase (ALPP) mRNA that promoted ER anchorage (Cui et al., 2013). We fused different regions of Sec61b to t-ftz (Fig. 2A), an artificial mRNA that encodes a secretory protein and requires translation for ER association (Cui et al., 2012). These constructs were expressed in COS7 cells. After 18–24 h, cells were treated with either control medium or HHT for 30 min to disrupt ribosomes, then extracted to remove non-ER-associated mRNAs, followed by FISH staining to visualize the chimeric mRNAs. To our surprise, versions of t-ftz containing either the 5′UTR (5′UTR-t-ftz) or 3′UTR (3′UTR-t-ftz) of Sec61b did not remain anchored to the ER after HHT treatment, resembling the original t-ftz mRNA (Fig. 2B, for a quantification of the fluorescence intensity, see Fig. 2C). In contrast, a version of t-ftz fused to the Sec61b open reading frame (ORF) (t-ftz-ORF) remained ER associated after HHT treatment (Fig. 2B). In fact, quantification of the FISH intensities revealed that the level of ER association did not significantly change between control and HHT-treated cells (Fig. 2C).
Fig. 2.
Overexpressed GFP-Sec61b mRNA is associated with the ER membrane. (A) Schematic diagram of constructs. All t-ftz sequences are shown in white, Sec61b sequences are shown in gray and EGFP sequences are shown as hatched boxes. (B,C) Chimera plasmids containing either the Sec61b 5′UTR, 3′UTR or the ORF fused to t-ftz were transfected into COS7 cells. At 18–24 h post-transfection, cells were treated with either control or HHT, followed by digitonin extraction to remove cytoplasmic contents. Cells were fixed, stained using FISH probes against ftz, and imaged. (D–F) Plasmids encoding GFP–Sec61b or H1B–GFP were transfected into U2OS cells. At 18–24 h post transfection, cells were either fixed directly (Unextracted) or after digitonin extraction (Extracted). GFP–Sec61b or H1B–GFP mRNAs were stained with FISH probes against the GFP-coding sequence and visualized. mRNAs in unextracted and digitonin-extracted cells are shown in D. Note that GFP–Sec61b, but not H1B–GFP mRNA, is resistant to digitonin extraction and exhibits a reticular staining pattern. (E) Distribution of GFP–Sec61β protein and mRNA in a digitonin-extracted U2OS cell. Both images are from a single field of view. Note the extensive colocalization of the mRNA with its encoded protein, which is localized to the ER (Rolls et al., 1999; Shibata et al., 2008). (F) Quantification of GFP–Sec61b and H1B–GFP mRNA cytoplasmic intensity signals. The ratio of fluorescence in the cytoplasms of extracted versus unextracted cells was determined. Each bar in C and F represents the mean±s.e.m. of three independent experiments, each containing at least 30 cells. Scale bars: 20 µm.
To further validate these findings, we examined the distribution of GFP–Sec61b, a construct that contains the ORF of the human Sec61b gene (Fig. 2A). In unextracted COS7 cells, the mRNA had a noticeable reticular-like distribution, suggesting that a large fraction of this mRNA was localized to the ER (Fig. 2D). In digitonin-treated cells, a large portion of the GFP–Sec61b mRNA was resistant to extraction (Fig. 2D). In these cells, GFP–Sec61b mRNA colocalized with its translated product, GFP–Sec61β protein (Fig. 2E), which is a well-established marker of the ER (Rolls et al., 1999). In contrast, H1B–GFP mRNA, which encodes a nuclear histone protein, was mostly extracted by digitonin treatment (Fig. 2D). When the FISH fluorescence levels in extracted and unextracted cells were compared, we observed that 60% of the GFP–Sec61b mRNA was resistant to extraction (Fig. 2F). This is comparable to what we have previously observed for other overexpressed mRNAs encoding secreted and membrane-bound proteins (Cui et al., 2012; Cui and Palazzo, 2012). In contrast, only ∼10% of H1B–GFP mRNA was resistant to digitonin extraction (Fig. 2F), which is also in line with our previous observations (Cui et al., 2012).
Next, we assessed whether ER association of GFP–Sec61b mRNA required translation. Neither puromycin+EDTA nor HHT treatment disrupted the ER association of GFP–Sec61b mRNA in COS7 cells, as assessed by digitonin extraction (Fig. 3A,B). HHT treatment only slightly decreased the ER localization of this mRNA in U2OS cells (Fig. 3C,D). To control for differences in mRNA expression and staining efficiency, we also measured the nuclear fluorescence, and this did not change under any of the tested conditions (Fig. 3B,C). The localization of GFP–Sec61b mRNA to the ER in HHT-treated U2OS cells was confirmed by colocalization of the mRNA with the ER marker Trapα (also known as SSR1) (Fig. 3E).
Fig. 3.
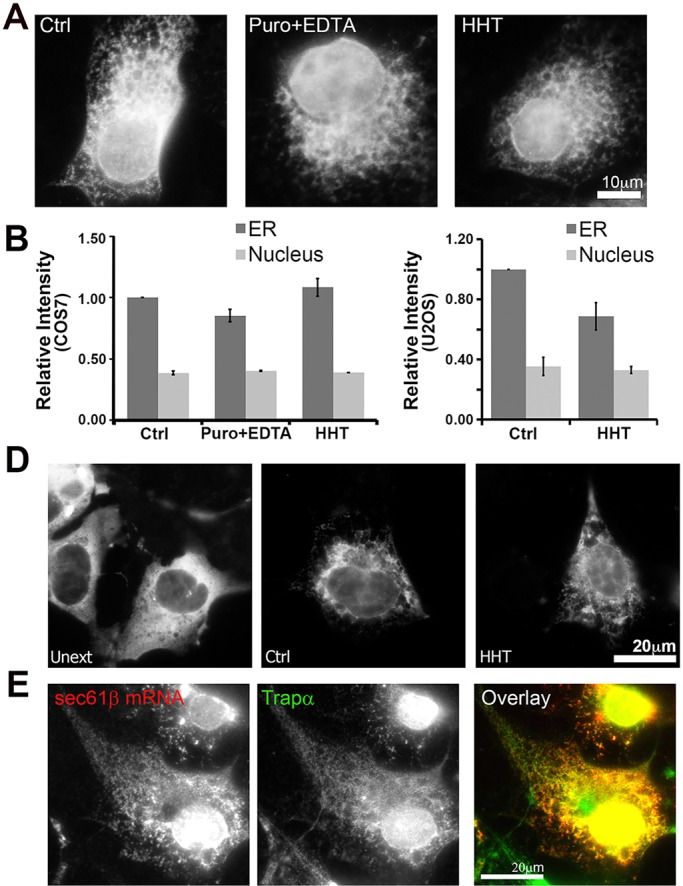
ER association of overexpressed GFP–Sec61b mRNA is partially independent of translation. (A,B) COS7 and U2OS (C,D) cells were transfected with plasmid encoding GFP–Sec61b and allowed to express mRNA for 18–24 h. Cells were then treated with DMSO (Ctrl), puromycin (Puro) or homoharringtonin (HHT) for 30 min, and then extracted with digitonin with or without EDTA. Cells were then fixed and stained for mRNAs using a specific FISH probe against the GFP-coding sequence. Cells were imaged (A,D), and the fluorescent intensities were quantified (B,C). To control for changes in staining, nuclear fluorescent intensities were also analyzed. Each bar represents the mean±s.e.m. of three independent experiments, with each experiment consisting of at least 30 cells. (E) U2OS cells expressing GFP–Sec61b were treated with HHT and then digitonin extracted. Cells were then stained for the GFP–Sec61b mRNA, and immunostained with the ER marker Trapα. Images in E are from a single field of view including a color overlay showing the GFP–Sec61b mRNA in red and Trapα in green. Scale bars: 20 µm.
