Abstract
Induction of mammalian heme oxygenase-1 and exposure of animals to carbon monoxide ameliorates experimental colitis. When enteric bacteria, including Escherichia coli, are exposed to low iron conditions, they express an heme oxygenase-like enzyme, chuS, and metabolize heme into iron, biliverdin and carbon monoxide. Given the abundance of enteric bacteria residing in the intestinal lumen, we hypothesized that commensal intestinal bacteria may be a significant source of carbon monoxide, with the consequence that enteric bacteria expressing chuS and other heme oxygenase -like molecules suppress inflammatory immune responses through release of carbon monoxide. Carbon monoxide exposed mice have altered enteric bacterial composition and increased E. coli 16S and chuS DNA by real-time PCR. Moreover, severity of experimental colitis correlates with increased E. coli chuS expression in IL-10 deficient mice. To explore functional roles, E. coli were genetically modified to overexpress chuS or the chuS gene was deleted. Co-culture of chuS-overexpressing E. coli with bone marrow derived macrophages results in decreased IL-12 p40 and increased IL-10 secretion compared to wild-type or chuS-deficient E. coli. Mice infected with chuS-overexpressing E. coli have increased levels of hepatic carbon monoxide and decreased serum IL-12 p40 compared to mice infected with chuS-deficient E. coli. Thus, carbon monoxide alters the composition of the commensal intestinal microbiota and expands E. coli populations harboring the chuS gene. These bacteria are capable of attenuating innate immune responses through expression of chuS. Bacterial heme oxygenase -like molecules and bacterial-derived carbon monoxide may represent novel targets for therapeutic intervention in inflammatory conditions.
Keywords: Heme oxygenase, chuS, enteric microbiota, carbon monoxide
INTRODUCTION
Carbon monoxide (CO) has anti-inflammatory effects in experimental models of varied inflammatory conditions(1-4). Mammalian cells generate CO endogenously during heme degradation by the heme oxygenase (HO) enzymes. Heme oxygenase-1 (HO-1) plays a critical role in defending the body against oxidant-induced injury(5). We have previously demonstrated that CO ameliorates active inflammation in, T helper (Th)1/Th17(6)- and a Th2-mediated(7) models of chronic inflammatory bowel disease (IBD) through HO-1 dependent pathways. In macrophages and in vivo, pharmacologic induction of HO-1 recapitulates the immunosuppressive effects of CO, abrogating expression of the pro-inflammatory cytokine, IL-12 p40(6), while increasing expression of the anti-inflammatory cytokine, IL-10(7).
Interestingly, enteric bacteria such as E. coli express HO-like enzymes. In E. coli this gene is named chuS (8, 9). Despite marked differences in amino acid sequence, these bacterial enzymes share a similar structure and heme-degrading function with mammalian HOs(8). In bacteria, HO-like molecules scavenge iron from heme as a nutrient source in low iron conditions(10). Consequently, bacterial HO-like enzymes are up- and down-regulated in low and high iron conditions, respectively(11, 10).
Mammalian HO-1 protects against immune-mediated damage regardless of whether expressed in mammalian cells or by genetically engineered luminal bacteria. Notably, administration of a commensal strain of Lactobacillus lactis that over-expresses mammalian HO-1 is associated with decreased mucosal injury and inflammation in a rat model of hemorrhagic shock(12).
We hypothesized that the HO-like activity of commensal enteric bacteria attenuates inflammatory responses via CO production. Here, we report that the severity of experimental colitis correlates with increased E. coli chuS expression, E. coli chuS expression attenuates inflammatory immune responses in macrophages, and infection of mice with E. coli that overexpress chuS is associated with increased CO production and decreased serum IL-12 p40 in a sepsis model.
MATERIALS AND METHODS
Mice
Wild type (WT) and IL-10 deficient (Il10−/−) mice on a C57BL/6 background were reared in a specific pathogen free environment (SPF) at the University of North Carolina and the Beth Israel Deaconess Medical Center Laboratory Animal Resources Facilities. All animals were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research. Protocols and experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina and Beth Israel Deaconess Medical Center (Permit Number: 10-091.3). Mice were euthanized as soon as they lost 20 percent in body weight, or showed signs of hunching, ruffled fur, immobility, decreased food or water intake, rectal prolapse or bloody diarrhea. Otherwise, all mice were euthanized at the end of the experiment. Mice were killed by exposure to CO2 followed by cervical dislocation.
Exposure of mice to carbon monoxide
WT male mice on a C57BL/6 background at the age of 10 weeks were exposed to carbon monoxide (CO) (250 ppm for one hour daily) for 14 days. Fecal pellets were collected and snap frozen in liquid nitrogen on day 0 (before first treatment), on day 14 (maximal exposure to CO), and 14 days after last CO treatment (washout) on day 28.
Monoassociation studies
Germ-free Il10−/− mice on the SvEv/129 genetic background were maintained on a normal iron diet in the National Gnotobiotic Rodent Resource Center at the University of North Carolina and selectively colonized (monoassociated) by oral gavage with 200μl of an overnight culture of NC101 E. Coli grown in LB broth. At the indicated time points, mice were euthanized and cecal contents were immediately snap-frozen in liquid nitrogen, and fragments of mid-colon were harvested for colonic explant culture.
Bacterial sepsis model
WT C57BL/6 mice were infected with one of three E. coli NC101 mutant strains through intraperitoneal injection of 2×108 bacteria/mouse. Mice were euthanized 15 hours later. Blood was withdrawn for bacterial cultures and cytokine serum level. Livers were harvested for CO content and mammalian cytokine mRNA expression.
Bacterial strains lysates and growth curves
The nonpathogenic murine E. coli strain designated NC101 was originally isolated from a randomly chosen colony from the feces of WT mice raised in SPF conditions(13, 14). Construction of E. coli NC101 mutants with deleted chuS gene (ΔchuS) and overexpressed chuS gene (pGEN-MCST5chuS) was done using standard molecular biology techniques (Supplementary Methods). For growth curves, Luria-Bertani broth was inoculated with an overnight bacterial culture and incubated at 37°C. The OD600 of the cultures was measured at the indicated time points. For Western blot analysis of ChuS and for infection of BMDMs with the different strains of E. coli NC101, Luria-Bertani broth was inoculated with an overnight bacterial culture and in the morning 100ul of each culture was incubated at 37°C for 3 hours in the presence of 250μM iron chelator (2,2 Bipyridil- (Sigma)). Bacteria were washed with PBS and concentrations were determined using OD600. To prepare bacterial lysates, anti-proteases (Roche) and Laemmli Buffer were added and the mixture was boiled for 5 minutes. Supernatants were frozen at -80°C for future use.
Bacterial RNA extraction
Bacterial cultures: bacteria were washed, pelleted and maintained in RNAprotect Bacteria Reagent (Qiagen) at -80°C. RNA extraction was performed using RiboPure™-Bacteria Kit (Ambion) according to manufacturer’s instructions. The presence of contaminating genomic DNA was assessed using no reverse transcriptase controls. Cecal content: approximately 300 mg of freshly-harvested cecal contents were snap frozen in N2 (l) and stored at -80°C until ready for use. Frozen samples were thawed into 1 ml of RNAprotect Bacteria Reagent (Qiagen) while vortexing, incubated at 25°C for 5 min, and bacterial RNA was isolated as described previously(14).
