Abstract
Methanosarcina species are the most metabolically versatile of the methanogenic Archaea and can obtain energy for growth by producing methane via the hydrogenotrophic, acetoclastic or methylotrophic pathways. Methanosarcina barkeri CM1 was isolated from the rumen of a New Zealand Friesian cow grazing a ryegrass/clover pasture, and its genome has been sequenced to provide information on the phylogenetic diversity of rumen methanogens with a view to developing technologies for methane mitigation. The 4.5 Mb chromosome has an average G + C content of 39 %, and encodes 3523 protein-coding genes, but has no plasmid or prophage sequences. The gene content is very similar to that of M. barkeri Fusaro which was isolated from freshwater sediment. CM1 has a full complement of genes for all three methanogenesis pathways, but its genome shows many differences from those of other sequenced rumen methanogens. Consequently strategies to mitigate ruminant methane need to include information on the different methanogens that occur in the rumen.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0038-5) contains supplementary material, which is available to authorized users.
Keywords: Methanogen, Methane, Ruminant, Methanosarcina barkeri
Introduction
Ruminants are foregut fermenters and have evolved an efficient digestive system in which microbes ferment plant fibre and provide fermentation end-products and other nutrients for growth of the animal [1]. A variety of methanogens can be found in the rumen [2] and ruminant derived methane (CH4) accounts for about one quarter of all anthropogenic CH4 emissions [3], and is implicated as a driver of global climate change. In terms of their metabolism the rumen methanogens fall into three groups, hydrogenotrophs (Methanobrevibacter, Methanomicrobium and Methanobacterium spp) which convert hydrogen and/or formate to CH4, methylotrophs (Methanosphaera spp and members of the order Methanomassiliicoccales) which produce CH4 from methyl compounds such as methanol and methylamines, and acetoclastic methanogens (Methanosarcina) which can utilise acetate to produce CH4 in addition to the hydrogenotrophic and methylotrophic pathways. Obtaining representative genome sequences from each of the above organisms will be important to understanding the metabolic capacity of these archaea and how they contribute to rumen fermentation processes. Currently, genome sequences are available for five rumen methanogens including strains of Methanobrevibacter ruminantium [4], M. boviskoreani [5, 6], Methanobacterium formicicum [7] and Thermoplasmatales archaeon BRNA1 [NCBI Reference Sequence: NC_020892.1]. Development of strategies to reduce CH4 emissions from farmed ruminant animals are currently being investigated with methanogen genome sequence information used to inform mitigation strategies based on vaccines and small-molecule inhibitors [8, 9]. Here we present the genome sequence from a rumen acetoclastic methanogen, Methanosarcina barkeri CM1.
Organism information
Classification and features
Methanosarcina sp. CM1 was isolated from the rumen of a New Zealand Friesian cow grazing a ryegrass/clover pasture [10]. CM1 grew as large cell aggregates in broth culture and showed the characteristic morphology associated with Methanosarcina barkeri [11] (Fig. 1). It was described as non-motile, and able to grow and produce methane from H2/CO2, acetate, methanol and methylamines. Growth occurred between 30° and 45 °C, and at pH 5.0 to 7.4. Rumen fluid was required for growth. The 16S rRNA gene from CM1 is 99 % similar to that of the Methanosarcina barkeri type strain MS (DSM 800) (Fig. 2) which was isolated from a sewage sludge digester [12, 13], and as such CM1 can be considered as a strain of M. barkeri. M. barkeri is found at high densities in anaerobic digesters and anoxic marine and freshwater sediments, but there have been several reports describing Methanosarcina from the rumen although these organisms were not characterized [14, 15]. In addition, non-rumen strains of M. barkeri have been used in co-culture studies with rumen anaerobic fungi [16] and ciliate protozoa [17, 18]. Characteristics of Methanosarcina barkeri CM1 are shown in Table 1 and Additional file 1.
Fig. 1.
