Abstract
Adaptive immunity in bacteria involves RNA-guided surveillance complexes that use CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas) proteins together with CRISPR RNAs (crRNAs) to target invasive nucleic acids for degradation. While Type I and Type II CRISPR-Cas surveillance complexes target double-stranded DNA, Type III complexes target single-stranded RNA. Near-atomic resolution cryo-electron microscopy (cryo-EM) reconstructions of native Type III Cmr (CRISPR RAMP module) complexes in the absence and presence of target RNA reveal a helical protein arrangement that positions the crRNA for substrate binding. Thumb-like β-hairpins intercalate between segments of duplexed crRNA:target RNA to facilitate cleavage of the target at 6-nt intervals. The Cmr complex is architecturally similar to the Type I CRISPR-Cascade complex, suggesting divergent evolution of these immune systems from a common ancestor.
Bacteria and archaea defend themselves against infection using adaptive immune systems comprising CRISPR (clustered regularly interspaced short palindromic repeats) arrays and CRISPR-associated (Cas) genes (1). A defining feature of CRISPR-Cas systems is the use of Cas proteins in complex with small CRISPR RNAs (crRNAs) to identify and cleave complementary target sequences in foreign DNA (2, 3). While Type I and Type II CRISPR–Cas systems recognize target sequences in double-helical DNA that is locally unwound to enable DNA target strand cleavage (4, 5), Type III systems bind and cleave single-stranded RNA target sequences (6, 7).
The effector complex of the Type III system from T. thermophilus (Cmr) is a 12-subunit assembly composed of six Cmr subunits (Cmr1–6) and a crRNA with a stoichiometry of Cmr112131445361:crRNA1 (7). The Cmr complex binds to target RNA that is complementary to the bound 40 or 46-nt crRNA and cleaves the target at 6-nt intervals measured from the 5' end of the crRNA sequence (7, 8). Although low-resolution structural studies revealed an overall capsule-like architecture of the Cmr complex (7), the molecular basis of subunit assembly, crRNA binding and ssRNA target recognition and cleavage by the intact surveillance complex remains unknown.
We performed cryo-electron microscopy (cryo-EM) of the intact ~350-kDa Cmr complex in the absence and presence of target ssRNA. We purified endogenous apo-Cmr (containing a crRNA) and used this sample for step-wise assembly with a 56-nt biotinylated ssRNA target followed by purification using streptavidin affinity chromatography. Frozen-hydrated samples of both apo-Cmr and target-bound Cmr were visualized using an FEI Titan Krios microscope equipped with a Gatan K2 Summit direct electron detector. Cryo-EM micrographs of both apo-Cmr and the ssRNA-bound complex showed mono-disperse, easily identifiable particles with sea worm-like features (fig. S1). Using LEGINON (9), we acquired ~7,000 and ~4,000 micrographs and automatically picked ~700,000 and ~300,000 apo- and target-bound Cmr particles, respectively, using Appion (10). After 3D classification and single-particle reconstruction (Supplementary Material and Methods) in RELION (11), we obtained structures of intact apo-Cmr and target-bound Cmr at ~4.1 and 4.4-Å resolution (fig. S1, S2) (using the 0.143 gold standard Fourier Shell Correlation – calculated from two independent half-sets – criterion) from a final set of 250,000 and 175,000 particles, respectively. Additionally, we obtained the structure of a smaller apo-Cmr species revealed during our 3D classification at ~4.4-Å resolution from a second class of ~100,000 particles.