Bacterial DNA isolation
Bacterial genomic (g)DNA isolation from E. coli was performed using a DNA purification kit (Wizard® Genomic DNA Purification Kit, Promega) according to manufacturer’s instructions. Bacterial gDNA isolation from fecal pellets: fecal pellets were snap frozen and maintained at -80°C until extraction. Samples were suspended in lysis buffer containing 20 mg/ml lysozyme and incubated for 30 minutes at 37°C and further treated chemically by SDS and proteinase K and mechanically homogenized using a bead beater (BioSpec Products). Finally, gDNA was extracted using a DNeasy DNA extraction kit (Qiagen) and brought to a concentration of 10 ng/ul.
Enteric microbial population analysis
To study bacterial compositional changes in fecal pellets, we used terminal restriction length polymorphism (T-RFLP) as described by Azcarate-Peril et al.(15). Briefly, amplification of the 16S rRNA gene was performed using the 16S universal primers 8F-Hex (5'-AGA GTT TGATC(A/C) TGG CTC AG-3'), and 1492R (5'-GGT TAC CTT GTT ACG ACT T-3'). PCR reactions were purified using a QIAquick PCR Purification kit (Qiagen). Each sample was digested with three different restriction enzymes HhaI, MspI and RsaI) (New England Biolabs) according to manufacturer’s instructions and mixed with size standards (Bioventures Map Marker 1000). Samples were run on an ABI 3130xl capillary sequencer for fragment detection of each of the enzymes. Peaks falling outside of the size standards (50–1,000 bp) were removed and only fragments with a relative peak area ratio (Pi) of ≥1% were considered for further analysis. To generate Principal-Coordinates graphs we used the Qiime (Quantitative Insights Into Microbial Ecology) module specifically developed for T-RFLP analysis.
Bone marrow-derived macrophages
Bone marrow derived macrophages (BMDMs) were harvested from C57BL/6 mice and stimulated with L929 cell media (as a source of M-CSF) for 6 days before their harvest, as previously described (16, 14).
Infection of bone marrow macrophages with E. coli
BMDMs were infected with E. coli (MOI 1:20) in a 12ml plate in RPMI 1640 medium supplemented with 10% FCS, 10μM Hemin (Frontier Bioscience) and 750 μM 2’,2’ Bipyridil (Sigma) for 3, 8 and 20 hours. Supernatants were collected and immediately frozen for cytokine concentration and RNA was extracted from adherent BMDMs to determine cytokine expression levels.
Bacterial real-time RT-PCR and PCR
Real-time PCR assays were performed with genomic DNA extracted from bacterial cultures and from enteric luminal contents as well as RT-PCR on bacterial complementary DNA (cDNA). A negative (no-template) control was included in every run. Amplification, detection, and analyses were performed in a HT-7900 machine (Applied Biosystems). Isolated bacterial genomic DNA (20 ng) was amplified using the following cycle profile: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds and 72°C for 30 seconds. A melting curve was included in all runs. CT was defined as the cycle at which the fluorescence became detectable above the background fluorescence. PCR primer sequences are provided (Supplemental Table 1).
Mammalian real time RT-PCR
RNA isolation and real time RT-PCR was performed as previously described(17). PCR primer sequences are available upon request.
Cytokine ELISAs
Murine IL-12 p40 and IL-10 (e-Bioscience) immunoassay kits were used according to the manufacturers’ instructions.
Western immunoblot
Specific polyclonal rabbit anti-mouse antibodies against the E. coli ChuS peptide (38.63 kD) were made by GeneScript and used at a 1:3000 dilution. Secondary antibody (donkey-anti-rabbit, GE Healthcare, NA934V) was used at 1:10000). Protein levels in supernatants, isolated from lysates of the three bacterial strains, were measured and equalized using the Coomassi Plus Assay Kit (Thermo Scientific).
Tissue carbon monoxide (CO) determination
Harvested livers were immediately removed from sacrificed mice and placed in iced buffer (0.1 M KPO4 buffer, pH 7.4) and flash frozen in liquid nitrogen and then stored at -80°C. Samples were then shipped in dry ice to the Stanford University School of Medicine (S.S. and R.W.). Upon arrival, specimens were diced with scissors, and washed with iced buffer. 100±2mg tissue were then sonicated at 50% power with an ultrasonic cell disruptor with a 1/8” microprobe (Model XL2000, Misonics Inc., Farmingdale NY) in 900 µL buffer using a 2.0mL polypropylene microfuge tube in an ice bath(18). Sonicates were kept on ice. CO levels in liver sonicates were analyzed as previously described(18). Briefly, 5µL of 30% sulfosalicylic acid (SSA) were pipetted into triplicate 2mL clear vials with 35µL ddH2O added. Two sets of triplicate blank vials containing 5µL of SSA and 55µL of water was also prepared. All vials were capped and then purged with CO-free air. In the tissue vial sets, 20µL of liver sonicate was then injected through the septa of vial caps and into the SSA. Vials were then vigorously shaken before incubation at 0°C. After 30 min, CO released into the vial headspace was quantitated by GC using a 60x0.53 cm (internal diameter) stainless-steel column packed with 5A molecular sieve, 60–80 mesh at a temperature of 125°C, and a reduction gas detector (RGA2, Trace Analytical Inc., Menlo Park CA) operated at 270°C(19, 20). The analyzer has a practical detection limit of 1 pmol CO/vial. Tissue CO concentrations were calculated as pmol CO produced/mg FW and expressed as fold change from controls.
Statistical Analyses
Statistical significance for data subsets from experiments performed was assessed by the two-tailed t-test using GraphPad Prism software version 5 (GraphPad Software, Inc. CA, USA). For non-parametric data a Mann-Whitney test was applied and a one-way ANOVA for comparison of multiple groups. For correlation analysis we used Pearson's correlation coefficient or Spearman's rank correlation coefficient for parametric or non-parametric data, respectively. Analysis of similarities (ANOSIM) between enteric bacterial communities generated through T-RFLP analysis, was calculated through the corresponding function in Qiime software. A value of P<0.05 was considered statistically significant.