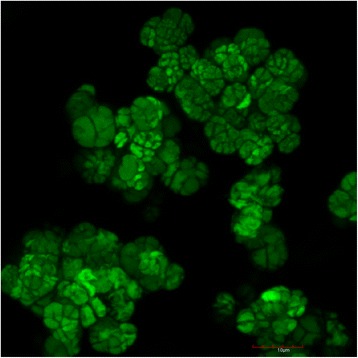
Morphology of Methanosarcina barkeri CM1. Micrograph showing aggregates of Methanosarcina barkeri CM1 cells captured with sectional depth scanning using an Olympus Fluoview FV1000D Spectral laser confocal scanning inverted microscope, with an UPLSAPO 60X oil objective (1.35 NA). Olympus Fluoview 10-ASW software was used to view fluorescent signals and to generate images. Emission at 635 nm wavelength shows methyl green stain incorporated into nucleic acids within cells and cell clusters. Bar is 10 μm
Fig. 2.
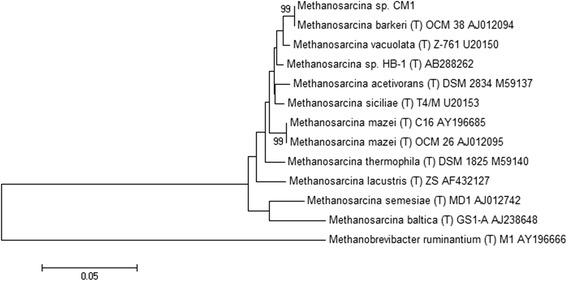
Phylogenetic tree showing the position of CM1 relative to type strains of other Methanosarcina species. The strains and their corresponding accession numbers are shown. The evolutionary history was inferred using the Neighbor-Joining method [45] with Methanobrevibacter ruminantium used as an outgroup. The optimal tree with the sum of branch length = 0.43777587 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [46]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method [47] and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 13 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1081 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [48]
Table 1.
Classification and general features of Methanosarcina barkeri CM1
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain: Archaea | TAS [49] | |
| Phylum: Euryarchaeota | TAS [50] | ||
| Class: Methanococci | TAS [51, 52] | ||
| Order: Methanosarcinales | TAS [51, 53] | ||
| Family: Methanosarcinaceae | TAS [24, 54] | ||
| Genus: Methanosarcina | TAS [55, 56] | ||
| Species: Methanosarcina barkeri | TAS [10] | ||
| strain: CM1 | |||
| Gram stain | Positive | TAS [12] | |
| Cell shape | Irregular | TAS [10] | |
| Motility | Non-motile | TAS [10] | |
| Sporulation | Not reported | IDA | |
| Temperature range | 30-45 °C | TAS [10] | |
| Optimum temperature | 40 °C | TAS [10] | |
| pH range; Optimum | 5.0-7.4; 6.8 | TAS [10] | |
| Carbon source | CO2, Acetate | IDA | |
| MIGS-6 | Habitat | Bovine rumen | TAS [10] |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen requirement | Anaerobic | IDA |
| MIGS-15 | Biotic relationship | Symbiont | TAS [10] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Palmerston North, New Zealand | IDA |
| MIGS-5 | Sample collection | Not reported | |
| MIGS-4.1 | Latitude | -40.35 (40°21'00"S) | IDA |
| MIGS-4.2 | Longitude | +175.61 (175°36'36"E) | IDA |
| MIGS-4.4 | Altitude | 30 M | IDA |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [57]
Genome sequencing information
Genome project history
Methanosarcina barkeri CM1 was selected for genome sequencing on the basis of its phylogenetic position relative to other methanogens isolated from the rumen. A summary of the genome project information is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality, closed genome |
| MIGS-28 | Libraries used | 454 3 kb mate paired-end library, Illumina paired-end 170 bp insert library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium chemistry, Illumina |
| MIGS-31.2 | Fold coverage | 97× (454), 224× (Illumina) |
| MIGS-30 | Assemblers | Newbler, Spades |
| MIGS-32 | Gene calling method | Glimmer and BLASTX |
| Locus tag | MCM1 | |
| Genbank ID | CP008746 | |
| Genbank date of release | June 3, 2015 | |
| GOLD ID | Gp0007672 | |
| MIGS 13 | Source material identifier | CM1 |
| Project relevance | Ruminant methane emissions |
Growth conditions and genomic DNA preparation
Methanosarcina barkeri CM1 was grown in BY medium [19] with added SL10 Trace Elements solution (1 ml added l−1 [20], 20 mM sodium acetate, 60 mM sodium formate and Vitamin 10 solution (0.1 ml added to 10 ml culture before inoculation) [4]. H2 was supplied as the energy source by pumping the culture vessels to 180 kPa over pressure with an 80:20 mixture of H2:CO2. Genomic DNA was extracted from freshly grown cells using a modified version of a liquid N2 and grinding method as described previously [6].