From these experiments, we conclude that the ORF of Sec61b mRNA can promote ER association and that this activity is largely independent of its ribosome association and active translation.
mRNAs encoding other exogenously expressed tail-anchored proteins are mainly localized to the cytosol
To determine whether the results obtained with GFP–Sec61b mRNA can be generalized to other mRNAs encoding tail-anchored proteins, we examined the localization of other overexpressed GFP fusion transcripts. In particular, we analyzed the distribution of mRNAs containing ORFs that encode tail-anchored proteins destined to be targeted to the ER (Sec22β and Sec61γ), peroxisome (Pex26) or mitochondria (FIS1). Previously, it has been shown that newly synthesized Pex26 protein is targeted to the peroxisome by Pex19 and thus is independent of the TRC40-dependent pathway (Yagita et al., 2013). For tail-anchored proteins destined for the mitochondria, they are thought to be recognized by a pre-targeting complex which then prevents their sorting to the ER and instead diverts these to the mitochondrial outer membrane (Wang et al., 2010). This sorting process is thought to be dictated by the relative hydrophobicity of the TMD and the presence of charged residues in the vicinity of the TMD (Borgese et al., 2001; Horie, 2003; Wang et al., 2010).
As expected, GFP–Sec22β and GFP–Sec61γ proteins were targeted to the ER in COS7 cells (data not shown). Likewise, GFP–FIS1 and GFP–Pex26 proteins were targeted, as expected, to the mitochondria (supplementary material Fig. S2A) and peroxisomes (data not shown), respectively. However, unlike GFP–Sec61b, all of the other tested mRNAs were efficiently removed by digitonin extraction (Fig. 4A, compare ‘Cyto/ER’ levels in unextracted and extracted cells), similar to what was seen for mRNAs encoding non-secretory proteins (H1B–GFP; Fig. 4A).
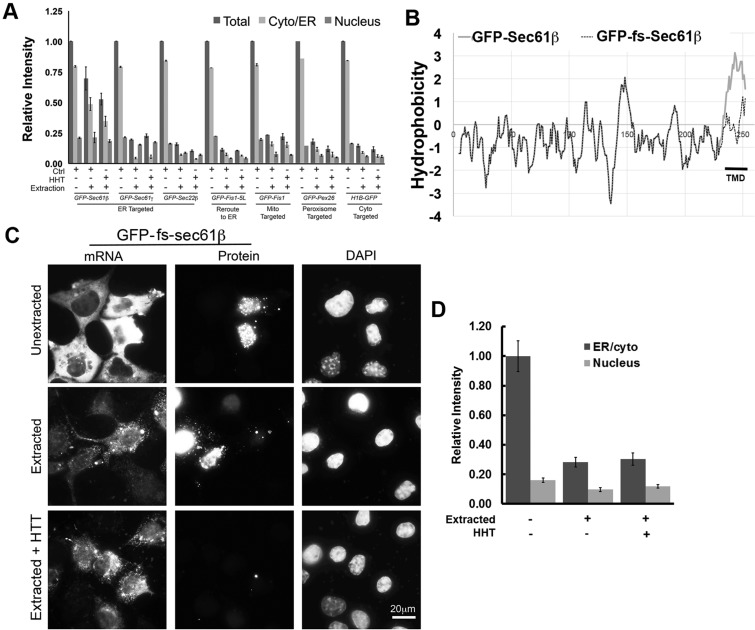
Fig. 4.
The coding potential of GFP-Sec61b is not required for its localization to the ER. (A) COS7 cells were transfected with plasmid encoding various GFP-tagged tail-anchored proteins and allowed to express for 18–24 h. The cells were treated with control medium or HHT for 30 min, then either directly fixed or extracted with digitonin and then fixed. Cells were stained for mRNAs using specific FISH probe against the GFP-coding sequence, which was then imaged and quantified. Fluorescent intensities in the cytoplasm and nucleus were quantified. All results were normalized to the cytoplasmic staining intensity in the unextracted cells. Each bar represents the mean±s.e.m. of three independent experiments, each consisting of at least 30 cells. (B) Hydrophobicity (y-axis, left) of the polypeptides encoded by GFP–Sec61b and GFP-fs-Sec61b was plotted against the peptide length (x-axis, bottom). Kyte–Doolittle hydropathy values were computed with ProtScale (http://web.expasy.org/protscale/), using a moving window size of 21 amino acids. Note the high hydrophobicity of the TMD region of GFP–Sec61β that is lost in GFP–fs-Sec61β. (C) COS7 cells were transfected with plasmid encoding GFP–fs-Sec61b and allowed to express mRNA for 18-–24 h. Cells were then treated with control medium or HHT for 30 min, and then either fixed (Unextracted) or extracted with digitonin and then fixed (Extracted). Cells were stained for mRNAs using a specific FISH probe against the GFP-coding sequence, and for DNA using DAPI. Each row represents a single field of view imaged for GFP–fs-Sec61b mRNA, GFP protein and DAPI. (D) Quantification of the cytoplasmic (in unextracted cells), ER (in extracted cells) and nuclear fluorescence intensities of GFP–fs-Sec61b mRNA. Each bar represents the mean±s.e.m. of 30 cells. Scale bar: 20 µm.
We next explored the idea of whether the targeting of a mitochondrial tail-anchored protein to the ER would also increase the amount of ER targeting of its mRNA. When we increased the hydrophobicity of the TMD of FIS1 (FIS1-5L; supplementary material Fig. S2B), the protein was successfully rerouted to the ER (supplementary material Fig. S2A). However, the mRNA of GFP–FIS1-5L was still sensitive to extraction and did not localize to the ER (Fig. 4A).
From these experiments, we conclude that ER targeting of the protein product is not sufficient for the ER localization of an mRNA.
The encoded TMD is not strictly required for the ER localization of GFP–Sec61b mRNA
Although ER targeting of the protein product did not correlate with the ER association of the mRNA, it was still possible that Sec61b mRNA localization was dependent on its encoded TMD. To further examine this possibility, we frame shifted the TMD of Sec61b by inserting a single cytosine nucleotide before the TMD coding region (to create GFP–fs-Sec61b). This mutation eliminated the hydrophobic region at the C-terminus of the coding protein (Fig. 4B). Only a few COS7 cells that expressed the GFP–Sec61b mRNA showed GFP protein synthesis (for example, see Fig. 4C first row). When it was present, GFP–fs-Sec61β was found in small aggregates that concentrated in the nucleus (see GFP protein localization in Fig. 4C). Consistent with the idea that the translation of the mRNA was not required for ER localization, a fraction of GFP–fs-Sec61b mRNA was anchored to the ER (Fig. 4C). When we quantified the amount of mRNA before and after extraction, we found that the amount of ER association in COS7 cells was ∼30% (Fig. 3D), which is about half of what we observed for GFP–Sec61b mRNA (see Fig. 2F). This level of ER association was not affected by HHT treatment (Fig. 4D), further confirming that this localization activity occurred independently of ribosomes and translation.
From these results, we conclude that the ER localization of the encoded protein is not required for the localization of GFP–Sec61b mRNA. However, given that the targeting of the frame-shifted mutant was clearly decreased from what we had seen with GFP–Sec61b mRNA, it is likely that translation of this mRNA into an ER-targeted protein might enhance mRNA localization.