RESULTS
Exposure of mice to CO alters the enteric microbial composition
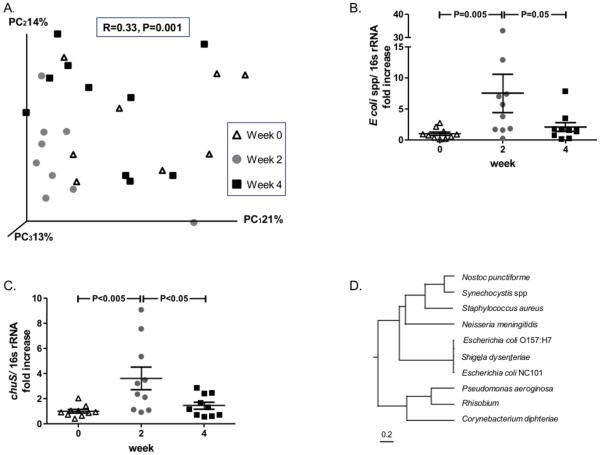
Wild type (WT) mice were exposed to CO and enteric bacteria compositional changes evaluated by T-RFLP analysis. Through this technique, each bacterial group is represented by a different length of the 16S ribosomal gene fragment generated by a restriction enzyme. The variety of all fragments in a fecal sample represents the enteric bacterial composition. One hour daily exposure of mice to CO for 2 weeks resulted in significant enteric bacterial compositional changes (analysis of similarities (ANOSIM): R=0.33, P=0.001) that returned closer to baseline composition after a washout period (no exposure to CO) of 14 days (Figure 1A). Targeted qPCR studies for common enteric bacterial species revealed a significant increase in the abundance of Escherichia coli (E. coli) spp. (7.52 fold increase, P<0.02) in fecal pellets of mice after two weeks of daily exposure to CO that also returned towards baseline within two weeks after CO withdrawal (Figure 1B). qPCR analysis of other bacterial groups (Lactobacillus spp, Bacteroides spp, Enterococcus faecalis and Clostridium coccoides) did not reveal similar changes (Supplemental Figure 1). ChuS is an enzyme with heme oxygenase activity. The chuS gene is found only in some E. coli strains. Interestingly, qPCR of the chuS gene revealed patterns of change in CO exposed mice as demonstrated for E. coli spp, with a significant increase in the abundance of chuS in fecal pellets of mice after two weeks of daily CO exposure (3.63 fold increase, P<0.005) that returned towards baseline two weeks after CO withdrawal (Figure 1C). These results led us to speculate that CO may exert immunomodulatory effects on the host, in part, through selection of bacteria that express HO-like genes.
Figure 1. Carbon monoxide (CO) exposure alters enteric microbial composition in mice.
Ten C57BL/6 mice were exposed to CO (250 ppm for one hour/day) for 14 days. Microbial composition alterations were analyzed before CO exposure, after 14 days of exposure, and 14 days after exposure by T-RFLP. (A) Principal coordinate analysis (PCoA) plot of bacterial populations demonstrates a significant effect of CO (gray circles, week 2) and CO withdrawal (black squares, week 4) on microbial populations compared to pre-exposure (triangles, week 0). (B) qPCR analysis of the 16S rRNA gene for E. coli spp. (C) and of chuS gene demonstrates an increase in abundance during CO exposure and a decrease towards baseline following withdrawal. Both were normalized to the abundance of total bacteria gDNA and represent a fold increase compared to day 0. (D) Multiple sequence alignment of bacterial heme oxygenase sequences. Alignment of the E. coli NC101 heme oxygenase, chuS, against heme oxygenases of other bacteria reveal >98% sequence similarities with phylogenetically proximal bacteria such as Shigela dysenteriae and E. coli O157:H7 as opposed to phylogenetically more distant bacteria such as Pseudomonas aeroginosa, Staphylococcus aureus and Neisseria meningitidis. Tree was built through clustalw software using rooted phylogenetic tree with branch length function on bacterial heme oxygenases gene sequences.
E. coli NC101 express a heme oxygenase-like gene
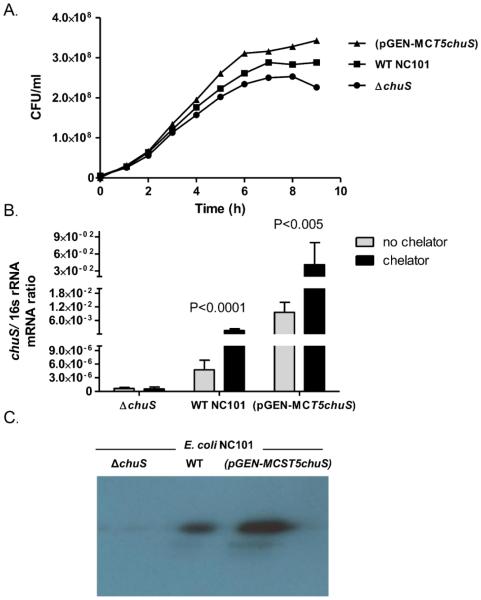
E. coli NC101 (NC101) is a resident commensal microorganism in mouse colon(8) and mono-association of germ-free (GF) Il10−/− mice with NC101 results in colitis. Given that not all E. coli strains utilize heme(21), we first verified that NC101 carries the chuS gene. The E. coli 0157:H7 chuS gene sequence shares 99% nucleotide sequence similarity with an un-annotated gene in E. coli NC101 with a putative heme degradation activity (E. coli NC101 contig6, whole genome shotgun sequence. ACCESSION AEFA01000022; REGION: 288646..289674, ECNC101_06049) (Supplemental Table 2). The presence of chuS in NC101 was definitively confirmed by sequencing the chuS PCR product using primers designed from the un-annotated sequence (data not shown- for primers see Supplemental Table 1). Furthermore, alignment of the NC101 chuS gene sequence with sequences of other bacterial HO-like genes, reveals high similarity to sequences of phylogenetically related bacteria as opposed to phylogenetically distant bacteria (Figure 1D). Functionally, incubation of NC101 with an iron chelator resulted in >300 fold increase in chuS mRNA expression, supporting a role for chuS in NC101 iron homeostasis (Figure 2).
Figure 2. Wild type (WT), chuS overexpressing (pGEN-MCST5chuS), and chuS deleted(ΔchuS) E. coli NC101 strains growth curves and chuS expression in response to changes in iron availability.
(A) A growth curve (representative of 3 experiments) of the three NC101 strains in the absence of iron and in the presence of heme (as an alternative iron source) demonstrates a growth advantage in NC101(pGEN-MCST5chuS) compared to the two other strains. (B) chuS mRNA expression normalized to total bacteria 16s rRNA expression is significantly higher in WT E. coli NC101 compared to ΔchuS and in (pGEN-MCST5chuS) compared to WT NC101. In low iron conditions, expression of chuS is significantly increased in NC101(pGEN-MCST5chuS) and WT E coli NC101 and is absent in ΔchuS. (C) Western immunoblot (representative gel displayed from 3 experiments with identical results) demonstrates increased ChuS protein expression in NC101(pGEN-MCST5chuS) and no expression in NC101 ΔchuS. Equal amounts of protein (3.4ug protein) were loaded on the gel.
E. coli chuS expression correlates with intestinal inflammation
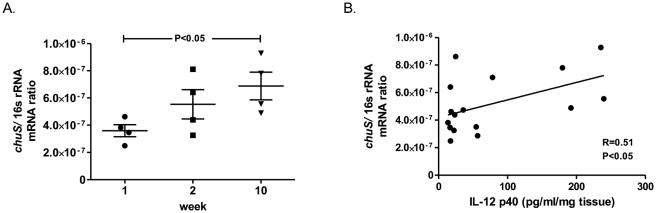
Colitis results in decreased enteric iron(22) and increased enteric heme availability(23), propagating an environment favorable for heme utilizing bacteria. Monoassociation of germ free Il10−/− mice with E. coli NC101 results in colitis that progresses over a period of 10 weeks(14). Interestingly, increased fecal chuS expression correlates (R=0.51, P<0.05) with progression of inflammation as assessed by IL-12 p40 levels from colonic explant cultures (Figure 3).