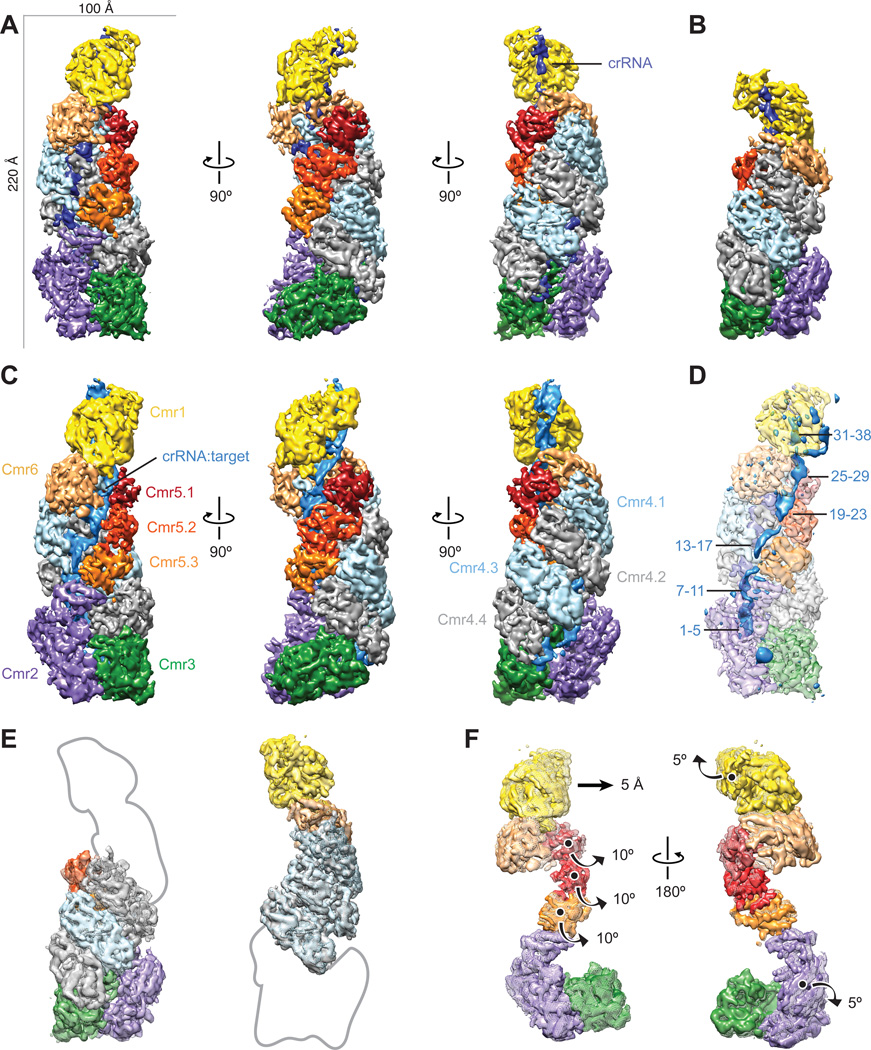
The structure of intact apo-Cmr resembles a capsule in which a central, double helical core of four Cmr4 subunits and three Cmr5 subunits is capped by a Cmr2–Cmr3 heterodimer at one end and Cmr1–Cmr6 at the other (Fig. 1A). The 5’-handle of the crRNA, derived from the CRISPR repeat sequence, is fixed in the Cmr2–Cmr3 heterodimer. An α-helical bundle within Cmr2 makes extensive contacts with the bottom Cmr5 subunit, while the body of Cmr2 engages Cmr3. The architecture of the smaller apo-Cmr species is strikingly similar to that of intact apo-Cmr and maintains the same inter-subunit interactions, despite lacking one Cmr4–Cmr5 subcomplex and being shorter by ~25 Å in the longest direction (Fig. 1B). This apo-Cmr complex likely represents the shorter (40-nt) crRNA-bound species. 3D classification of the target ssRNA-bound Cmr complex showed the sample to be homogeneous, lacking a detectable amount of the smaller complex. There are several segments of additional rod-shaped density along the helical backbone of the complex engaged with target ssRNA that are absent from the apo-Cmr structure (Fig. 1C, D).
Fig. 1. Architecture of the native crRNA-bound (apo) and ssRNA target-bound Cmr.