The initial targeting of GFP–Sec61b mRNAs to the ER is partially independent of translation and ribosomes
Although our data indicated that most ER-targeted GFP–Sec61b mRNA could be maintained on the ER independently of translation and ribosomes, we wanted to investigate whether these processes were required for the initial targeting of this mRNA to the membrane. This could potentially explain why more of the GFP–Sec61b mRNA was associated with the ER in comparison to GFP–fs-Sec61b mRNA. To test this, we microinjected plasmid encoding GFP–Sec61b into the nuclei of U2OS cells that were pretreated with either control solution or the translation inhibitor HHT, and examined the targeting of the newly synthesized transcript. As these mRNAs would have never encountered a ribosome, their initial targeting would be strictly mediated by RNA localization pathways. In unextracted cells, mRNAs were efficiently exported out of the nucleus (Fig. 5A). As expected, GFP–Sec61β protein was only made in the control and not the HHT-treated cells, indicating that the translation inhibitor efficiently blocked protein synthesis (Fig. 5A). In extracted cells, GFP–Sec61b mRNA was still observed on the ER (Fig. 5A), and by comparing the difference between FISH intensity in unextracted and extracted cells we estimate that ∼70% of the cytosolic mRNA was targeted to the ER. After HHT treatment, ER targeting of the Sec61b mRNAs decreased by two thirds (Fig. 5A,B). It is possible that this number underestimates the level of ER targeting, as in the absence of ribosome association, newly synthesized mRNAs might be more efficiently degraded.
Fig. 5.
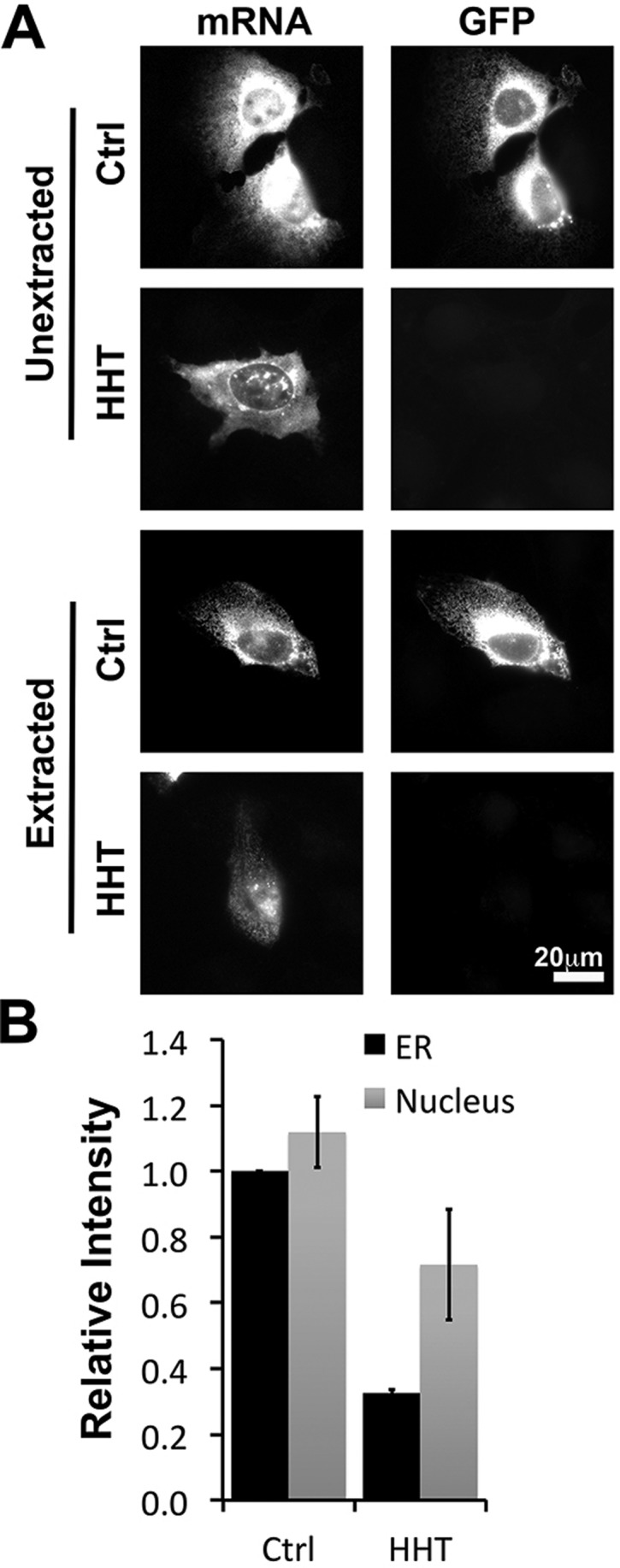
The initial targeting of Sec61b mRNA to the ER is partially dependent on ribosomes and translation. U2OS cells were pretreated with control medium (Ctrl) or HHT for 15 min, then microinjected with plasmids containing GFP–Sec61b and allowed to express mRNAs for 2 h in the presence of medium with or without HHT. The cells were then extracted with digitonin, fixed and stained with FISH probe against the GFP-coding sequence, and imaged. (A) Representative images, with each row representing a single field of view imaged for GFP–Sec61b mRNA (mRNA) and GFP fluorescence (GFP). (B) Quantification of the fluorescence intensities of mRNAs in the ER and nucleus of extracted cells. Each bar represents the mean±s.e.m. of three independent experiments, each experiment consisting of at least 30 cells. Scale bar: 20 µm.
In conclusion, these results suggest that although the initial targeting of Sec61b mRNA can occur to a certain extent in the absence of translation, it is clearly enhanced in the presence of translating ribosomes.
p180 is not required for the localization of either GFP–Sec61b mRNA or its encoded protein
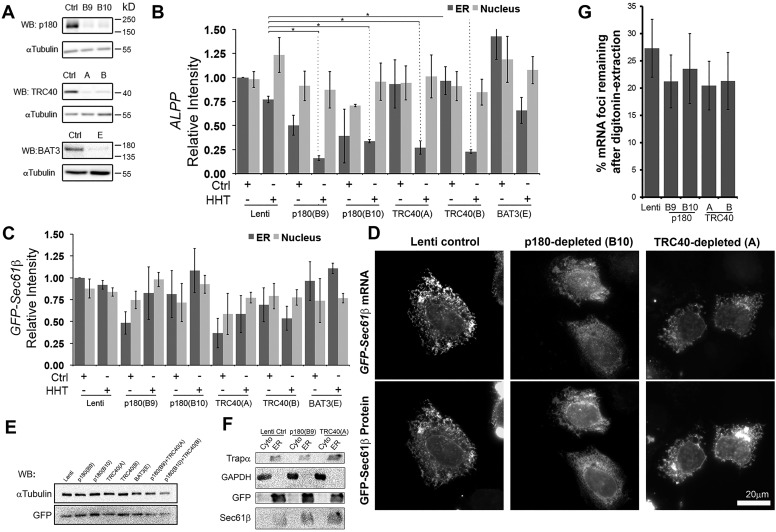
We previously identified p180 as an mRNA receptor that promoted the ER anchoring of several mRNAs to the ER independently of ribosomes and translation (Cui et al., 2012), and we next tested whether it was required for the localization of GFP–Sec61b mRNA. p180 was depleted from U2OS cells using two separate lentiviral-delivered shRNAs (B9 and B10, Fig. 6A). As a positive control, we tested the ER localization of the ALPP mRNA. This transcript, which encodes a glycosylphosphatidylinositol (GPI)-anchored protein, can be targeted and maintained on the surface of the ER by the action of p180 (Cui et al., 2012, 2013). Indeed, depletion of p180 with either shRNA construct decreased the ER association of ALPP mRNA in both control and HHT-treated cells (Fig. 6B), as we have previously published (Cui et al., 2012). In contrast, depletion of p180 did not consistently decrease the amount of GFP–Sec61b mRNA on the ER (Fig. 6C). p180 depletion did not affect ER localization (Fig. 6D) or the overall levels (Fig. 6E) of GFP–Sec61β protein. When various cell fractions were assayed, GFP–Sec61β protein was present in the ER (Fig. 6F), which was consistent with the localization data (Fig. 6D). When we measured the number of individual endogenous Sec61b mRNA foci (as in Fig. 1), we observed that p180 depletion did not have a significant impact on the percentage of ER-associated mRNAs (Fig. 6G).
