Abstract
Background
Host pathogen relationships can be classified as allopatric, when the pathogens originated from separate, non-overlapping geographic areas from the host; or sympatric, when host and pathogen shared a common ancestral geographic location. It remains unclear if host–pathogen relationships, as defined by phylogenetic lineage, influence clinical outcome. We sought to examine the association between allopatric and sympatric phylogenetic Mycobacterium tuberculosis lineages and pulmonary impairment after tuberculosis (PIAT).
Methods
Pulmonary function tests were performed on patients 16 years of age and older who had received ≥20 weeks of treatment for culture-confirmed M. tuberculosis complex. Forced Expiratory Volume in 1 min (FEV1) ≥80%, Forced Vital Capacity (FVC) ≥80% and FEV1/FVC >70% of predicted were considered normal. Other results defined pulmonary impairment. Spoligotype and 12-locus mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) were used to assign phylogenetic lineage. PIAT severity was compared between host–pathogen relationships which were defined by geography and ethnic population. We used multivariate logistic regression modeling to calculate adjusted odds ratios (aOR) between phylogenetic lineage and PIAT.
Results
Self-reported continental ancestry was correlated with Mycobacterium. tuberculosis lineage (p < 0.001). In multivariate analyses adjusting for phylogenetic lineage, age and smoking, the overall aOR for subjects with allopatric host–pathogen relationships and PIAT was 1.8 (95% confidence interval [CI]: 1.1, 2.9) compared to sympatric relationships. Smoking >30 pack-years was also associated with PIAT (aOR: 3.2; 95% CI: 1.5, 7.2) relative to smoking <1 pack-years.
Conclusions
PIAT frequency and severity varies by host–pathogen relationship and heavy cigarette consumption, but not phylogenetic lineage alone. Patients who had disease resulting from allopatric–host–pathogen relationship were more likely to have PIAT than patients with disease from sympatric–host–pathogen relationship infection. Further study of this association may identify ways that treatment and preventive efforts can be tailored to specific lineages and racial/ethnic populations.
Keywords: Tuberculosis, Pulmonary impairment, Genotypes, Race/ethnicity, Lineage
1. Background
Approximately 2 million persons die annually from tuberculosis (TB) and among those that survive, at least half will experience pulmonary impairment after TB (PIAT) that can be debilitating (Pasipanodya et al., 2007, 2010; Vecino et al., 2011; World Health Organization, 2010). Previous studies have described host, pathogen, environmental, and lifestyle factors associated with outcomes after TB exposure, after latent Mycobacterium tuberculosis infection, or after TB disease (Blaser and Kirschner, 2007; Caws et al., 2008; Coscolla and Gagneux, 2010; Kato-Maeda et al., 2001; Malik and Godfrey-Faussett, 2005; Pasipanodya et al., 2012b; van Crevel et al., 2009). Clinical studies have not demonstrated clear relationships between host genetic susceptibility factors, M. tuberculosis lineages, and TB outcomes (Caws et al., 2008; van Crevel et al., 2009).
PIAT may vary by race/ethnicity (Pasipanodya et al., 2010). Non-Hispanic Whites suffer more frequent and severe PIAT relative to other racial groups. Increased risk for PIAT remained after controlling for cigarette smoking, healthcare access, treatment, and socioeconomic status, suggesting that other factors contributed to PIAT (Pasipanodya et al., 2010). However, pathogen factors including M. tuberculosis phylogenetic differences were not included in these studies. Evidence suggests hosts and pathogens co-evolution and some mycobacterial species, including M. tuberculosis, might adapt to certain human populations (Blaser and Kirschner, 2007; Gagneux, 2012). While the theory of co-evolution remains unproven, molecular characterization of the M. tuberculosis complex has allowed genotypes to be grouped into distinct lineages in a way that correlates with ethnic populations and geography (Blaser and Kirschner, 2007; Coscolla and Gagneux, 2010; Gagneux, 2012). This host–pathogen relationship can be classified as sympatric, when host and pathogen shared a common ancestral geographic location or allopatric, when the pathogens originated from different, non-overlapping geographic areas from the host (Blaser and Kirschner, 2007).
Our study aims to examine whether co-evolution, as determined by host–pathogen relationships, is associated with clinical outcomes in patients treated for pulmonary TB. We hypothesized that persons with pulmonary TB disease caused by sympatric genotypes are less likely to have pulmonary impairment (because of evolutionary coadaptation) as compared to persons with allopatric genotypes.