Genome sequencing and assembly
The complete genome sequence of CM1 was determined using pyrosequencing of 3Kb mate paired-end sequence libraries using a 454 GS FLX platform with Titanium chemistry (Macrogen, Korea). Pyrosequencing reads provided 97× coverage of the genome and were assembled using the Newbler assembler version 2.7 (Roche 454 Life Sciences, USA). The Newbler assembly resulted in 85 contigs across 9 scaffolds. Gap closure was managed using the Staden package [21] and gaps were closed using additional Sanger sequencing by standard and inverse PCR based techniques. In addition, CM1 genomic DNA was sequenced using the Illumina HiSeq 2000 platform (Beijing Genomics Institute, China) which provided 223× genome coverage. Illumina reads were assembled using the Spades assembler version 3.0 [22] and combined with the Newbler assembly using the Staden package. Assembly validation was confirmed by pulsed-field gel electrophoresis as described previously [6].
Genome annotation
The procedure for genome annotation was as described previously for Methanobrevibacter sp. [4, 6], and the CM1 genome sequence was prepared for NCBI submission using Sequin. The adenine residue of the start codon of the Cdc6-1 replication initiation protein A (MCM1_0001) gene was chosen as the first base for the CM1 genome. The nucleotide sequence of the Methanosarcina barkeri CM1 chromosome has been deposited in Genbank under accession number CP008746.
Genome properties
The genome of Methanosarcina barkeri CM1 consists of a single 4,501,171 basepair (bp) circular chromosome with an average G + C content of 39 %. A total of 3656 genes were predicted, 3523 of which were protein-coding genes, representing 70 % of the total genome sequence. A COG category was assigned to 2267 of the protein-coding genes. The properties and statistics of the genome are summarized in Tables 3 and 4. As with the other sequenced Methanosarcina strains CM1 has dual origins of replication (MCM1_001 and MCM1_3593, 95 kb apart) surrounded by conserved genes [23]. The CM1 genome has neither plasmid nor prophage sequences, but does contain three clusters of CRISPR genes associated with CRISPR repeat regions, and three type I restriction/modification systems.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,501,171 | 100.00 |
| DNA coding (bp) | 3,149,919 | 69.98 |
| DNA G + C (bp) | 1,763,740 | 39.18 |
| DNA scaffolds | 1 | |
| Total genes | 3,655 | 100.00 |
| Protein coding genes | 3,523 | 96.39 |
| RNA genes | 69 | 1.89 |
| Pseudo genes | 63 | 1.72 |
| Genes with function prediction | 2,410 | 65.94 |
| Genes assigned to COGs | 2,267 | 64.35 |
| Genes with Pfam domains | 2,953 | 80.79 |
| Genes with signal peptides | 358 | 10.16 |
| Genes with transmembrane helices | 881 | 25.01 |
| CRISPR repeats | 3 |
Table 4.
Number of genes associated with the 25 general COG functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 158 | 4.48 | Translation |
| A | 1 | 0.03 | RNA processing and modification |
| K | 112 | 3.18 | Transcription |
| L | 126 | 3.58 | Replication, recombination and repair |
| B | 2 | 0.06 | Chromatin structure and dynamics |
| D | 15 | 0.43 | Cell cycle control, mitosis and meiosis |
| Y | - | - | Nuclear structure |
| V | 76 | 2.16 | Defense mechanisms |
| T | 63 | 1.79 | Signal transduction mechanisms |
| M | 99 | 2.81 | Cell wall/membrane biogenesis |
| N | 16 | 0.45 | Cell motility |
| Z | - | - | Cytoskeleton |
| W | - | - | Extracellular structures |
| U | 18 | 0.51 | Intracellular trafficking and secretion |
| O | 96 | 2.72 | Posttranslational modification, protein turnover, chaperones |
| C | 223 | 6.33 | Energy production and conversion |
| G | 81 | 2.30 | Carbohydrate transport and metabolism |
| E | 221 | 6.27 | Amino acid transport and metabolism |
| F | 54 | 1.53 | Nucleotide transport and metabolism |
| H | 109 | 3.09 | Coenzyme transport and metabolism |
| I | 30 | 0.85 | Lipid transport and metabolism |
| P | 138 | 3.92 | Inorganic ion transport and metabolism |
| Q | 48 | 1.36 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 368 | 10.44 | General function prediction only |
| S | 213 | 6.04 | Function unknown |
| - | 1256 | 35.67 | Not in COGs |
aThe total is based on the total number of protein coding genes in the annotated genome
Insights from the genome sequence
The genome of Methanosarcina barkeri CM1 is compared with genomes of other sequenced methanogens from the genus Methanosarcina in Table 5. Overall, the gene content of the CM1 genome is very similar to that of Methanosarcina barkeri Fusaro, but gene organization shows very little synteny.