Fig. 6.
p180, TRC40 and BAT3 are not required for the ER association of Sec61b mRNA and protein. (A–C) U2OS cells were infected with lentivirus carrying control shRNA (Lenti), or shRNAs against p180 (clones B9 or B10), TRC40 (clones A or B), or BAT3 (clone E). The control and shRNA-infected cells were transfected with plasmids containing either the ALPP or GFP–Sec61b constructs and allowed to express these mRNAs for 18–24 h. Cells were then treated with either control medium or HHT for 30 min, digitonin extracted, fixed and stained with specific FISH probes, and imaged. (A) Cell lysate was collected on the day of transfection, separated by SDS-PAGE and immunoblotted against p180, TRC40, BAT3 and αtubulin. (B,C) Quantification of the fluorescence intensities of ALPP (B) and GFP–Sec61b (C) mRNAs, in the ER and nucleus. The results were normalized to the ER staining intensity of cells infected with control shRNA and treated with control medium. Each bar represents the mean±s.e.m. of three or four independent experiments, each experiment consisting of at least 30 cells. *P<0.05 (Student's unpaired t-test). (D) shRNA-infected U2OS cells were transfected with plasmids containing GFP–Sec61b and allowed to express mRNAs for 18–24 h. The cells were then treated with or HHT for 30 min. Cells were digitonin-extracted, fixed and stained for GFP–Sec61b mRNA using FISH probe against GFP-coding region. Each column represents a single field of cells imaged for GFP protein and GFP mRNA. (E,F) shRNA-infected U2OS cells were transfected with plasmids containing GFP–Sec61b and allowed to express mRNAs for 18–24 h. Cells were either lysed directly (E) or fractionated into cytosolic (Cyto) and ER fractions (F). The total lysate (E) and fractionated samples (F) were analyzed by immunoblotting using antibody against GFP (GFP–Sec61β), endogenous Sec61β, Trapα (an ER marker) and GAPDH (a cytosolic marker). Depletion of p180 or TRC40 either alone or together did not affect the levels or ER localization of GFP–Sec61β or endogenous Sec61β protein. (G) shRNA-infected U2OS cells that were either digitonin-extracted, or directly fixed were stained with a pool of FISH probes to visualize individual endogenous human Sec61b mRNAs. The percentage of cytoplasmic foci remaining in digitonin-extracted versus unextracted cells was calculated. Quantitative results represents the mean±s.e.m. of 30 unextracted and 30 extracted cells. Scale bar: 20 µm.
From these observations, we conclude that p180 is dispensable for the ER association of GFP–Sec61b mRNA and protein. It formally remains possible that p180 might still play a role, but that other compensatory pathways exist for the ER localization of this mRNA.
TRC40 and BAT3 are not required for the localization of either GFP–Sec61b mRNA or its encoded protein to the ER
As the initial ER targeting of GFP–Sec61b mRNA was partially dependent on translation (Fig. 5A,B) and GFP–fs-Sec61b mRNA was not as efficiently localized to the ER as GFP-Sec61b, it was possible that mRNA localization might be partially coupled to the proper targeting of the encoded protein. In light of this, we assessed whether components of the TRC pathway were required for GFP–Sec61b mRNA localization to the ER.
TRC40 and BAT3 were depleted in U2OS cells by lentiviral delivered shRNAs (Fig. 6A), but to our surprise these treatments did not significantly interfere with the ER localization of the GFP–Sec61β protein (for TRC40-depleted cells, see Fig. 6D; for BAT3-depleted cells the data is not shown). Depletion of TRC40 might have had some effect on the amount of ER localization of GFP–Sec61b mRNA; however, this varied greatly between experiments (Fig. 6C). TRC40 depletion did not affect GFP–Sec61β protein levels (Fig. 6E) and did not mislocalize the protein to mitochondria (Fig. 6D) or the cytosol (Fig. 6F). Even when both p180 and TRC40 were co-depleted, levels of GFP–Sec61β protein remained constant relative to the loading control (Fig. 6E). Consistent with our observations on overexpressed GFP–Sec61β mRNA and protein, depletion of TRC40 did not have a significant impact on the amount of endogenous Sec61b mRNA that was associated to the ER (Fig. 6G).
Unexpectedly, depletion of TRC40 affected the ER localization of ALPP mRNA in the HHT-treated cells (Fig. 6B). Interestingly, it has been previously shown that the S. cerevisiae ortholog of TRC40, Get3, is required for the ER targeting of GPI-anchored proteins in SRP-disrupted yeast cells (Ast et al., 2013). Our new results suggest that cells depleted of TRC40 might have defects in the localization of certain mRNAs, and this might explain these previous results.
Depletion of BAT3 had no effect on the ER localization of GFP–Sec61b and ALPP mRNA (Fig. 6B,C).
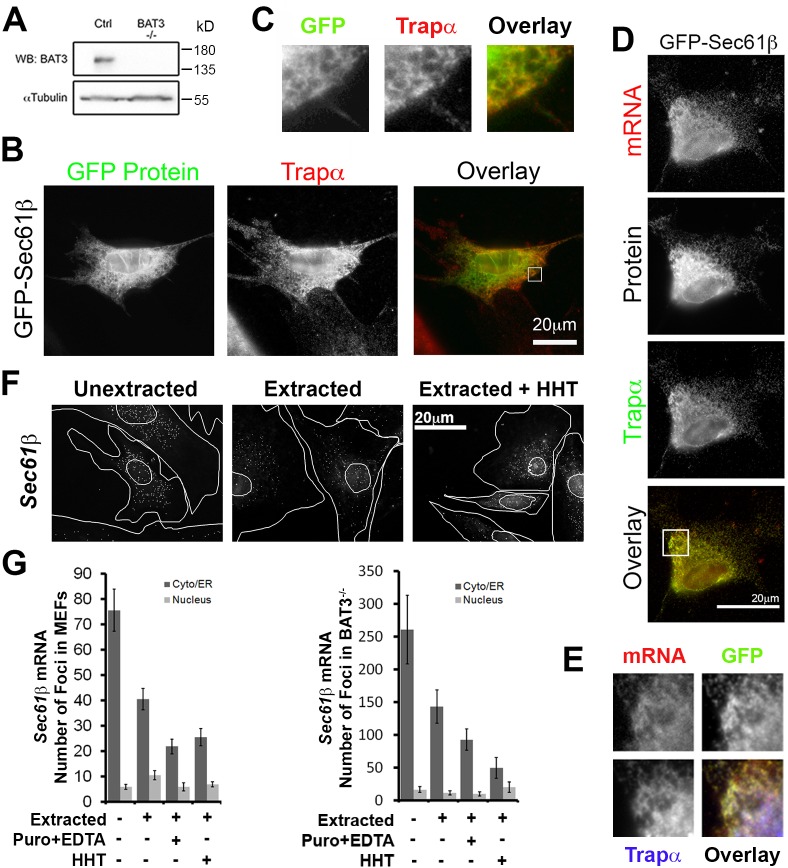
To confirm the observation that the ER localization of GFP–Sec61b mRNA and its encoded protein are mostly independent of the TRC pathway, we repeated these experiments in BAT3-knockout mouse embryonic fibroblasts (MEFs; Fig. 7A). In unextracted cells, exogenously expressed GFP–Sec61β protein clearly colocalized with the ER marker Trapα (Fig. 7B, see high magnification of the boxed area in Fig. 7C), indicating that BAT3 was not required for the ER targeting of this protein. In extracted cells, both the Sec61b mRNA and protein colocalized with Trapα (Fig. 7D, high magnification images of the boxed area are shown in Fig. 7E).