2. Materials and methods
2.2. Patients and setting
This was a prospective study of patients 16 years of age and older treated for culture-confirmed M. tuberculosis at Tarrant County Public Health (TCPH) during July 2005–December 2010. TCPH has jurisdiction as the health authority for Tarrant County, Texas, an urban county with a population of 1,789,900 persons (Tarrant County Public Health, 2010). TCPH provides subsidized treatment to all persons with TB within this jurisdiction, using universal direct observation of therapy (DOT) delivered to the patient’s preferred location (Tarrant County Public Health, 2010; Weis et al., 1994). Standard treatments for TB and TB/HIV recommended by the American Thoracic Society (ATS) and U.S. Centers for Diseases and Prevention Control (CDC) were used for all patients (Blumberg et al., 2003). Patients with at least three consecutive negative cultures, and who had completed at least 20 weeks of DOT (i.e., non-infectious) were eligible for enrolment. Patients with exclusive extra-pulmonary tuberculosis (i.e., disease outside of the lung parenchyma) or those treated for, or cured of, drug resistant TB were excluded from this study. Ever smokers were patients who gave a history of current or past cigarette smoking. Lifetime cigarette exposure was estimated using pack-years. Similarly, exposure to solid fuel smoke (i.e., biomass fuel exposure) and duration of biomass fuel exposure was compared between groups. Body mass index (BMI) was calculated using patient height and weight at the time of enrolment.
The Institutional Review Board of the University of North Texas Health Science Center at Fort Worth approved the study. All subjects gave written informed consent.
2.3. Pulmonary function testing
Spirometric pulmonary function tests (PFTs) were performed on each patient. Spirometry was conducted according to ATS guidelines for maneuver, technique, and quality control using the Spirotouch® device (Spirotouch®, Spirometry System 086578; Spacelabs Burdick; Deerfield, WI) (Miller et al., 2005). Patients with a history of bronchodilator use received nebulized albuterol 15 min before the test. Results were considered consistent if there was ≤5% variation between results on three separate tests (Miller et al., 2005; Cocchiarella and Andersson, 2000). Once consistency was attained, the best of three results determined pulmonary impairment grading.
An interpretive algorithm from the American Medical Association (AMA), which is consistent with the global initiative for chronic obstructive lung disease (GOLD) classification of chronic obstructive pulmonary disease (COPD), was used to categorize the degree of pulmonary impairment associated with non-normal PFT values (Miller et al., 2005; Cocchiarella and Andersson, 2000). Forced Expiratory Volume in 1 min (FEV1) ≥80%, Forced Vital Capacity (FVC) ≥80% and FEV1/FVC >70% of predicted were considered normal. Other results defined pulmonary impairment. Impairment was categorized as none, mild (if FEV1 or FVC was >60% but <80%), moderate (if FEV1 or FVC was 41–59%) or severe (if FEV1 or FVC was ≤40%). Analyses compared any impairment versus no impairment and examined stratification by grade of impairment.
2.4. Genotyping methods
Mycobacterial isolates were characterized using a standardized protocol for spacer oligotyping (spoligotyping) and 12-locus mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) as part of routine public health program activities at the TCPH through CDC’s National TB Genotyping Service (Centers for Diseases Control and Prevention, 2005; Ghosh et al., 2012).
2.4.1. Genotypic definitions
Four major phylogenetic lineages for M. tuberculosis [East African Indian, East Asian (i.e., Beijing strains), Euro-American, Indo-Oceanic], along with speciation of Mycobacterium africanum, were assigned using a set of rules correlating spoligotype with lineages defined by large sequence polymorphisms (LSP) (Click et al., 2012b; Supply et al., 2006). In cases that did not meet the full requirement for phylogenetic lineage assignment based on spoligotype, 12-locus MIRU-VNTR results was used in addition to spoligotype to assign lineage, as described elsewhere (Click et al., 2012b; Shabbeer et al., 2012). We excluded patients with M. africanum or Mycobacterium bovis isolates.
2.5. Race/ethnicity and ancestral origins
Race was based on a self-reported continental ancestry (i.e., African/Black, European/Caucasian, Asian, Pacific Islander, and Native American) established during interview (Lee et al., 2008; Lin and Kelsey, 2000; Risch and Ziv, 2002; Rosenberg et al., 2002; Zhivotovsky et al., 2003). Africans denote persons whose primary ancestry is sub-Saharan Africa including African-American and Afro-Caribbean. Caucasians includes those with ancestry in Europe and West Asia including the Middle East and North Africa. Hispanics, who represent a recently admixed group between Native Americans, Caucasian and Africans (Baena et al., 2002; Mountain and Risch, 2004; Risch and Ziv, 2002; Zhivotovsky et al., 2003), were considered a separate and distinct group.