Figure 3. chuS expression in E. coli NC101 monoassociated Il10−/− mice correlates with colonic IL-12 p40 secretion.
129S6/SvEv germ free Il10−/− mice were colonized with E. coli NC101. Four mice were sacrificed at 3 different time points during a 10 week time course. (A) chuS expression normalized to total bacteria 16s rRNA expression increased significantly from week 1 to week 10 post colonization of mice. (B) Colonic IL-12 p40 secretion from colonic explants correlate with E. coli NC101 chuS expression from cecal contents (R=0.51, P<0.05).
chuS expression modulates innate immune responses
While colonic inflammation in Il10−/− mice correlates with increased chuS expression it is unknown whether chuS can affect the host inflammatory response, akin to mammalian HO-1. To begin to address this question, chuS was deleted (NC101 ΔchuS) and overexpressed [NC101(pGEN-MCST5chuS)] from wild type E. coli NC101 (WT NC101). chuS expression was 2000 times higher in NC101 (pGEN-MCST5chuS) compared to WT NC101, and was not expressed in NC101 ΔchuS (Figure 2). Exposure of the three bacterial strains to an iron chelator resulted in significant increase (P<0.005) of chuS expression in WT NC101 and NC101(pGEN-MCST5chuS) but not in NC101 ΔchuS (Figure 2). Similarly, ChuS protein was not expressed by NC101 ΔchuS and was highly expressed by NC101(pGEN-MCST5chuS) (Figure 2C). Moreover, while all strains have similar growth curves when iron is abundant (Supplemental Figure 2), NC101(pGEN-MCST5chuS) strain demonstrates a growth advantage when heme is the source of iron (Figure 2), supporting the physiological role of chuS in heme metabolism and iron homeostasis.
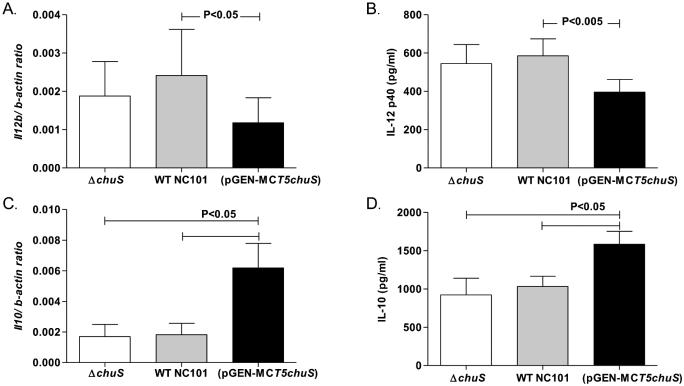
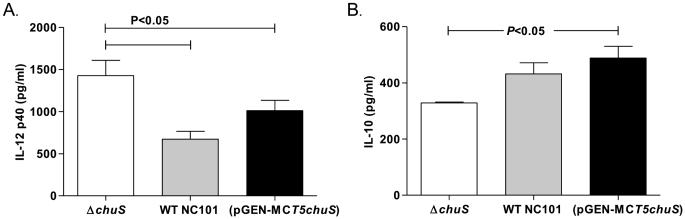
Bone marrow derived macrophages (BMDMs) were cultured with the three NC101 strains. IL-12 p40, a pro-inflammatory cytokine, was significantly decreased, while IL-10, a central anti-inflammatory cytokine, was significantly increased in BMDMs infected with NC101(pGEN-MCST5chuS) (P<0.05) compared to BMDMs infected with WT NC101 or NC101 ΔchuS (Figure 4). Similarly, in BMDMs infected with NC101(pGEN-MCST5chuS), mRNA expression of Il12b and Il10 was decreased and increased, respectively, compared to BMDMs infected with WT NC101 or NC101 ΔchuS(Figure 4). In BMDMs infected with WT NC101 and NC101 ΔchuS, differences in IL-12 p40 and IL-10 expression were not detected.
Figure 4. Expression of cytokines in bone marrow derived macrophages (BMDMs) cultured with genetically altered chuS and WT E. coli NC101 strains.
BMDMs were cultured with WT, chuS overexpressing (pGEN-MCST5chuS),and chuS deleted (ΔchuS) E. coli NC101. Total RNA was extracted from cells and mRNA was assessed by qPCR and normalized to β-actin. Cytokine protein level was assayed by ELISA from supernatants. Expression of Il12b mRNA was assessed by qPCR after 3.5 hours, IL-12 p40 protein level was assessed after 8.5 hours, Il10 mRNA was assessed by qPCR after 8.5 hours and IL-10 protein level was assayed by ELISA from supernatants after 20 hours of incubation. Il12b mRNA expression normalized to β-actin (A) and IL-12 p40 protein level (B) production were significantly decreased in BMDMs incubated with NC101(pGEN-MCST5chuS) compared to WT NC101 and NC101 ΔchuS. Il10 mRNA expression normalized to β-actin (C) and IL-10 protein (D) production were significantly increased in BMDMs incubated with NC101(pGEN-MCST5chuS) compared with WT NC101 and NC101 ΔchuS. Each result represents the mean± SEM from 5-7 independent experiments.
To examine whether alteration of macrophage cytokine expression by NC101 (pGEN-MCST5chuS) is mediated by a bacterial product requiring ongoing active bacterial metabolism, BMDMs were cultured with heat killed bacteria. There was no difference in IL-12 p40 or IL-10 expression (Supplemental Figure 3), supporting the requirement of live NC101 (pGEN-MCST5chuS) to modulate cytokine expression. Furthermore, incubation of NC101 strains with BMDMs separated by a membrane non-permeable to bacteria (0.4um), resulted in decreased IL-12 p40 and increased IL-10 induction by NC101 (pGEN-MCST5chuS) compared with NC101 ΔchuS (Figure 5), implicating a soluble substance released by live bacteria. Bacterial numbers of NC101 (pGEN-MCST5chuS) in the upper chambers were similar or higher compared to the two other strains (Supplemental Figure 4), excluding the possibility that immunologic differences detected between strains are a consequence of reduced bacterial numbers.
Figure 5. IL-12 p40 and IL-10 regulation in BMDMs by NC101(pGEN-MCST5chuS) is mediated by a soluble factor secreted by live bacteria.
(A) Culture of BMDMs with WT, chuS overexpressing (pGEN-MCST5chuS), and chuS deleted(ΔchuS) E. coli NC101 in two chambers separated by a membrane permeable to substances <0.4um, resulted in decreased IL-12 p40 production and (B) increased IL-10 production by BMDMs incubated with the NC101(pGEN-MCST5chuS) compared to the other strains. Results are presented as mean±SEM from 3-4 independent experiments.