Fig. 7.
BAT3 is not required for the ER association of Sec61b mRNA and protein. (A) Western blot of BAT3 protein in control and BAT3−/− MEFs. (B,C) BAT3−/− MEFs expressing GFP–Sec61b for 18–24 h were fixed and immunostained for the ER marker Trapα. Images in B are from a single field of view including a color overlay showing the GFP–Sec61b mRNA in green and Trapα in red. Higher magnification images of the boxed area in B are shown in C. (D,E) BAT3−/− MEFs expressing GFP–Sec61b for 18–24 h were extracted and stained for the GFP mRNA by FISH and immunostained for the ER marker Trapα. (D) A single field of view showing GFP mRNA, GFP protein, Trapα and an overlay of the GFP mRNA (red) and Trapα (green). Higher magnification images of the boxed area are shown in E with an overlay of GFP–Sec61b mRNA (red), GFP–Sec61β protein (green) and Trapα (blue). (F) BAT3−/− MEFs were either directly fixed (Unextracted), first extracted with digitonin and then fixed (Extracted), or pre-treated with homoharringtonine (HHT) for 30 min, extracted with digitonin and then fixed. Cells were stained with a pool of FISH probes to visualize individual endogenous mouse Sec61b mRNA molecules. (G) The number of cytoplasmic (i.e. non-nuclear) foci were determined for each condition in control MEFs and BAT3−/− cells. Each bar is the mean±s.e.m. of 30 cells. Scale bars: 20 µm.
We then investigated whether the endogenous Sec61b mRNA was associated with the ER. As we had seen previously with U2OS cells, a sizeable number of Sec61b mRNA foci were resistant to digitonin extraction in both BAT3−/− cells and wild-type MEFs (∼50%, Fig. 7F,G). The number of foci decreased after ribosomes were disrupted with either HHT or puromycin+EDTA treatments, but was still substantial.
From these results, we conclude that the ER targeting of GFP–Sec61b mRNA and its encoded protein was largely independent of the TRC pathway components TRC40 and BAT3. Although it is possible that the small amount of TRC40 remaining after shRNA depletion might be sufficient for the correct targeting of mRNA and/or protein, the fact that these processes are unaltered in BAT3−/− cells suggests that this component is dispensable. Although TRC pathway components might promote mRNA and protein targeting to the ER, our data suggests that other parallel pathways should exist. The presence of these alternative pathways for tail-anchored protein insertion, beyond the TRC pathway, would explain how BAT3-knockout cells are able to survive, despite the fact that certain tail-anchored proteins, such as Sec61γ, are required for cell viability.
GFP–Sec61b mRNA competes with other mRNAs for ribosome binding sites on the ER
In order to understand how different mRNAs associate with the ER and whether they share similar binding sites, we investigated whether two mRNAs could compete with each other (i.e. whether an increase in the levels of one would displace the other from the ER).
We co-expressed GFP–Sec61b with two different mRNAs, t-ftz and ALPP. The first mRNA requires translation for both its targeting and maintenance on the surface of the ER (Cui et al., 2012); thus, we presume that it is anchored to the ER by virtue of the fact that it is being translated by translocon-bound ribosomes. As mentioned above, we have previously demonstrated that >50% of ALPP mRNA is associated with the ER in a translation-dependent manner and that the remaining fraction is largely dependent on p180 (Cui et al., 2012).
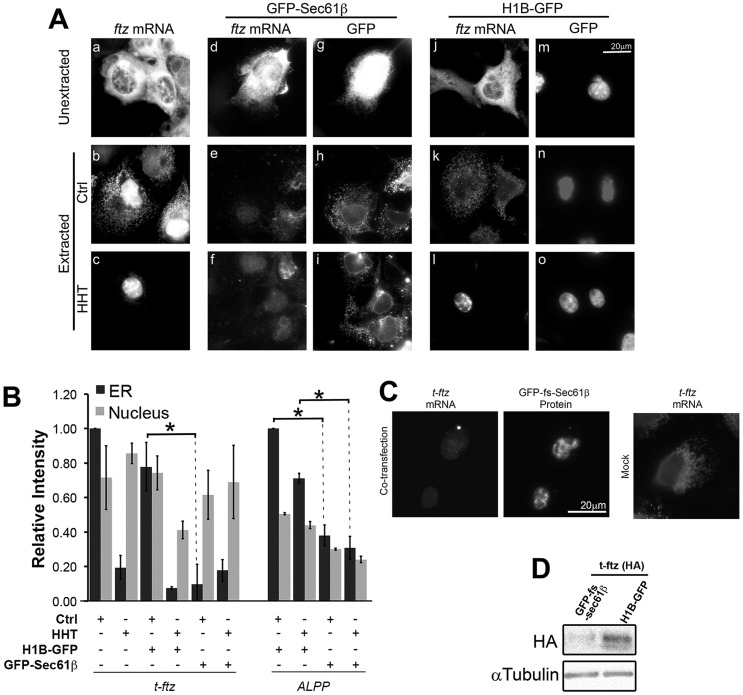
Interestingly, cells expressing GFP–Sec61β had a significant decrease in the amount of t-ftz mRNA on the ER in comparison to cells either expressing t-ftz alone or in combination with a control gene (H1B–GFP) (Fig. 8A,B). In most cases, no t-ftz mRNA could be detected on the ER (Fig. 8A, panel e). In agreement with our previous published results (Cui et al., 2012), t-ftz mRNAs were also displaced from the ER upon HHT treatment (Fig. 8A, compare panels c, f and l to panels b, e and k; see Fig. 8B for quantifications), further underscoring the fact that this mRNA requires active translation for ER association. Note that nuclear levels of t-ftz remained largely unaltered by GFP–Sec61β expression (see quantification in Fig. 8B).
Fig. 8.
GFP–Sec61b mRNA competes with t-ftz mRNA for the ribosome-binding sites on the ER. (A,B) COS7 cells were transfected with plasmid containing a test gene (t-ftz or ALPP) alone or in combination with plasmid containing a competitor gene (GFP–Sec61b or H1B–GFP). The cells were then treated with either control medium (Ctrl) or HHT for 30 min, then digitonin extracted, fixed and stained with specific FISH probes, and imaged. (A) Representative images of COS7 cells expressing t-ftz mRNA alone (a–c) or in combination with GFP–Sec61b (d–i) or H1B-–GFP (j–o). Panels a–c are stained for t-ftz mRNA, whereas each pair of panels in d–o represents a single field of view imaged for t-ftz mRNA and GFP fluorescence. (B) Quantification of the ER and nuclear staining intensity of either t-ftz mRNA or ALPP mRNA in transfected cells. All data was normalized to the ER staining intensities in the control treated group for each construct. Each bar represents the mean±s.e.m. of three independent experiments, each consisting of at least 30 cells. (C) COS7 cells were transfected with t-ftz alone or in combination of GFP–fs-Sec61b. At 18–24 h post transfection, cells were digitonin extracted to remove cytoplasmic contents. GFP–fs-Sec61b mRNAs were stained with FISH probe against the GFP-coding sequence and visualized. (D) Cell lysates of COS7 cells cotransfected with t-ftz in combination with either H1B–GFP or GFP–fs-Sec61β were analyzed by western blotting. t-ftz protein expression was examined using HA antibody against an HA epitope present in the t-ftz protein and antibodies against α-tubulin to control for loading. Scale bar: 20 µm.
When GFP–Sec61b was co-expressed with ALPP, we again observed a decrease in the amount of ALPP mRNA on the ER in comparison to cells expressing ALPP alone (Fig. 8B). However unlike t-ftz, the amount of ER-associated ALPP dropped by only 60%. When the co-expressing cells were treated with HHT, the level of ALPP mRNA on the ER did not decrease further (Fig. 8B), suggesting the decrease was mainly due to competition between GFP–Sec61b and ALPP mRNAs for translocon-associated ribosomes.