(A–C) Cryo-EM reconstructions of intact apo-Cmr (crRNA-bound) (A), a smaller apo-Cmr (B), and ssRNA target-bound Cmr (C), at 4.1-, 4.5- and 4.4-Å resolution (using the 0.143 gold standard Fourier Shell Correlation criterion), respectively. Subunits are segmented and colored as indicated. (D) Difference map between intact apo-Cmr and target-bound Cmr at 10-σ (solid, blue) superimposed on the apo-Cmr structure (transparent). (E) Aligning the smaller (surface) and intact (mesh) apo-Cmr complexes based on Cmr2–Cmr3 (left) or Cmr1–Cmr6 (right), shows that the helical geometry is preserved. (F) Aligning apo-Cmr (surface) and target-bound Cmr complexes (transparent mesh) based on the Cmr4 backbone (removed for clarity), reveals this surveillance complex undergoes concerted conformational changes upon ssRNA substrate recognition. Details of these movements can be seen in Movie S1, S2.
To analyze whether the helical geometry in the smaller apo-Cmr structure is perturbed, we aligned the Cmr2–Cmr3 base of both apo-Cmr structures and found that the Cmr4 backbone subunits from this smaller species fit perfectly into their respective subunits in the intact complex (Fig. 1E). Furthermore, when we aligned the two apo-Cmr structures based on the Cmr1–Cmr6 head, the equivalent Cmr4 backbone subunits were again superimposable (Fig. 1E), suggesting that the overall geometry of the complex and the nature of the subunit interactions are preserved in the smaller apo-Cmr structure. To study potential conformational changes that result from ssRNA target binding by the Cmr complex, we aligned the Cmr4 backbone from both structures, whose position remains relatively unchanged (cross-correlation coefficient of 0.92). This superposition shows that the remaining subunits undergo a concerted rearrangement. Upon substrate binding, the Cmr1 and Cmr2 subunits at either end rotate by ~5° in opposite directions (in addition to a 5 Å translation in the head) along axes perpendicular to the long axis of the complex (Fig. 1F). The 3-subunit Cmr5 filament opens (~10° rotation) away from the center of the complex by this ratchet-like motion at the ends (Fig. 1F), exposing the crRNA and forming an elongated channel to accommodate the crRNA:target ssRNA duplex.
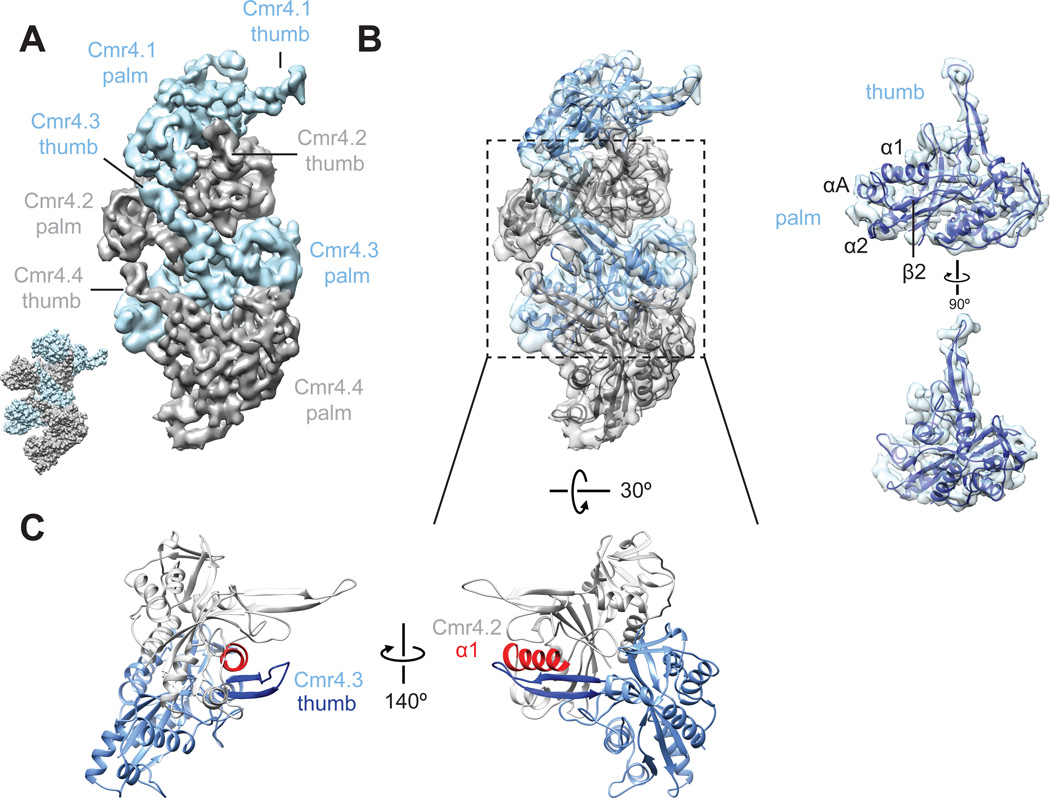