E. coli NC101 chuS demonstrates immunomodulatory effects and increases CO in vivo
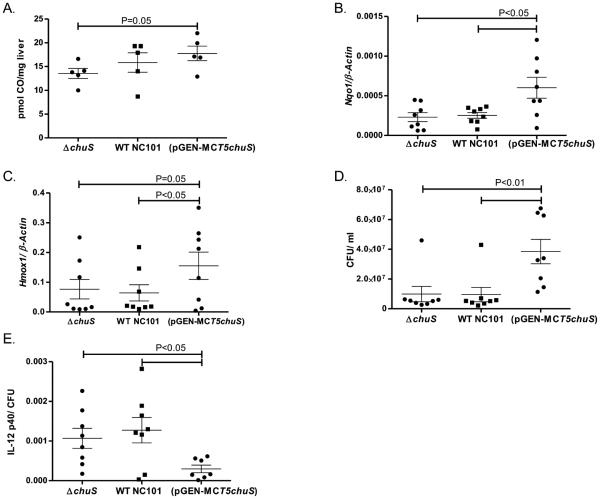
We hypothesized that chuS expression mediates immune modulation through release of CO, since CO is a soluble gas and a product of HO activity. To test this hypothesis, WT mice were injected intraperitoneally with equal numbers of each E. coli strain. There was a "dose response" correlation with increased hepatic CO and chuS abundance (Figure 6A). However, the only difference that reached statistical significance was that increased hepatic CO was detected in mice infected with NC101 (pGEN-MCST5chuS) compared to those infected with NC101 ΔchuS (17.8±3.4 vs 13.5±2.7 pmol CO/mg liver, respectively; P<0.05). Since CO has been shown to increase expression of mammalian HO-1 predominantly through recruitment of the transcription factor Nrf2(24, 25), the expression of mammalian Hmox1 and Nqo1, target genes of Nrf2, was determined. Hmox1 and Nqo1 expression was significantly increased in the livers of mice infected with NC101 (pGEN-MCST5chuS) compared to the other strains (Figure 6B, C). Despite significantly increased bacterial numbers in the blood of mice infected with NC101 (pGEN-MCST5chuS) (Figure 6), IL-12 p40 was not increased in sera from these mice (7023±3840 pg/ml; 5661±3447 pg/ml; 6006±2946 pg/ml) and IL-10 was marginally higher (6447±3190 pg/ml; 4212±2244 pg/ml; 5640±2890 pg/ml) compared with WT- NC101 and NC101 ΔchuS, respectively. Moreover, the ratio of IL-12 p40/ bacteria in the NC101 (pGEN-MCST5chuS) injected mice- was significantly lower compared to the two other groups (Figure 6). Together, these data support in vivo immune modulation mediated by E. coli NC101 that overexpresses the chuS gene.
Figure 6. chuS expression results in increased hepatic CO and expression of Nrf2 dependent genes in a sepsis model.
Mice (n=8/group) were inoculated intraperitoneally with WT, chuS overexpressing (pGEN-MCST5chuS),and chuS deleted(ΔchuS) E. coli NC101 strains. (A) Hepatic CO was measured as described in the methods 15 hours after infection and presented as pmol CO per mg hepatic tissue for individual mice. (B) Hepatic Nqo1 and (C) Hmox1 expression was determined by real time RT-PCR and presented normalized to β-actin for individual mice. (D) Blood bacterial load was determined by plating blood samples from each of the mice on LB gel plates and counting number of CFUs after an overnight incubation period. Bacterial CFU/ml was significantly higher in the blood of mice infected with (pGENMCST5chuS) compared with the two other bacterial strains. (E) Serum IL-12 p40 levels assayed by ELISA normalized to number of CFU/ml were significantly lower in mice infected with NC101(pGEN-MCST5chuS). Results are presented as mean± SEM.
DISCUSSION
The importance of mammalian HO activity in modulating immune responses has been demonstrated in multiple models of inflammatory disease(5, 4). Furthermore, enzymatic activity of HO on its substrate heme results in production of carbon monoxide (CO), which also exerts anti-inflammatory effects in cells and in vivo, in part through HO dependent pathways(6). Our group has shown that induction of mammalian HO-1 ameliorates colitis in several experimental models of intestinal inflammation(6, 7, 17). Moreover, we recently demonstrated that the enteric microbiota induce colonic expression of HO-1 in WT but not colitis-prone Il10−/− mice and that HO-1 expression inversely correlated with colonic inflammation and IL-12 p40 and TNF expression. Pharmacologic induction of HO-1 protected Il10−/− mice from microbiota induced colitis when transitioned from germ free (GF) to conventional conditions(17). These experiments highlight the importance of mammalian HO-1 and CO in the maintenance of intestinal homeostasis. Consequently, HO-1, with pleiotropic anti-inflammatory effects, is a potential therapeutic molecule of interest in various inflammatory diseases including IBD
The metabolic capacity of the enteric microbiome is large and diverse, having profound effects on the host's health. Various enteric bacteria (e.g. E. coli, Shigella dysenteriae) express genes with HO activity. Given pleiotropic anti-inflammatory effects of CO, we hypothesized that CO exposure may alter the enteric microbial composition. Wild type mice (as opposed to mice with experimental IBD) were utilized to assess CO-induced changes in enteric microbial communities to avoid the confounding factor of microbiome alterations as a consequence of inflammation, as CO ameliorates intestinal inflammation(6, 7) and likewise, inflammation shapes microbial composition(26). Indeed, significant enteric microbial compositional changes were demonstrated following CO exposure that were reversed two weeks after exposure was discontinued (Figure 1A).
Interestingly, abundance of E. coli spp and chuS DNA increased with CO administration, while abundance of other bacteria, that do not express chuS or heme oxygenase genes, such as Lactobacillus spp, Bacteroides spp, Enterococcus faecalis, Clostridium coccoides and Segmented filamentous bacteria did not change (Supplemental Figure 1), suggesting that CO exposure of mice may result in selection for bacteria that can potentially produce CO endogenously. This may be partially supported by our observation that two weeks after withdrawal of CO exposure, mice enteric bacterial population (Figure 1A), E. coli gDNA level (Figure 1B) and chuS gDNA level (Figure 1C) returned to near baseline levels (before CO exposure). Similarly, others have demonstrated that a temporal exposure to an environmental factor, such as an antibiotic, results in an alteration of bacterial population composition and a quick return to baseline upon withdrawal(27). An alternative explanation for the alterations in the composition of luminal bacterial population may be a shift of bacteria from the mucosal to the luminal niche. We believe this option is less probable given the total number of bacteria in the lumen did not changed (data not shown).