Thus, it is clear that the expression of GFP–Sec61b disrupts the ER localization of other mRNAs. The displacement of t-ftz by GFP-Sec61b suggests that both of these mRNAs occupy the same ER attachment site, namely translocon-bound ribosomes. It is, however, possible that expression of GFP–Sec61b caused some other indirect effects that ultimately resulted in a reduction of mRNA–ER association.
We next tested whether expression of GFP–fs-Sec61β would also displace t-ftz mRNA. Unfortunately, many of the cells expressing GFP–fs-Sec61b mRNA could not be identified, as few cells express visible levels of protein (for an example, see Fig. 4C). As such, we could not readily identify cells co-expressing both constructs. However, we observed that very few of the cells contained detectable levels of t-ftz mRNA in the cytosol after extraction, whether they expressed GFP–fs-Sec61β protein or not (Fig. 8C). If GFP–fs-Sec61b mRNA was displacing t-ftz mRNA off of the ER, we would also expect that the level of t-ftz protein should decrease in the co-transfected cells. To test this we co-expressed t-ftz with either GFP–fs-Sec61b or H1B–GFP, to control for non-specific competition of translation factors by an overexpressed protein. We found that expression of GFP–fs-Sec61β completely disrupted the production of t-ftz protein (Fig. 8D).
From these results, we conclude that overexpressed GFP–Sec61b disrupts the ER localization of other mRNAs and likely perturbs their translation into secretory proteins.
DISCUSSION
In this paper, we provide evidence that at least one mRNA that encodes a tail-anchored protein is efficiently targeted and then maintained on the surface of the ER. Although our data suggests that mRNA localization does not strictly require active translation and/or ribosomes, it appears that these processes might contribute to the association of this mRNA to the ER.
Overall, our results suggest that multiple pathways exist to target tail-anchored proteins to the ER, including the direct localization of certain mRNAs, such as Sec61b, to the surface of the organelle. The encoded protein either might then be spontaneously inserted into the membrane or might use some protein-conducting channel. In agreement with our findings, other groups have also observed that certain mRNAs encoding tail-anchored proteins are associated with the ER (Reid and Nicchitta, 2012; Kraut-Cohen et al., 2013). In particular, Reid and Nichitta observed that 20% of Sec61b mRNA is associated with the ER in HEK293 cells (Reid and Nicchitta, 2012), a figure that is close to our measurements (Fig. 1B).
Our data also suggests that once the GFP–Sec61b mRNA is at the ER, it might be able to access translocon-bound ribosomes. This finding raises the possibility that the encoded protein of GFP–Sec61b mRNA might use translocons to insert into the membrane. Interestingly, the insertion of Sec61β protein into the ER of extracted cells is not affected by translocon depletion (Lang et al., 2012). Moreover, the insertion of this protein, along with most other tail-anchored proteins, into ER-derived rough microsomes requires components of the TRC pathway (Stefanovic and Hegde, 2007). However, these in vitro and ex vivo results contrast sharply with the in vivo situation in which the deletion of components in this pathway is compatible with cellular viability in both yeast (Schuldiner et al., 2005) and mammalian tissue culture cells (Sasaki et al., 2007) despite the fact that many protein substrates are essential. Many of these crucial tail-anchored proteins, such as Sec61γ, whose mRNA does not appear to be ER associated (at least through overexpression), must be able to be correctly inserted into the ER independently of BAT3, as BAT3−/− cells are viable. Intriguingly, newly synthesized Sec61β and other tail-anchored proteins can interact with SRP and translocon components in vitro (Abell et al., 2003, 2004). In addition, other HSP40 or HSC70 chaperone systems might also act to promote membrane insertion of these proteins (Rabu et al., 2008). It should also be pointed out that certain tail-anchored proteins, such as cytochrome B5, can spontaneously insert into membranes (Brambillasca et al., 2005; Borgese and Fasana, 2011), and it is possible that localization of the mRNA to the membrane might facilitate this activity. Thus, many pathways likely act redundantly to insert tail-anchored proteins into the ER in vivo.
Our final finding that the overexpression of GFP–Sec61b displaces other mRNAs off of the ER has one major caveat. Although we have interpreted this observation as being due to the action of the GFP–Sec61b mRNA, we cannot totally rule out the possibility that this is due to the expression of the GFP–Sec61β protein. In particular, it is possible that this protein might be incorporated into native Sec61 translocons, which are composed of α, β and γ subunits. These altered translocons would have an additional GFP on their cytosolic face, which would likely prevent the binding of ribosomes and thus impede all translation- or ribosome-dependent anchoring of mRNAs to the ER. We, however, believe that this is unlikely for several reasons. First, GFP–Sec61β protein diffuses in the membrane of the ER at a rate compatible with that of membrane-tethered GFP and not of a large complex such as the translocon (Shibata et al., 2008) (for diffusion measurements of translocons, see Nikonov et al., 2002). Second, translocon disruption is extremely toxic to mammalian tissue culture cells (Lang et al., 2012), whereas the expression of GFP–Sec61β has little to no effect on cell viability. Third, translocons are typically distributed to the perinuclear sheets of the ER and are excluded from both the nuclear envelope and the peripheral tubes (Shibata et al., 2006, 2010), whereas in contrast, GFP–Sec61β is distributed to all three regions (nuclear envelope, sheets and tubes) and does not show a preference for the sheets even when expressed at very low levels (Shibata et al., 2008) (X.A.C. and A.F.P., unpublished observations). It is possible that a minority of translocons incorporate GFP–Sec61β; however, this would not explain why the majority of ER-bound t-ftz mRNA would be prevented from accessing translocon-bound ribosomes. Finally, direct perturbation of translocons would not explain why the expression of GFP–fs-Sec61b mRNA, which does not encode an ER-targeted protein, also displaces t-ftz mRNA from the ER (Fig. 8C).
Finally, it is interesting to note that Sec61β is required for efficient secretion and is an integral part of the endomembrane system. Work from the Nicchitta laboratory has found that mRNAs that encode endomembrane system components have an enhanced, translation-independent affinity for the ER (Chen et al., 2011). Additionally, the association between Sec61b mRNA and translocon-bound ribosomes might provide an opportunity for feedback regulation. In this way, translocon availability could potentially be linked to the translation of the Sec61b mRNA in order to regulate the production of new translocons and boost secretory capacity.