We observed long thumb-like β-strand extensions emerging from the palm of each Cmr4 subunit and engaged with the adjacent subunit (Fig. 2A), a feature that is remarkably reminiscent of the interactions seen in the atomic models of the E. coli (Ec) Type I CRISPR-Cascade DNA targeting complex (12, 13). Docking of the crystal structure of P. furiosus (Pf) Cmr4 (PDB 4Y8W) (14) into the density for each individual Cmr4 subunit of the Cmr4 filament shows that nearly the entire density for the thumb is unaccounted for by this crystal structure. Interestingly, when we aligned the structure of PfCmr4 with the core (RRM) of the structure of Cascade EcCas7 (12) (PDB 1VY9) (RMSD 2.087 Å) (fig. S3), the segments of the structure immediately preceding the unresolved stretch of residues in PfCmr4 (residues 206–227) align well with the segments where the thumb extends from the Cas7 structure (residues 198–217) and fit with the β-strand feature in our cryo-EM density for this subunit (fig. S3). We combined these two sets of atomic coordinates to create a complete, chimeric homology model of Cmr4 and used Rosetta (15) to relax the resulting atomic model into the cryo-EM density (Fig. 2B). This model of an individual Cmr4 subunit can be docked unambiguously into each of the Cmr4 backbone subunits to show a thumb-to-palm interaction network (Fig. 2B) mediated by association of the β-hairpin of the lower Cmr4 with the α1 helix of the neighboring Cmr4 subunit (Fig. 2C).
Fig. 2. Thumb-to-palm interactions between adjacent Cmr4 subunits form the Cmr backbone.
(A) The Cmr4 backbone subunits (other subunits have been removed for clarity), form a helical arrangement by interaction of a thumb-like density from one subunit with the palm of the subunit above. This layout is strikingly similar to the Cas7 subunits (PDB 1VY9) (12) within E. coli Cascade (inset). (B) Homology model of Cmr4 (right) and the Cmr4 helical oligomer (left) shows the Cmr4 thumbs easily accommodate the β-hairpin extension (C) Close-up view of Cmr4.2 and 4.3 showing that the β-hairpin of Cmr4 (dark blue) associates with the α1 helix (red) of the neighboring Cmr4 subunit.
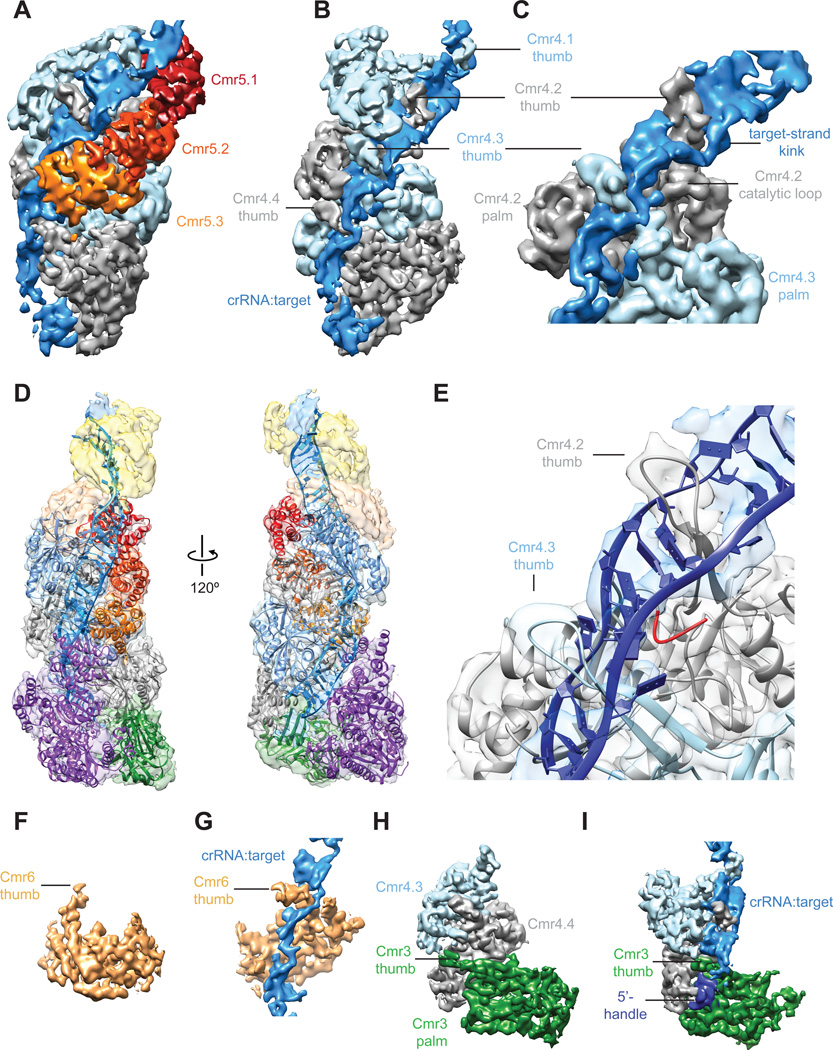
The Cmr4 backbone in the context of the ssRNA target-bound complex shows segments of ~20-Å wide additional density anchored rigidly by the Cmr4 backbone and the other helical array of Cmr5 subunits (Fig. 3A). The thumb-like β-hairpin domains of the Cmr4 filament intercalate between segments of duplexed crRNA:target RNA, distorting the crRNA:target RNA duplex after every 5-bp segment and disrupting the formation of an extended A-form double helix (Fig. 3B). This arrangement places regions of the distorted or kinked ssRNA target in proximity to an adjacent