Table 5.
Genomes of Methanosarcina species from various anaerobic environments
| Species | Isolation source | Genome size (Mb) | Accession # | CDS | % GC | Reference |
|---|---|---|---|---|---|---|
| Methanosarcina barkeri CM1 | Bovine rumen | 4.50 | CP008746 | 3,524 | 39.2 | This report |
| Methanosarcina barkeri Fusaro | Freshwater sediment | 4.87 | NC_007355 | 3,758 | 39.2 | [23] |
| Methanosarcina acetivorans C2A | Marine sediment | 5.75 | AE010299 | 4,721 | 42.7 | [32] |
| Methanosarcina mazei Go1 | Sewage | 4.10 | AE008384 | 3,398 | 41.5 | [58] |
| Methanosarcina mazei Tuc01 | Freshwater sediment | 3.42 | CP004144 | 3,395 | 42.5 | [59] |
Methanogenesis
Methanosarcina species are the most metabolically versatile of the methanogenic archaea [24] and can obtain energy for growth by producing methane via three different pathways (Fig. 3). Methane can be derived from the reduction of CO2 with hydrogen (hydrogenotrophic pathway), from the methyl group of acetate (acetoclastic pathway), or from the methyl group of methanol, methylamines or methylthiols (methylotrophic pathway). Each pathway culminates in the transfer of a methyl group to coenzyme M and the subsequent reduction to methane. The bioenergetics of aceticlastic methanogens have been recently reviewed [25, 26], and a metabolic reconstruction presented for M. barkeri Fusaro [27]. There is evidence that the genes essential to both the acetoclastic and methyoltrophic pathways were horizontally acquired during evolution of the Methanosarcinaceae [28–30]. Although acetoclastic methanogenesis contributes approximately two-thirds of the methane in the biosphere [31], acetate is not metabolized to methane to any significant extent in the rumen [2]. CM1 has a full complement of genes for all three methanogenesis pathways and as with other Methanosarcina species several genes are present as multiple copies [32]. Unlike many hydrogenotrophic methanogens, CM1 does not have the gene for [Fe]-hydrogenase dehydrogenase (hmd), or the genes that encode methyl coenzyme M reductase II (mrt), however it does have genes for formate dehydrogenase (MCM1_3047-3048) although CM1 and other M. barkeri strains are unable to use formate [10, 23]. The pathway for coenzyme M biosynthesis differs from that found in other sequenced rumen methanogens which belong to the order Methanobacteriales [33]. Consequently methanogen inhibitors targeting coenzyme M biosynthesis would not be expected to work against all rumen methanogen species. M. barkeri is the organism in which the 22nd amino acid (pyrrolysine) was discovered during examination of the methyltransferases required for methane formation from methylamines [34]. Biosynthesis of this amino acid requires specialized enzymes together with a specific aminoacyl-tRNA synthetase [35], and the genes encoding these (pylSBCD) are found in CM1 (MCM1_2535-2538).
Fig. 3.