Given the large biomass of enteric bacteria, some of which express HO-like genes, we became interested in whether enteric bacterial heme oxygenases physiologically participate in intestinal immune homeostasis. CO producing bacteria have been shown to be protected from direct bactericidal activity mediated by CO(28). While the exact mechanisms for bacterial resistance to CO are not clear, they may be involved in downstream signaling of CO, such as redox sensing (soxS and oxyR) or through genes that are associated with biofilm formation (tqsA and bhsA)(28). Specifically, it has been shown that resistance of Mycobacterium tuberculosis to CO is dependent on the cor gene, whose function is not entirely clear(29). Some of these mechanisms possibly allow survival of chuS expressing bacterial species in environments toxic to other enteric bacterial species. To verify that bacterial heme oxygenases have a role in the inflammatory process, the bacterial strain E. coli NC101 (NC101) was utilized. NC101 is a commensal enteric bacteria in mice with functional similarities to adherent-invasive E. coli (AIEC) strain LF82 isolated from patients with chronic ileal Crohn's disease(30). Monoassociation of germ-free Il10−/− mice with NC101 induces colitis by 5 to 7 weeks(13). We demonstrated that NC101 carries and expresses chuS, which is 99% homologous to a gene in E. coli 0157:H7 with HO activity. chuS expression in NC101 in vivo correlated with inflammation in monoassociated germ-free Il10−/− mice. During inflammation, competition for iron is increased(22) and intraluminal heme becomes more available(23), both conditions favor increased expression of chuS(10). Moreover, in another murine model of IBD, a low iron diet was associated with decreased ileal inflammation(31). Altogether, these findings may support a survival compensatory mechanism of these bacteria, aimed to decrease the hostile inflammatory environment by increasing chuS expression to reduce the inflammatory response (and IL-12 production) by the host. Similar mechanisms have been previously demonstrated for E. coli that expressed stress response genes aimed to protect it from oxidative stress upon intestinal inflammation(14). However, whether HO-like enzyme expression by enteric bacteria mediates anti-inflammatory properties in the setting of low intestinal iron availability in experimental IBD and whether expression of chuS gene by E. coli ameliorates intestinal inflammation in vivo remains to be determined by performing a formal mono-association study of germ-free Il10−/− and WT mice with WT NC101 or ΔchuS E. coli bacteria.
We have previously shown that in macrophages activated by bacterial products, induction of mammalian HO-1 in BMDMs (through exposure to CO or protoporphyrins), resulted in inhibition of inflammatory mediators including IL-12 p40 and induction of anti-inflammatory pathways such as IL-10 and IL-22(7). However, whether bacterial expression of chuS mediates anti-inflammatory effects, akin to mammalian HO-1, has not been previously investigated. In BMDMs, overexpression of chuS in NC101 resulted in decreased production of IL-12 p40 and increase of IL-10, compared to BMDMs infected with WT NC101 or ΔchuS. Although the major anti-inflammatory effect of chuS over-expression is probably mediated through IL-10 production, there seems to be also a direct anti-inflammatory effect of chuS over-expression by lowering IL-12 p40 levels through IL-10 independent pathways. This was demonstrated when we infected BMDMs, isolated from Il10−/− deficient mice, with the three strains and found that exposure to the NC101 (pGEN-MCST5chuS) strain resulted in a trend towards lower IL-12 p40 levels (Supplemental Figure 5). Further, expression of TNF-α by the infected BMDMs did not change (data not shown) among the three groups, supporting the importance bacterial HO modulation of IL-10 and IL-12 p40 in BMDMs. These experiments were conducted in a low iron (through the addition of an iron chelator to the media) and heme-rich environment as an alternative iron source and chuS substrate. Hence, bacterial heme oxygenases may have similar immunomodulatory effects to mammalian HO-1. Since heat killed bacteria did not demonstrate similar activities, this process is dependent on an active secretion and/or metabolism of immunologically active substances. Immunomodulatory effects of live bacteria were apparent when BMDMs were separated from the bacteria by a membrane that allowed only products <0.4μm to diffuse freely between chambers, demonstrating that physical interaction between bacteria and BMDMs or bacterial internalization is not necessary for modulation of cytokine expression. Bacterial presence from the BMDMs chamber was absent at the end of the incubation period and the number of bacteria of the NC101 (pGEN-MCST5chuS) strain were equal or higher in the bacterial chamber compared to WT NC101 and ΔchuS NC101, suggesting that demonstrated differences in cytokine expression are not due to differences in bacterial number(Supplemental Figure 4). Therefore, release of a soluble factor by NC101 (pGEN-MCST5chuS) mediated the described immunologic effects. In vivo, CO have been previously demonstrated to be protective against sepsis through increased clearance of bacteria(32, 33). Our findings support a decreased immune response mediated also by a soluble factor released by bacteria. We speculate this factor to be CO, although we have not directly shown that NC101 (pGEN-MCST5chuS) bacteria release more CO than the other two strains.
There were no significant differences in IL-12 p40 or IL-10 expression in BMDMs incubated with WT or ΔchuS NC101. In addition, despite decreased growth of ΔchuS under low iron conditions (Figure 2A), this bacterial strain was fully capable of replication, perhaps through recovery of iron through alternate pathways(34). Hence, the immunomodulatory effects of chuS may be apparent only when there is a significant expression of the gene (as found in NC101 (pGEN-MCST5chuS)) and sufficient substrate in the environment.
Finally, following in vivo infection of mice, despite significantly increased numbers of NC101 (pGEN-MCST5chuS) recovered from the blood, IL-12 p40 levels per bacteria (CFU) were significantly lower compared to mice infected with WT ΔchuS NC101. One of the limitations of this study is that the NC101 (pGEN-MCST5chuS) has a growth advantage in substrate rich (and possibly substrate poor) environments compared to the other two strains. Nevertheless, this finding also strengthens our hypothesis that this strain has specific immunomodulatory properties mediated by chuS, as blood IL-12 p40 levels per bacteria were significantly lower in vivo compared to the other strains. Moreover, hepatic CO content of mice infected with NC101 (pGEN-MCST5chuS) was increased compared to other strains, leading to speculation that CO released by bacteria is responsible for attenuating the inflammatory response. However, we also demonstrate that NC101 (pGEN-MCST5chuS) induces hepatic Hmox1 expression to a greater extent than the WT NC101 and NC101 ΔchuS, suggesting that mammalian HO-1 induction could also contribute to increased hepatic CO and immunomodulation. Another limitation of the current study is the lack of experiments using a NC101 ΔchuS complemented by the deleted gene, that should have demonstrated reversed effects for this strain. Nevertheless, the main aim of this work was to examine whether over expression of chuS by the NC101 (pGEN-MCST5chuS) is associated with a decreased inflammatory response compared to the WT-NC101, as has been demonstrated and eventually further supported by the use of the ΔchuS NC101.
In addition, these findings add another layer to the understanding of the importance of CO as a regulator of the immune system. Similar to our findings, that bacteria capable of releasing CO, can modulate the innate immune response, others have demonstrated that CO-releasing molecules (CORMs) may reduce endotoxic shock response (35, 36) and modulate systemic IL-12 level. These actions have been demonstrated to be mediated modulation of TLR4 expression on dendritic cells(37). Other mechanisms for immune tolerance induced by dendritic cells include heme oxygenase(38) and CO(39) dependent induction of T regulatory cells, modulation of TLR4 expression(40), and inhibition of antigen presentation to T cells(41). Moreover, CORMs can directly affect the adaptive immune response through modulation of pro-inflammatory Th1/Th17 and anti-inflammatory Th2 cells(42). These wide range of anti-inflammatory properties of CO and heme oxygenase have been found to be effective in various animal models. CORMs improved clinical and histological signs of experimental allergic encephalomyelitis(43) and exerted strong protective effect in type 1 diabetes in mice(44) and CO exposure ameliorated the immune response and damage in a systemic lupus erythematosus model in mice(39).