loop density containing several of the catalytic residues (H16 and D27) (14) from the Cmr4 subunit, positioning it for productive cleavage (Fig. 3C). This rearranging of target nucleic acid is strikingly reminiscent of that occurring in Cascade. In fact, the atomic coordinates of the nucleic acid from the ssDNA target-bound Cascade (PDB 4QYZ) (13) can be accommodated easily within this additional density, placing the flipped out base of the target strand near the catalytic loop of Cmr4 (Fig. 3D-E).
Fig. 3. The Cmr4 backbone and Cmr5 subunits position target ssRNA for segmented cleavage.
(A) The channel formed between Cmr4 and Cmr5 backbones creates the binding cleft for target RNA. (B) Cmr4 β-hairpins (thumbs) intercalate between every fifth base pair of duplex crRNA:target RNA, disrupting the helix. (C) Expanded view of (B) shows how the thumb-like domain of the lower Cmr4 positions the kinked target near the catalytic loop. (D) Pseudo-atomic model created by docking available crystal structures of Cmr subunits and our homology model of Cmr4 into the target-bound Cmr structure. (E) Model of target recognition and cleavage shows that the loop (red) of the homology model (presented here) containing catalytic residues H16 and D27 previously identified (14) is positioned near the target. (F, G) The Cmr6 subunit (gold) also contains a long thumb-like extension (F), which disrupts base-paring between the crRNA and target RNA (G) for the 5’-most cleavage event. (H, I) The thumb of the Cmr3 subunit (H) engages the crRNA 5’-handle and bottom target:crRNA segment (I). For a full overview and description of the target-bound Cmr complex see Movie S3 and Supplementary Materials and Methods.
Previously, we showed that the Cmr effector complex cleaves target ssRNAs at five sites in vitro (7), despite containing only four Cmr4 subunits. Reanalyzing our structures, we noticed a thumb-like extension, nearly identical to those observed in individual Cmr4 subunits, originating in Cmr6 (Fig. 3F). In the context of the target-bound Cmr structure, this thumb places the target strand in a position for cleavage of the 5’-most site on the target RNA (Fig. 3G); however, the active site within and mechanism by which the subunit cleaves this end of the target remain to be determined. Similarly, there is a thumb-like domain in Cmr3 that stretches into the palm of the bottom Cmr4 subunit (Fig. 3H), which stacks on top of the 5’-handle and scaffolds the 3’-most discontinuous segment of crRNA:target (Fig. 3I).