MATERIALS AND METHODS
DNA constructs
The various fragments of the Sec61b cDNA were inserted into t-ftz pCNDA3 (Palazzo et al., 2007) using restriction-free cloning (van den Ent and Löwe, 2006). For the 5′UTR insertion, the primer pair 5′-CAAGCTTGTCGACGCCGCCACCGCCAGCTGCCGGTCTTTC-3′ and 5′-GGAGCAGCGTGCACGGTACCATATTGGAGATGAGGGTGGCAA-3′ was used. For the ORF insertion, the primer pair 5′-GATGTTCCAGATTACGTCCTGCAGATGCCTGGTCCGACCCCCAG-3′ and 5′-TGGGACAGCAAGAAAGCGAGCTTACGAACGAGTGTACTTGCCCCAAATG-3′ was used. For the 3′UTR insertion, the primer pair 5′-GTTCCAGATTACGTCCTGCAGTAAATTCAGTTACATCCATCTGTCATC-3′ and 5′-AATTGGGACAGCAAGAAAGCGAGCCAGTATAAGTGAATTAAAAAGTTTAT-3′ was used. FIS1 ORF was amplified from a U2OS cDNA library with forward primer 5′-AGATCTATGGAGGCCGTGCTGAACG-3′ and reverse primer 5′-GAATTCCTTGCTGTGTCCAAGTCCAAATCCTGA-3′. The amplified ORFs were then inserted into the pEGFP-C1 multi-cloning site (MCS) using the EcoRI and BglII sites. To alter the TMD of the FIS1 (GGMALGCAG to LLMALLVLL, see supplementary material Fig. 2B), restriction enzyme-free cloning was performed as previously described (van den Ent and Löwe, 2006) to incorporate five leucine residues into the TMD (forward primer, 5′-TTACTTATGGCCCTGTTGGTGCTTTTGCTGGCCGGACTCATCGGACTTGC-3′ and reverse primer, 5′-CAAAAGCACCAACAGGGCCATAAGTAACACGATGGCCATGCCCACGAGTC-3′). All other genes were amplified from a cDNA library prepared from U2OS cells. For GFP–Sec22β, the forward primer 5′-ATGGTGTTGCTAACAATGATCGCC and reverse primer 5′-GTCCGATTCTGGTGGCTGTGA-3′ were used to amplify the Sec22β ORF, which was inserted into the TOPO cloning vector (Invitrogen) and subsequently cloned into the pEGFP-C1 vector using the BglII cloning site. For Sec61γ, the forward primer 5′-GGCAGAAACCCGGA-3′ and reverse primer- 5′-TTCATTTACTTTGAAATTACTTTAATTTAG-3′ were used to amplify the gene including the UTRs which were subsequently inserted into the MCS of pcDNA3.1 vector. The GFP ORF was then inserted at the N-terminal of the Sec61γ sequence using restriction enzyme-free cloning with forward primer, 5′-GGTTGGGTAGGCAGTCATGGTGAGCAAGGGC-3′ and the reverse primer, 5′-CAAACTGCATTACCTGATCCATAGATCTGAGTCCGGACTTG-3′. GFP–Sec61β (Rolls et al., 1999) was obtained from Tom Rapoport (Harvard University, Cambridge, MA), and GFP-Pex26 was obtained from Peter Kim (University of Toronto, Toronto, Canada). To construct GFP–fs-Sec61b, a single cytosine was inserted using restriction enzyme-free cloning (van den Ent and Löwe, 2006) with forward primer, 5′-CGATTCTACACAGAAGATTCACCTGG-3′ and reverse primer, 5′-GCTCAAAGCTTGGCCCTGT-3′ using GFP–Sec61b as template.
Cell culture, fractionation, transfection, microinjection, FISH and immunofluorescence
Cell culture, DNA transfection or microinjection, digitonin extraction, FISH staining and immunostaining were performed as previously described (Gueroussov et al., 2010; Cui and Palazzo, 2012; Cui et al., 2012). BAT3-knockout MEF cells were obtained from Hitoshi Okada (University of Toronto, Toronto, Canada) (Sasaki et al., 2007). BAT3−/− cells were grown in DMEM supplemented with 10% FBS and 2-mercaptoethanol. U2OS and COS7 cells were transfected using GenJet Transfection Reagent (SignaGen Laboratories). BAT3−/− and MEFs cells were transfected using JetPrime Polyplus (Invitrogen) transfection reagent. Cell fractionation was performed as previously described (Cui et al., 2013). Samples were separated by SDS-PAGE and analyzed by western blotting using rabbit polyclonal antibodies against Trapα (dilution 1:1000; Görlich et al., 1990), Sec61β (dilution 1:1000; Görlich et al., 1992), GAPDH (dilution 1:1000; Abgen) and GFP (dilution 1:1000; Molecular Probes), and monoclonal mouse antibody against α-tubulin (dilution 1:1000; Sigma). To detect t-ftz protein, which contains an HA epitope (Palazzo et al., 2007), samples were immunoblotted with anti-HA mouse monoclonal antibody (GeneTex; dilution 1:1000).
For FISH staining, the deoxyoligonucleotides used to recognize ftz (5′-GTCGAGCCTGCCTTTGTCATCGTCGTCCTTGTAGTCACAACAGCCGGGACAACACCCCAT-3′), ALPP (5′-CAGCTTCTTGGCAGCATCCAGGGCCTCGGCTGCCTTTCGGTTCCAGAAG-3′), GFP (5′-CTCCATCTTATTGCCCAGGATGTTGCCATCCTCCTTGAAATCGGTGCCGG-3′) were conjugated at the 5′ end with Alexa Fluor 546 or Alexa Fluor 647 (Integrated DNA Technologies). Polyclonal rabbit ATP5A antibody was obtained from Angus McQuibban (Abcam, ab14748). Polyclonal rabbit anti-Trapα antibody was obtained from Tom Rapoport (Görlich et al., 1990). For immunofluorescence staining, permeabilized cells were stained with primary antibodies at 1:200 dilution for Trapα, and 1:2000 for ATP5A for 1 h at room temperature. The secondary goat anti-rabbit-IgG antibody (conjugated to Alexa Fluor 647; Molecular Probes) was used at 1:500 dilution for 30 min at room temperature. All reagents were purchased from Sigma-Aldrich unless otherwise specified.
Fluorescence imaging and FISH quantification were performed as described previously (Gueroussov et al., 2010; Cui et al., 2012; Cui and Palazzo, 2012). All P-values were calculated using a Student's unpaired t-test.
Lentiviral-mediated shRNA knockdown
Lentiviral-mediated shRNA knockdown was performed as previously described (Cui et al., 2012). Plasmids encoding shRNA against p180 (clone B9, TRCN0000117407 and clone B10, TRCN0000117408, Sigma), BAT3 (TRCN0000007357, Sigma), TRC40 (clone A, TRCN0000042959 and clone B, TRCN0000042960, Sigma), nesprin-2 (also known as SYNE2; TRCN0000303799, Sigma) or empty vector (pLKO.1) were transfected into the HEK293T cells together with the accessory plasmids, VSVG and Δ8.9, to generate lentivirus carrying specific shRNA plasmids. U2OS cells were infected with lentivirus for 3–4 days and selected using 2 ng/μl puromycin. The level of knockdown was examined using western blotting analysis and was performed as described previously (Cui et al., 2012). Antibodies against BAT3 and TRC40 were obtained from Manu Hegde (Mariappan et al., 2010) and used at 1:1000 dilution.
Visualization and quantification of endogenous mRNA
The localization of endogenous Sec61b, nesprin-2 (SYNE2) and GAPDH mRNA was visualized using customized or cataloged Stellaris probe arrays (Biosearch Technologies, Petaluma, CA) against human and mouse genes. U2OS or MEF cells were grown on coverslips, either treated with control or HHT-containing medium for 30 min, then fixed directly or after digitonin extraction. The staining was performed as per the manufacturer's protocol with the following exception: after overnight staining with FISH probes, the cells were washed three times with 2× SSC solution containing 10% formamide at room temperature. After washing, the cells were mounted and visualized. After cells were imaged using phase microscopy, the number of endogenous mRNA foci in each cell was quantified using NIS Element software (Nikon Corporation, Tokyo, Japan). Briefly, cell and nuclear peripheries were selected to generate regions of interest (ROIs). Then, the number of endogenous mRNA foci was counted using the ‘spot detection’ function, selecting for bright spots that were about 0.32 μm in diameter.
Supplementary Material
Acknowledgements
We would like to thank T. A. Rapoport for antibodies against Trapα and Sec61β, A. McQuibban for antibody against ATP5A, R. Hegde for antibodies against BAT3 and TRC40, H. Okada for BAT3−/− and control MEFs, P. Kim and R. Hua for wild-type MEFs and GFP–Pex26 plasmids. We also thank J. Wan and D. Williams for helpful comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Experiments were conducted by X.A.C., H.Z., L.I., A.X.L. and I.K. The manuscript was written by X.A.C. and A.F.P.