Together, these findings suggest that enteric bacterial heme oxygenase expression may be involved in regulation of the enteric immune response. Our work as an aggregate begins to elucidate the cross-talk between mammalian HO-1 and the enteric microbiota. The enteric microbiota induce mammalian HO-1 and HO-1-derived CO(17), which in turn, may shape the enteric microbiota perhaps through selection for bacteria that express HO-like molecules. Thus, we speculate that host-microbe interactions in the intestine through mammalian and bacterial heme oxygenases are central participants in the in maintenance of homeostasis. These findings are particularly relevant to the pathophysiology of IBD. The prevalence of IBD and of other immune mediated diseases has been continuously increasing during the last few decades. Alterations in bacterial populations and function and in the immune response towards them have been shown to occur in these patients and have been regarded as a possible consequence of a cleaner environment and lack of early exposure and education of the immune response ('the Hygiene theory') or from loss of certain important bacterial groups that may have beneficial effects for the host ('the Old Friends theory')(45). Our findings may add an important evidence of how alterations in bacterial function may affect the immune response and may pave the way for future therapeutic interventions. More generally, modulation of bacterial HOs, should be examined in other models of systemic immune mediated diseases where CO have been shown to be beneficial, such as diabetes and multiple sclerosis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01 DK54452 (S.E.P.); a Crohn’s and Colitis Foundation of America Research Fellowship Award (N.M.); Gastroenterology Research Training Grant T32 DK007737 (J.C.O.); and the Center for Gastrointestinal Biology and Disease P30 DK034987 (Immunotechnologies and Histology Core).
Abbreviations
- BMDMs
bone marrow derived macrophages
- CO
carbon monoxide
- CORMs
CO releasing molecules
- E. coli
Escherichia coli
- GF
germ-free
- HO
heme oxygenase
- IL
interleukin
- IBD
inflammatory bowel diseases
- SPF
specific pathogen free
- T-RFLP
terminal restriction length polymorphism
- WT
wild type
Footnotes
Disclosure: None of the authors have conflicts of interest.
Supplemental methods:
- Construction of E. coli NC101 ΔchuS.
- Construction of E. coli NC101 (pGEN-MCST5chuS).
REFERENCES
- 1.Jick H, Walker AM. Cigarette smoking and ulcerative colitis. N Engl J Med. 1983;308:261–3. doi: 10.1056/NEJM198302033080507. [DOI] [PubMed] [Google Scholar]
- 2.Moore BA, Otterbein LE, Turler A, Choi AM, Bauer AJ. Inhaled carbon monoxide suppresses the development of postoperative ileus in the murine small intestine. Gastroenterology. 2003;124:377–91. doi: 10.1053/gast.2003.50060. [DOI] [PubMed] [Google Scholar]
- 3.Ryter SW, Choi AM. Cytoprotective and anti-inflammatory actions of carbon monoxide in organ injury and sepsis models. Novartis Found Symp. 2007;280:165–75. discussion 75-81. [PubMed] [Google Scholar]
- 4.Sato K, Balla J, Otterbein L, Smith RN, Brouard S, Lin Y, Csizmadia E, Sevigny J, Robson SC, Vercellotti G, Choi AM, Bach FH, Soares MP. Carbon monoxide generated by heme oxygenase-1 suppresses the rejection of mouse-to-rat cardiac transplants. J Immunol. 2001;166:4185–94. doi: 10.4049/jimmunol.166.6.4185. [DOI] [PubMed] [Google Scholar]
- 5.Otterbein LE, Soares MP, Yamashita K, Bach FH. Heme oxygenase-1: unleashing the protective properties of heme. Trends Immunol. 2003;24:449–55. doi: 10.1016/s1471-4906(03)00181-9. [DOI] [PubMed] [Google Scholar]
- 6.Hegazi RA, Rao KN, Mayle A, Sepulveda AR, Otterbein LE, Plevy SE. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 2005;202:1703–13. doi: 10.1084/jem.20051047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheikh SZ, Hegazi RA, Kobayashi T, Onyiah JC, Russo SM, Matsuoka K, Sepulveda AR, Li F, Otterbein LE, Plevy SE. An anti-inflammatory role for carbon monoxide and heme oxygenase-1 in chronic Th2-mediated murine colitis. J Immunol. 2011;186:5506–13. doi: 10.4049/jimmunol.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suits MD, Pal GP, Nakatsu K, Matte A, Cygler M, Jia Z. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16955–60. doi: 10.1073/pnas.0504289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegele R, Tasler R, Zeng Y, Rivera M, Frankenberg-Dinkel N. The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J Biol Chem. 2004;279:45791–802. doi: 10.1074/jbc.M408303200. [DOI] [PubMed] [Google Scholar]
- 10.Frankenberg-Dinkel N. Bacterial heme oxygenases. Antioxid Redox Signal. 2004;6:825–34. doi: 10.1089/ars.2004.6.825. [DOI] [PubMed] [Google Scholar]
- 11.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 12.Pang QF, Ji Y, Bermudez-Humaran LG, Zhou QM, Hu G, Zeng Y. Protective effects of a heme oxygenase-1-secreting Lactococcus lactis on mucosal injury induced by hemorrhagic shock in rats. J Surg Res. 2009;153:39–45. doi: 10.1016/j.jss.2008.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, Hansen JJ. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011;141:1842–51. doi: 10.1053/j.gastro.2011.06.064. e1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azcarate-Peril MA, Foster DM, Cadenas MB, Stone MR, Jacobi SK, Stauffer SH, Pease A, Gookin JL. Acute necrotizing enterocolitis of preterm piglets is characterized by dysbiosis of ileal mucosa-associated bacteria. Gut microbes. 2011;2:234–43. doi: 10.4161/gmic.2.4.16332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, Zhu C, Li F, Hegazi R, He K, Babyatsky M, Bauer AJ, Plevy SE. Inhibition of Interleukin-12 p40 Transcription and NF-{kappa}B Activation by Nitric Oxide in Murine Macrophages and Dendritic Cells. J Biol Chem. 2004;279:10776–83. doi: 10.1074/jbc.M313416200. [DOI] [PubMed] [Google Scholar]
- 17.Onyiah JC, Sheikh SZ, Maharshak N, Steinbach EC, Russo SM, Kobayashi T, Mackey LC, Hansen JJ, Moeser AJ, Rawls JF, Borst LB, Otterbein LE, Plevy SE. Carbon Monoxide and Heme Oxygenase-1 Prevent Intestinal Inflammation in Mice by Promoting Bacterial Clearance. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vreman HJ, Wong RJ, Kadotani T, Stevenson DK. Determination of carbon monoxide (CO) in rodent tissue: effect of heme administration and environmental CO exposure. Analytical biochemistry. 2005;341:280–9. doi: 10.1016/j.ab.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Vreman HJ, Stevenson DK. Heme oxygenase activity as measured by carbon monoxide production. Analytical biochemistry. 1988;168:31–8. doi: 10.1016/0003-2697(88)90006-1. [DOI] [PubMed] [Google Scholar]