Our previous work revealed that the Type I CRISPR-Cascade complex undergoes a concerted rearrangement upon target binding (16, 17). Here, we show that the Type III CRISPR-Cmr complex undergoes an analogous conformational change upon target recognition. While the rearrangement in Cascade most likely permits the docking of the trans-acting Cas3 nuclease, in Type III complexes this rearrangement likely regulates propagation of crRNA:target base-pairing into duplex segments and substrate recognition by the thumbs of Cmr4 subunits. Since the amino acid sequence identity between E. coli Cas7 and T. thermophilus Cmr4 is ~22% and they share conserved structural features (palm and thumb), at least the core of Type I and Type III CRISPR-Cas surveillance complexes likely diverged from a common ancestor. While Type I evolved thumb-like domains for recognition of the non-target strand, Type III repurposed these thumbs for distorting a ssRNA substrate for cleavage. Importantly, we also show here that apo-Cmr complexes of different sizes (corresponding to the presence or absence of one Cmr4–Cmr5) preserve their overall geometry, suggesting that this architecture is used for productive substrate recognition by crRNAs of different sizes. The functional advantage of this built-in plasticity remains to be determined.
These Type I and Type III complexes both use thumb-mediated local disruption of duplex geometry in their interactions with substrate sequences, leading to a lack of continuous double-helix formation between guide RNA and target strands. The fact that RecA employs similar discontiguous DNA-DNA interactions for homology searches (18) hints at a common mode of substrate recognition among CRISPR-Cas surveillance complexes. Although Type II CRISPR-Cas9-RNA-ssDNA crystal structures contain a canonical crRNA-DNA helix (19, 20), crRNA could form a discontinuous helix with one strand of dsDNA targets during sequence interrogation, consistent with tolerance of large 5’ end extensions on the crRNA (21). In the related Type III CRISPR-Csm complex, discontinuous helix formation might occur during association with topologically constrained R-loops formed during transcription (22). Future structural and functional studies will be required to test these target recognition possibilities from this mode of binding by CRISPR-Cmr.
Supplementary Material
One Sentence Summary.
Molecular structures of CRISPR-Cmr complexes show how thumb-like domain intercalation between segments of duplexed crRNA:target RNA mediates target cleavage at 6-nt intervals.
Acknowledgements
The structures of intact apo-Cmr, smaller apo-Cmr, and target-bound Cmr have been deposited into the EMDataBank with accession codes EMD-2898, EMD-2899, and EMD-2900, respectively. We thank R. Louder, S. Howes, E. Kellogg, R. Zhang, P. Grob, Y. He, T. Houweling, Z. Yu and M.J. de la Cruz for expert electron microscopy assistance. D.W.T is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2218-15). R.H.J.S. was supported by the University of Otago’s Division of Health Sciences Career Development postdoctoral fellowship. Y.Z. and J.O. received financial support from the Netherlands Organisation for Scientific Research (NWO), via a Gravitation grant to the Soehngen Institute for Anaerobic Microbiology (024.002.002) and an ALW-TOP project (854.10.003), respectively. This work was supported in part by JSPS KAKENHI Grant Number 25440013 (to A.S.). J.A.D and E.N. are Howard Hughes Medical Institute Investigators.
Footnotes
Supplementary Materials:
Materials and Methods
References and Notes
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staals RH, et al. Structure and Activity of the RNA-Targeting Type III-B CRISPR-Cas Complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson RN, et al. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulepati S, Héroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benda C, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 15.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 19.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimasu H, et al. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 24.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Cocozaki AI, et al. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao Y, et al. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto K, et al. X-ray crystal structure of a CRISPR-associated RAMP module Cmr5 protein from Thermus thermophilus HB8. Proteins. 2009;75:528–532. doi: 10.1002/prot.22358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.