Funding
This work was funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) fellowship to X.A.C. and an NSERC grant to A.F.P. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.168583/-/DC1
References
- Abell B. M., Jung M., Oliver J. D., Knight B. C., Tyedmers J., Zimmermann R. and High S. (2003). Tail-anchored and signal-anchored proteins utilize overlapping pathways during membrane insertion. J. Biol. Chem. 278, 5669-5678. 10.1074/jbc.M209968200 [DOI] [PubMed] [Google Scholar]
- Abell B. M., Pool M. R., Schlenker O., Sinning I. and High S. (2004). Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23, 2755-2764. 10.1038/sj.emboj.7600281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T., Cohen G. and Schuldiner M. (2013). A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134-1145. 10.1016/j.cell.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Borgese N. and Fasana E. (2011). Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808, 937-946. 10.1016/j.bbamem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Borgese N., Gazzoni I., Barberi M., Colombo S. and Pedrazzini E. (2001). Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12, 2482-2496. 10.1091/mbc.12.8.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S., Yabal M., Soffientini P., Stefanovic S., Makarow M., Hegde R. S. and Borgese N. (2005). Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 24, 2533-2542. 10.1038/sj.emboj.7600730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Jagannathan S., Reid D. W., Zheng T. and Nicchitta C. V. (2011). Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol. Biol. Cell 22, 2646-2658. 10.1091/mbc.E11-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coassin S. R., Orjalo A. V., Semaan S. J. and Johansson H. E. (2014). Simultaneous detection of nuclear and cytoplasmic RNA variants utilizing Stellaris® RNA fluorescence in situ hybridization in adherent cells. Methods Mol. Biol. 1211, 189-199. 10.1007/978-1-4939-1459-3_15 [DOI] [PubMed] [Google Scholar]
- Cui X. A. and Palazzo A. F. (2012). Visualization of endoplasmic reticulum localized mRNAs in mammalian cells. J. Vis. Exp. 70, e50066 10.3791/50066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. A. and Palazzo A. F. (2014). Localization of mRNAs to the endoplasmic reticulum. Wiley Interdiscip. Rev. RNA 5, 481-492. 10.1002/wrna.1225 [DOI] [PubMed] [Google Scholar]
- Cui X. A., Zhang H. and Palazzo A. F. (2012). p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 10, e1001336 10.1371/journal.pbio.1001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. A., Zhang Y., Hong S. J. and Palazzo A. F. (2013). Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J. Biol. Chem. 288, 29633-29641. 10.1074/jbc.M113.482505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Hartmann E., Herz J., Otto A., Kraft R., Wiedmann M., Knespel S., Dobberstein B. and Rapoport T. A. (1990). The signal sequence receptor has a second subunit and is part of a translocation complex in the endoplasmic reticulum as probed by bifunctional reagents. J. Cell Biol. 111, 2283-2294. 10.1083/jcb.111.6.2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Hartmann E., Kalies K.-U. and Rapoport T. A. (1992). A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71, 489-503. 10.1016/0092-8674(92)90517-G [DOI] [PubMed] [Google Scholar]
- Gueroussov S., Tarnawsky S. P., Cui X. A., Mahadevan K. and Palazzo A. F. (2010). Analysis of mRNA nuclear export kinetics in mammalian cells by microinjection. J. Vis. Exp. 46, 2387 10.3791/2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R. S. and Keenan R. J. (2011). Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 12, 787-798. 10.1038/nrm3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. E. and Bullock S. L. (2009). Subcellular mRNA Localization in Animal Cells and Why It Matters. Science 326, 1212-1216. 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie C. (2003). Targeting and assembly of mitochondrial tail-anchored protein Tom5 to the TOM complex depend on a signal distinct from that of tail-anchored proteins dispersed in the membrane. J. Biol. Chem. 278, 41462-41471. 10.1074/jbc.M307047200 [DOI] [PubMed] [Google Scholar]
- Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S. et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693-1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut-Cohen J., Afanasieva E., Haim-Vilmovsky L., Slobodin B., Yosef I., Bibi E. and Gerst J. E. (2013). Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 24, 3069-3084. 10.1091/mbc.E13-01-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hartmann E. and Rapoport T. A. (1993). A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72-75. 10.1016/0962-8924(93)90066-A [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B. and Rapoport T. A. (1995). Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Benedix J., Fedeles S. V., Schorr S., Schirra C., Schauble N., Jalal C., Greiner M., Hassdenteufel S., Tatzelt J. et al. (2012). Different effects of Sec61, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 125, 1958-1969. 10.1242/jcs.096727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R. S., Seiser R. M., Zheng T., Lager P. J., Reedy M. C., Keene J. D. and Nicchitta C. V. (2003). Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9, 1123-1137. 10.1261/rna.5610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., Clancy A., Schwappach B. and High S. (2010). Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 123, 2170-2178. 10.1242/jcs.066738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., Warwicker J. and High S. (2011). A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem. J. 436, 719-727. 10.1042/BJ20101737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G. W. G., Gomes E. R., Folker E. S., Vintinner E. and Gundersen G. G. (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956-959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J. and Hegde R. S. (2010). A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120-1124. 10.1038/nature09296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. C. and Ephrussi A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719-730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov A. V., Snapp E. L., Lippincott-Schwartz J. and Kreibich G. (2002). Active translocon complexes labeled with GFP–Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 158, 497-506. 10.1083/jcb.200201116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A. F., Springer M., Shibata Y., Lee C.-S., Dias A. P. and Rapoport T. A. (2007). The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 5, e322 10.1371/journal.pbio.0050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B., Zheng T., Lager P. J., Keene J. D., Reedy M. C. and Nicchitta C. V. (2008). Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 14, 445-453. 10.1261/rna.721108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C., Wipf P., Brodsky J. L. and High S. (2008). A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J. Biol. Chem. 283, 27504-27513. 10.1074/jbc.M804591200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C., Schmid V., Schwappach B. and High S. (2009). Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 122, 3605-3612. 10.1242/jcs.041210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D. W. and Nicchitta C. V. (2012). Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287, 5518-5527. 10.1074/jbc.M111.312280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M. M., Stein P. A., Taylor S. S., Ha E., McKeon F. and Rapoport T. A. (1999). A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell Biol. 146, 29-44. 10.1083/jcb.146.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Gan E. C., Wakeham A., Kornbluth S., Mak T. W. and Okada H. (2007). HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21, 848-861. 10.1101/gad.1534107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F. et al. (2005). Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123, 507-519. 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B. and Weissman J. S. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634-645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Voeltz G. K. and Rapoport T. A. (2006). Rough sheets and smooth tubules. Cell 126, 435-439. 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J. M., Hu J., Rapoport T. A., Prinz W. A. and Voeltz G. K. (2008). The reticulon and Dp1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283, 18892-18904. 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W. A., Palazzo A. F., Kozlov M. M. and Rapoport T. A. (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774-788. 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S. and Hegde R. S. (2007). Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147-1159. 10.1016/j.cell.2007.01.036 [DOI] [PubMed] [Google Scholar]
- van den Ent F. and Löwe J. (2006). RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67-74. 10.1016/j.jbbm.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Vilardi F., Lorenz H. and Dobberstein B. (2011). WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 124, 1301-1307. 10.1242/jcs.084277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F., Stephan M., Clancy A., Janshoff A. and Schwappach B. (2014). WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE 9, e85033 10.1371/journal.pone.0085033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Brown E. C., Mak G., Zhuang J. and Denic V. (2010). A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40, 159-171. 10.1016/j.molcel.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Chan C., Weir N. R. and Denic V. (2014). The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 512, 441-444. 10.1038/nature13471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y., Hiromasa T. and Fujiki Y. (2013). Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200, 651-666. 10.1083/jcb.201211077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y. and Sakisaka T. (2012). Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 48, 387-397. 10.1016/j.molcel.2012.08.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.