- 20.Vreman HJ, Stevenson DK. Detection of Heme Oxygenase Activity by Measurement of CO. Current protocols in toxicology / editorial board, Mahin D Maines. 2001 doi: 10.1002/0471140856.tx0902s00. Chapter 9: Unit9 2. [DOI] [PubMed] [Google Scholar]
- 21.Wyckoff EE, Duncan D, Torres AG, Mills M, Maase K, Payne SM. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–52. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 22.Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 2012;7:e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sydora BC, Mcfarlane SM, Doyle JS, Fedorak RN. Neonatal exposure to fecal antigens reduces intestinal inflammation. Inflamm Bowel Dis. 2011;17:899–906. doi: 10.1002/ibd.21453. [DOI] [PubMed] [Google Scholar]
- 24.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–8. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Cao W, Biswal S, Dore S. Carbon monoxide-activated Nrf2 pathway leads to protection against permanent focal cerebral ischemia. Stroke; a journal of cerebral circulation. 2011;42:2605–10. doi: 10.1161/STROKEAHA.110.607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–37. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobre LS, Al-Shahrour F, Dopazo J, Saraiva LM. Exploring the antimicrobial action of a carbon monoxide-releasing compound through whole-genome transcription profiling of Escherichia coli. Microbiology. 2009;155:813–24. doi: 10.1099/mic.0.023911-0. [DOI] [PubMed] [Google Scholar]
- 29.Zacharia VM, Manzanillo PS, Nair VR, Marciano DK, Kinch LN, Grishin NV, Cox JS, Shiloh MU. cor, a novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis. mBio. 2013;4:e00721–13. doi: 10.1128/mBio.00721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SC, Tonkonogy SL, Jarvis HW, Darfeuille-Michaud A, Sartor RB. Escherichia coli Strains Differentially Induce Colitis in IL-10 Gene Deficient Mice. Gastroenterology. 2008;134:A–23. [Google Scholar]
- 31.Werner T, Wagner SJ, Martinez I, Walter J, Chang JS, Clavel T, Kisling S, Schuemann K, Haller D. Depletion of luminal iron alters the gut microbiota and prevents Crohn's disease-like ileitis. Gut. 2011;60:325–33. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Lee SJ, Coronata AA, Fredenburgh LE, Chung SW, Perrella MA, Nakahira K, Ryter SW, Choi AM. Carbon monoxide confers protection in sepsis by enhancing beclin 1-dependent autophagy and phagocytosis. Antioxidants & redox signaling. 2014;20:432–42. doi: 10.1089/ars.2013.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otterbein LE, May A, Chin BY. Carbon monoxide increases macrophage bacterial clearance through Toll-like receptor (TLR)4 expression. Cellular and molecular biology. 2005;51:433–40. [PubMed] [Google Scholar]
- 34.Zheng T, Bullock JL, Nolan EM. Siderophore-mediated cargo delivery to the cytoplasm of Escherichia coli and Pseudomonas aeruginosa: syntheses of monofunctionalized enterobactin scaffolds and evaluation of enterobactin-cargo conjugate uptake. J Am Chem Soc. 2012;134:18388–400. doi: 10.1021/ja3077268. [DOI] [PubMed] [Google Scholar]
- 35.Shen WC, Wang X, Qin WT, Qiu XF, Sun BW. Exogenous carbon monoxide suppresses Escherichia coli vitality and improves survival in an Escherichia coli-induced murine sepsis model. Acta pharmacologica Sinica. 2014;35:1566–76. doi: 10.1038/aps.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin W, Zhang J, Lv W, Wang X, Sun B. Effect of carbon monoxide-releasing molecules II-liberated CO on suppressing inflammatory response in sepsis by interfering with nuclear factor kappa B activation. PLoS One. 2013;8:e75840. doi: 10.1371/journal.pone.0075840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riquelme SA, Bueno SM, Kalergis AM. Carbon monoxide down-modulates Toll-like receptor 4/MD2 expression on innate immune cells and reduces endotoxic shock susceptibility. Immunology. 2015;144:321–32. doi: 10.1111/imm.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreau A, Hill M, Thebault P, Deschamps JY, Chiffoleau E, Chauveau C, Moullier P, Anegon I, Alliot-Licht B, Cuturi MC. Tolerogenic dendritic cells actively inhibit T cells through heme oxygenase-1 in rodents and in nonhuman primates. FASEB J. 2009;23:3070–7. doi: 10.1096/fj.08-128173. [DOI] [PubMed] [Google Scholar]
- 39.Mackern-Oberti JP, Llanos C, Carreno LJ, Riquelme SA, Jacobelli SH, Anegon I, Kalergis AM. Carbon monoxide exposure improves immune function in lupus-prone mice. Immunology. 2013;140:123–32. doi: 10.1111/imm.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remy S, Blancou P, Tesson L, Tardif V, Brion R, Royer PJ, Motterlini R, Foresti R, Painchaut M, Pogu S, Gregoire M, Bach JM, Anegon I, Chauveau C. Carbon monoxide inhibits TLR-induced dendritic cell immunogenicity. J Immunol. 2009;182:1877–84. doi: 10.4049/jimmunol.0802436. [DOI] [PubMed] [Google Scholar]
- 41.Tardif V, Riquelme SA, Remy S, Carreno LJ, Cortes CM, Simon T, Hill M, Louvet C, Riedel CA, Blancou P, Bach JM, Chauveau C, Bueno SM, Anegon I, Kalergis AM. Carbon monoxide decreases endosome-lysosome fusion and inhibits soluble antigen presentation by dendritic cells to T cells. European journal of immunology. 2013;43:2832–44. doi: 10.1002/eji.201343600. [DOI] [PubMed] [Google Scholar]
- 42.Nikolic I, Vujicic M, Stojanovic I, Stosic-Grujicic S, Saksida T. Carbon monoxide-releasing molecule-A1 inhibits Th1/Th17 and stimulates Th2 differentiation in vitro. Scandinavian journal of immunology. 2014;80:95–100. doi: 10.1111/sji.12189. [DOI] [PubMed] [Google Scholar]
- 43.Fagone P, Mangano K, Quattrocchi C, Motterlini R, Di Marco R, Magro G, Penacho N, Romao CC, Nicoletti F. Prevention of clinical and histological signs of proteolipid protein (PLP)-induced experimental allergic encephalomyelitis (EAE) in mice by the water-soluble carbon monoxide-releasing molecule (CORM)-A1. Clinical and experimental immunology. 2011;163:368–74. doi: 10.1111/j.1365-2249.2010.04303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolic I, Saksida T, Mangano K, Vujicic M, Stojanovic I, Nicoletti F, Stosic-Grujicic S. Pharmacological application of carbon monoxide ameliorates islet-directed autoimmunity in mice via anti-inflammatory and anti-apoptotic effects. Diabetologia. 2014;57:980–90. doi: 10.1007/s00125-014-3170-7. [DOI] [PubMed] [Google Scholar]
- 45.Rook GA, Lowry CA, Raison CL. Microbial 'Old Friends', immunoregulation and stress resilience. Evolution, medicine, and public health. 2013;2013:46–64. doi: 10.1093/emph/eot004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.