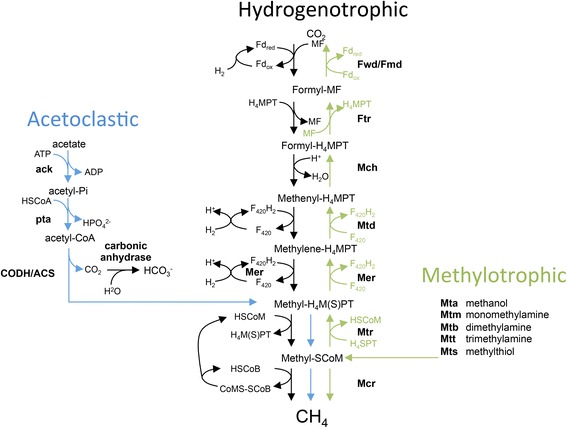
The three methanogenesis pathways inferred from the genome of Methanosarcina barkeri CM1
Cell envelope
The majority of rumen methanogens belong to the family Methanobacteriaceae and have a characteristic pseudomurein-containing cell wall [4]. The cell surface of Methanosarcina sp. is different and electron microscopy shows a protein S-layer surrounding the cytoplasmic membrane. A major S-layer protein has been identified in three Methanosarcina species and used to define a family of proteins [36, 37]. All paralogs possess signal peptides and one or two DUF1608 (Pfam accession PF07752) domains. Both M. barkeri Fusaro and CM1 possess nine proteins containing this domain. Gene expression studies show that a single DUF1608 domain-containing protein is abundantly expressed in both M. acetivorans (MA0829) and M. barkeri (Mbar_A1758), and is among the most highly expressed of all proteins in the cell [37]. A similar protein is predicted from the CM1 genome (MCM1_2018, 84 % amino acid identity to the product of Mbar_A1758), and likely to be the major S-layer protein for this strain. The DUF1608 domain from MA0829 has been crystallised and was used to propose an elegant model of the Methanosarcina S-layer structure [38]. Methanosarcina cells can form large aggregates (Fig. 1) mediated by the production of methanochondroitin, a polymer composed of uronic acid and N-acetylgalactosamine residues [39]. While the steps in methanochondroitin biosynthesis have been determined, the genes involved have yet to be identified. These aggregates are observed to separate into single cells and CM1 encodes eleven proteins with disaggregatase-related domains (Pfam accession PF08480). The CM1 genome has four oligosaccharyl transferase genes, three of which (MCM1_1841-1843) are at the end of a large gene cluster that contains 14 glycosyl transferases and is likely to be involved in polysaccharide biosynthesis (MCM1_1841-1889). The fourth oligosaccharyl transferase is associated with a smaller gene cluster containing glycosyl transferases, methyltransferases and transporters (MCM1_2113-2123). A third cluster of polysaccharide biosynthesis genes is found at MCM1_2831-2857. CM1 also encodes a secreted protein (MCM_2974) containing a glycoside hydrolase family 18 (chitinase) domain that is not found in M. barkeri Fusaro which may be involved in mediating interaction with rumen anaerobic fungi. Like many other archaea, CM1 has an identifiable archaella (archaeal flagella) operon (FlaB-FlaJ, MCM1_1947-1953), together with a cluster of chemotaxis genes (MCM1_3655-3662) [40, 41]. However, motility has never been observed in any Methanosarcina species and thus the function of these genes remains unknown.
Methanosarcina barkeri has been reported to fix nitrogen [42] and sets of nitrogenase genes are found in Methanosarcina genomes. CM1 contains two different nif operons comprising nitrogenase and nitrogenase cofactor biosynthesis genes that match to those reported from M. barkeri strain 227 [43]. These are a molybdenum- and iron-containing nitrogenase (MCM1_2924-2930) and a vanadium- and iron-containing nitrogenase (MCM1_1063-1072). However, it does not have the genes for the third type, the iron-only nitrogenase that is found in M. acetivorans and M. barkeri Fusaro. Electron micrographs of M. barkeri log phase cells [11] show the presence of numerous electron-dense granules in the cytoplasm. In M. thermophila similar granules were found to contain glycogen [44], and CM1 has several genes predicted to encode the enzymes necessary for the biosynthesis and degradation of this reserve polysaccharide. CM1 does not have the genes for gas vesicle biosynthesis that are found in the M. barkeri Fusaro genome [23], but it does have genes for the two-subunit acetyl-CoA synthetase (MCM1_1658 and 2708) that have been lost from the Fusaro strain.