2.6. Host–pathogen relationships
A conservative, and more ancient, phylogeographic distribution of M. tuberculosis lineages that precedes recent human global population migrations, first described by Gagneux (2012), were used in this study. Thus, a sympatric host–pathogen relationship was considered if the lineage of the M. tuberculosis isolated from the patient was similar to the designated ancestral geographic origin of the patient. If the person and the lineage of the isolate originated from non-overlapping geographic locale, the host–pathogen relation was defined as allopatric. For example: East Asians infected with the East Asian lineage, European/Caucasians infected with Euro-American lineage, and South East Asians or East Africans infected with the East African Indian lineage were considered sympatric.
2.7. Statistical analysis
PIAT, the primary outcome variable, was dichotomized by combining mild, moderate and severe impairment versus no impairment. Multivariate logistic regression models were built to examine PIAT and host–pathogen relationships. Smoking and age have been are associated with greater pulmonary impairment and, therefore, were included in multivariate models for all patients enrolled in study regardless of statistical significance (Boelaert et al., 2003; Davies et al., 2006; Kerstjens et al., 1997). Additional potential confounders included in the ‘full’ or initial model were sex, body mass index, biomass exposure and lineage. We built the final model by eliminating non-significant independent variables with p-values <0.1 and using the backward likelihood ratio test procedure at each elimination step. The Hosmer–Lemeshow goodness-of-fit was used to examine and compare the models. Separate within lineage multivariate logistic regression analyses were also examined. In view of the small size, particularly for certain lineage groups, to gain power we also dichotomized lineage as ancient versus modern to examine if modern lineages were more ‘aggressive/destructive’ in allopatric relationships. Pearson’s chi-square, symmetric lambda statistic, or Fisher’s exact tests were used to compare proportions wherever appropriate. Analysis of variance (ANOVA) or Kruskall–Wallis tests were used to compare continuous variables. The Mantel–Haenszel test was used to examine the trend across pulmonary impairment levels.
3. Results
3.1. Study participants and phylogenetic lineages
Phylogenetic lineage of the M. tuberculosis-complex isolates collected was determined for 317 (95%) of 332 patients with valid pulmonary function tests, including 311 (98%) M. tuberculosis isolates, 5 (2%) M. bovis isolates and 1 (<1%) M. africanum isolate. Of the 311 M. tuberculosis isolates assigned to lineages, 182 (59%) belonged to the Euro-American lineage, 72 (23%) to the East Asian lineage, 6 (2%) to the East African Indian lineage and 51 (16%) to the Indo-Oceanic lineage (Table 1).
Table 1.
Demographic, Clinical and Epidemiological characteristics of all 211 patients enrolled in the study.
| Characteristic | Total | Host-Pathogen relationship |
||||
|---|---|---|---|---|---|---|
| Demograhic | Variable | All n (%) | Allopatric n=127 (%) |
Sympatric n=184 (%) |
Crude (OR) | P- value |
| Sex | Male | 216 (69) | 78 (61) | 138 (75) | 0.53 (0.33, 0.87) | 0.01 |
| Female | 95 (31) | 49 (39) | 46 (25) | referent | -- | |
| Country of birth | US-Born | 142 (46) | 85 (67) | 57 (31) | 4.51 (2.78, 7.32) | <0.01 |
| Foreign-born | 169 (54) | 42 (33) | 127 (69) | referent | -- | |
| Age groups | <30 | 56 (18) | 19 (15) | 37 (20) | referent | -- |
| (years) | 30 – <40 | 50 (16) | 26 (21) | 24 (13) | 2.11 (0.96, 4.69) | 0.06 |
| 40 – <50 | 65(21) | 30 (24) | 35 (19) | 1.67 (0.80, 3.49) | 0.17 | |
| 50 – <60 | 73 (24) | 32 (25) | 41 (22) | 1.52 (0.74, 3.13) | 0.25 | |
| 60 – <70 | 37 (12) | 14 (11) | 23 (12) | 1.19 (0.50, 2.82) | 0.70 | |
| >70 | 30 (10) | 6 (5) | 24 (13) | 0.49 (0.17, 1.39) | 0.22 | |