Conclusion
The genome of Methanosarcina barkeri CM1 is very similar to that of the freshwater sediment isolate M. barkeri Fusaro, but markedly different from the dominant rumen methanogens, most of which are members of the family Methanobacteriaceae. CM1 has a much larger genome and its sequence provides new insights into the metabolic versatility of rumen methanogens. With its ability to use three different methanogenesis pathways, M. barkeri appears to be a generalist able to occupy a range of different environments but is not particularly at home in the rumen. This is in contrast to the more specialised rumen methanogens, such as the Methanobrevibacter species which dominate the rumen environment. Analysis of the methanogenesis pathway and the cell envelope have been important for the design of methane mitigation strategies targeting rumen methanogens, but differences highlighted from the CM1 genome stress the need to include information from all rumen methanogens in the design of mitigation approaches.
Acknowledgements
The CM1 genome sequencing project was funded by the New Zealand Pastoral Greenhouse Gas Research Consortium.
Additional file
Associated MIGS record for CM1, which links to the SIGS supplementary content website.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WJK, SCLe, GTA, EA, conceived and designed the experiments. SCLa, SCLe, DL, KR performed the sequencing and assembly experiments. WJK, EA, SCL performed the genome annotation and comparative studies. TAM, ERV performed the fluorescence microscopy. WJK, SCLa, SCLe wrote the manuscript. All authors commented on the manuscript before submission. All authors read and approved the final manuscript.
Contributor Information
Suzanne C. Lambie, Email: suzannejames.lambie@gmail.com
William J. Kelly, Email: bill.kelly@agresearch.co.nz
Sinead C. Leahy, Email: sinead.leahy@agresearch.co.nz
Dong Li, Email: dong.li@agresearch.co.nz.
Kerri Reilly, Email: kerri.reilly@agresearch.co.nz.
Tim A. McAllister, Email: tim.mcallister@agr.gc.ca
Edith R. Valle, Email: edith.valle@agr.gc.ca
Graeme T. Attwood, Email: graeme.attwood@agresearch.co.nz
Eric Altermann, Email: eric.altermann@agresearch.co.nz.
References
- 1.Clauss M, Hume ID, Hummel J. Evolutionary adaptations of ruminants and their potential relevance for modern production systems. Animal. 2010;4:979–92. doi: 10.1017/S1751731110000388. [DOI] [PubMed] [Google Scholar]
- 2.Janssen PH, Kirs M. Structure of the archaeal community of the rumen. Appl Environ Microbiol. 2008;74:3619–25. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorpe A. Enteric fermentation and ruminant eructation: the role (and control?) of methane in the climate change debate. Climate Change. 2009;93:407–31. doi: 10.1007/s10584-008-9506-x. [DOI] [Google Scholar]
- 4.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One. 2010;5:e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Rhee MS, Kumar S, Lee GH, Chang DH, Kim DS, et al. Genome sequence of Methanobrevibacter sp. strain JH1, isolated from rumen of Korean native cattle. Genome Announc. 2013;1:e00002–13. doi: 10.1128/genomeA.00002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leahy SC, Kelly WJ, Li D, Li Y, Altermann E, Lambie SC, et al. The complete genome sequence of Methanobrevibacter sp. AbM4. Stand Genomic Sci. 2013;8:215–27. doi: 10.4056/sigs.3977691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly WJ, Leahy SC, Li D, Perry R, Lambie SC, Attwood GT, et al. The complete genome sequence of the rumen methanogen Methanobacterium formicicum BRM9. Stand Genomic Sci. 2014;9:15. doi: 10.1186/1944-3277-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leahy SC, Kelly WJ, Ronimus RS, Wedlock N, Altermann E, Attwood GT. Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal. 2013;7(Suppl 2):235–43. doi: 10.1017/S1751731113000700. [DOI] [PubMed] [Google Scholar]
- 9.Wedlock N, Janssen PH, Leahy S, Shu D, Buddle BM. Progress in the development of vaccines against rumen methanogens. Animal. 2013;7(Suppl 2):244–52. doi: 10.1017/S1751731113000682. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis GN, Strömpl C, Burgess DM, Skillman LC, Moore ERB, Joblin KN. Isolation and identification of ruminal methanogens from grazing cattle. Curr Microbiol. 2000;40:327–32. doi: 10.1007/s002849910065. [DOI] [PubMed] [Google Scholar]
- 11.Zeikus JG, Bowen VG. Comparative ultrastructure of methanogenic bacteria. Can J Microbiol. 1975;21:121–9. doi: 10.1139/m75-019. [DOI] [PubMed] [Google Scholar]
- 12.Bryant MP, Boone DR. Emended description of strain MST (DSM 800T), the type strain of Methanosarcina barkeri. Int J Syst Bacteriol. 1987;37:169–70. doi: 10.1099/00207713-37-2-169. [DOI] [Google Scholar]
- 13.Maestrojuán GM, Boone DR. Characterization of Methanosarcina barkeri MST and 227, Methanosarcina mazei S-6T, and Methanosarcina vacuolata Z-761T. Int J Syst Bacteriol. 1991;41:267–74. doi: 10.1099/00207713-41-2-267. [DOI] [Google Scholar]
- 14.Rowe JB, Loughnan ML, Nolan JV, Leng RA. Secondary fermentation in the rumen of a sheep given a diet based on molasses. Br J Nutr. 1979;41:393–7. doi: 10.1079/BJN19790048. [DOI] [PubMed] [Google Scholar]
- 15.McInerney MJ, Mackie RI, Bryant MP. Syntrophic association of a butyrate-degrading bacterium and Methanosarcina enriched from bovine rumen fluid. Appl Environ Microbiol. 1981;41:826–8. doi: 10.1128/aem.41.3.826-828.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mountfort DO, Asher RA, Bauchop T. Fermentation of cellulose to methane and carbon dioxide by a rumen anaerobic fungus in a triculture with Methanobrevibacter sp. strain RA1 and Methanosarcina barkeri. Appl Environ Microbiol. 1982;44:128–34. doi: 10.1128/aem.44.1.128-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillman K, Lloyd D, Williams AG. Interactions between the methanogen Methanosarcina barkeri and rumen holotrich ciliate protozoa. Lett Appl Microbiol. 1988;7:49–53. doi: 10.1111/j.1472-765X.1988.tb01250.x. [DOI] [Google Scholar]
- 18.Ushida K, Newbold CJ, Jouany J-P. Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J Gen Appl Microbiol. 1997;43:129–31. doi: 10.2323/jgam.43.129. [DOI] [PubMed] [Google Scholar]
- 19.Joblin KN, Naylor GE, Williams AG. Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl Environ Microbiol. 1990;56:2287–95. doi: 10.1128/aem.56.8.2287-2295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widdel F, Kohring G, Mayer F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol. 1983;134:286–94. doi: 10.1007/BF00407804. [DOI] [Google Scholar]
- 21.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–30. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, et al. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol. 2006;188:7922–31. doi: 10.1128/JB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–96. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlegel K, Müller V. Evolution of Na+ and H+ bioenergetics in methanogenic archaea. Biochem Soc Trans. 2013;41:421–6. doi: 10.1042/BST20120294. [DOI] [PubMed] [Google Scholar]
- 26.Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta. 1837;2014:1130–47. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Gonnerman MC, Benedict MN, Feist AM, Metcalf WW, Price ND. Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotech J. 2013;8:1070–9. doi: 10.1002/biot.201200266. [DOI] [PubMed] [Google Scholar]
- 28.Fournier GP, Huang J, Gogarten JP. Horizontal gene transfer from extinct and extant lineages: biological innovation and the coral of life. Phil Trans R Soc B. 2009;364:2229–39. doi: 10.1098/rstb.2009.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothman DH, Fournier GP, French KL, Alm EJ, Boyle EA, Cao C, et al. Methanogenic burst in the end-Permian carbon cycle. Proc Natl Acad Sci USA. 2014;111:5462–7. doi: 10.1073/pnas.1318106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrel G, Gaci N, Peyret P, O’Toole PW, Gribaldo S, Brugère J-F. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea. 2014;2014:ID 374146. doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferry JG. How to make a living exhaling methane. Ann Rev Microbiol. 2010;64:453–73. doi: 10.1146/annurev.micro.112408.134051. [DOI] [PubMed] [Google Scholar]
- 32.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, Fitzhugh W, et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–42. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham DE, Taylor SM, Wolf RZ, Namboori SC. Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem J. 2009;424:467–78. doi: 10.1042/BJ20090999. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–62. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 35.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature. 2011;471:647–50. doi: 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francoleon DR, Boontheung P, Yang Y, Kin U, Ytterberg AJ, Denny PA, et al. S-layer, surface-accessible, and concanavalin A binding proteins of Methanosarcina acetivorans and Methanosarcina mazei. J Proteome Res. 2009;8:1972–82. doi: 10.1021/pr800923e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohlin L, Leon DR, Kim U, Loo JA, Loo RRO, Gunsalus RP. Identification of the major expressed S-layer and cell surface-layer-related proteins in the model methanogenic archaea: Methanosarcina barkeri Fusaro and Methanosarcina acetivorans C2A. Archaea. 2012;2012:ID 873589. doi: 10.1155/2012/873589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arbing MA, Chan S, Shin A, Phan T, Ahn CJ, Rohlin L, et al. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc Natl Acad Sci USA. 2012;109:11812–7. doi: 10.1073/pnas.1120595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claus H, König H. Cell envelopes of methanogens. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds. Structure and Biochemistry. Heidelberg: Springer; 2010. pp. 231–51. [Google Scholar]
- 40.Desmond E, Brochier-Armanet C, Gribaldo S. Phylogenomics of the archaeal flagellum: rare horizontal gene transfer in a unique motility structure. BMC Evol Biol. 2007;7:106. doi: 10.1186/1471-2148-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarrell KF, Albers S-V. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–12. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Murray PA, Zinder SH. Nitrogen fixation by a thermophilic methanogenic archaebacterium. Nature. 1984;312:284–6. doi: 10.1038/312284a0. [DOI] [Google Scholar]
- 43.Chien Y-T, Auerbuch V, Brabban AD, Zinder SH. Analysis of genes encoding an alternative nitrogenase in the archaeon Methanosarcina barkeri 227. J Bacteriol. 2000;182:3247–53. doi: 10.1128/JB.182.11.3247-3253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray PA, Zinder SH. Polysaccharide reserve material in the acetotrophic methanogen, Methanosarcina thermophila strain TM-1: accumulation and mobilization. Arch Microbiol. 1987;147:109–16. doi: 10.1007/BF00415270. [DOI] [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrity GM, Holt JG. Phylum AII. Euryarchaeota phy. nov. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. Volume 1. Second Edition. New York: Springer; 2001. pp. 211–355. [Google Scholar]
- 51.Validation List no. 85 Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2002;52:685–90. doi: 10.1099/ijs.0.02358-0. [DOI] [PubMed] [Google Scholar]
- 52.Boone DR. Class II. Methanococci class. nov. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. Volume 1. Second Edition. New York: Springer; 2001. pp. 235–94. [Google Scholar]
- 53.Boone DR, Whitman WB, Koga Y. Order III. Methanosarcinales ord. nov. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey’s Manual of Systematic Bacteriology, volume 1. Second Edition. New York: Springer; 2001. p. 268–94.
- 54.Sowers KR, Johnson JL, Ferry JG. Phylogenetic relationships among the methylotrophic methane-producing bacteria and emendation of the family Methanosarcinaceae. Int J Syst Bacteriol. 1984;34:444–50. doi: 10.1099/00207713-34-4-444. [DOI] [Google Scholar]
- 55.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 56.Opinion 63 Rejection of the type species Methanosarcina methanica (approved lists, 1980) and conservation of the genus Methanosarcina (approved lists, 1980) emend. Mah and Kuhn 1984 with Methanosarcina barkeri (approved lists, 1980) as the type species. Int J Syst Bacteriol. 1986;36:492. doi: 10.1099/00207713-36-3-492. [DOI] [Google Scholar]
- 57.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol. 2002;4:453–61. [PubMed] [Google Scholar]
- 59.Assis das Graças D, Thiago Jucá Ramos R, Vieira Araújo AC, Zahlouth R, Ribeiro Carneiro A, Souza Lopes T, et al. Complete genome of a Methanosarcina mazei strain isolated from sediment samples from an Amazonian flooded area. Genome Announc. 2013;1:e00271–13. doi: 10.1128/genomeA.00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