| Clinical | ||||||
| Impairment | Some | 154 (50) | 73 (58) | 81 (44) | 1.72 (1.09, 2.71) | 0.02 |
| None | 157 (50) | 54 (42) | 103 (56) | referent | -- | |
| Disease Site | Pulmonary (PTB) only | 292 (94) | 117 (92) | 175 (95) | 0.60 (0.24, 1.53) | 0.28 |
| PTB & EPTB | 19 (6) | 10 (8) | 9 (5) | referent | -- | |
| Smoking Status | Never smokers | 141 (45) | 57 (46) | 84 (46) | 0.97 (0.61, 1.53) | 0.89 |
| Ever smokers | 170 (54) | 70 (54) | 100 (54) | referent | -- | |
| Smoking Volume | <1 | 153 (49) | 62 (49) | 91 (50) | referent | -- |
| (Pack-years) | 1 – 5 | 38 (12) | 18 (14) | 20 (11) | 1.32 (0.65, 2.70) | 0.44 |
| 6 – 15 | 54 (17) | 21 (16) | 33 (18) | 0.93 (0.49, 1.76) | 0.83 | |
| 16 – 29 | 26 (8) | 10 (8) | 16 (9) | 0.92 (0.39, 2.15) | 0.84 | |
| >30 | 40 (13) | 16 (12) | 24 (13) | 0.98 (0.48, 1.99) | 0.95 | |
| HIV status | Negative | 271 (87) | 106(84) | 165 (90) | referent | -- |
| Positive | 31 (10) | 17 (13) | 14 (8) | 1.89 (0.89, 4.00) | 0.09 | |
| Not Done | 9 (3) | 4 (3) | 5(3) | 1.25 (0.33, 4.74) | 0.74 | |
| BMI^ | <18.5 Underweight | 41 (14) | 21 (17) | 20 (12) | 1.46 (0.73, 2.89) | 0.30 |
| 18.5 – <25 (Healthy weight) | 167 (58) | 70 (59) | 97 (57) | referent | -- | |
| 25 – <30 (Over weight) | 62 (22) | 20 (17) | 42 (25) | 0.66 (0.36, 1.22) | 0.18 | |
| >30 (Obese) | 17(6) | 7 (6) | 10 (6) | 0.97 (0.35, 2.67) | 1.00 | |
| Epidemiological | ||||||
| Biomass exposure^^ | None | 185 (74) | 85 (83) | 100 (67) | 2.45 (1.31, 4.57) | |
| Some | 66(26) | 17 (17) | 49 (33) | referent | -- | |
| Lineage | East-Asian | 72 (23) | 46 (36) | 26 (14) | 5.17 (2.34, 11.42) | <0.01 |
| Euro-American | 182 (59) | 68 (54) | 114 (62) | 1.74 (0.87, 3.50) | 0.12 | |
| Indo-Oceanic | 51 (16) | 13 (10) | 38 (21) | referent | -- | |
| East Africa-Indian | 6 (2) | 0 | 6 (3) | 0.22 (0.01, 4.16) | 0.32 | |
OR (95%CI): crude odds ratio (95% confidence interval [CI]);
data available for 293 (94%);
data available for 256 (81%) of patients.
Table 1 shows the demographic and clinical characteristics of all 311 patients enrolled in the study, comprising of 184 (59%) sympatric and 127 (41%) allopatric host–pathogen relationships. Males were significantly less likely to be in allopatric host–pathogen relationships while U.S.-born persons were more likely; crude odds ratios (OR) were 0.5 (95% confidence interval [CI] 0.3, 0.8) and 4.5 (95% CI 2.8, 7.3), respectively. Body mass index, age, and exposure to cigarette smoke and biomass fuels; factors known to influence pulmonary function test results, were not significantly different between host–pathogen relationships.
Among all patients, a significant correlation between self-reported continental ancestry and M. tuberculosis lineage was observed (Lambda = 0.24; p < 0.01). This correlation remained significant when self-reported Hispanics were excluded from analysis (Lambda = 0.26; p < 0.01). Euro-American lineage was the most geographically distributed lineage and was identified in persons from all regions of the world. Eighty percent of the Euro-American isolates were collected from patients born in the United States, Mexico, or Europe. The majority (114/182) of patients with Euro-American lineage isolates identified as European/Caucasian ancestry either directly (i.e., self-reported race/ethnicity) or indirectly (i.e., country of birth) suggesting geographical association between this lineage and infected ethnic populations. However, the majority (67%) of East Asian isolates were collected from patients who did not self-identify as Asian and were born in countries outside East Asia. Hence, relative to the Indo-Oceanic, the East Asian lineage was more likely to be found in allopatric relationship (crude OR: 5.2; 95% CI: 2.3, 11.4). The East African Indian and the Indo-Oceanic lineages were predominantly sympatric, 6/6 (100%) and 38/51 (75%), respectively (Table 1).
3.2. Phylogenetic lineages, host–pathogenic relationships and PIAT
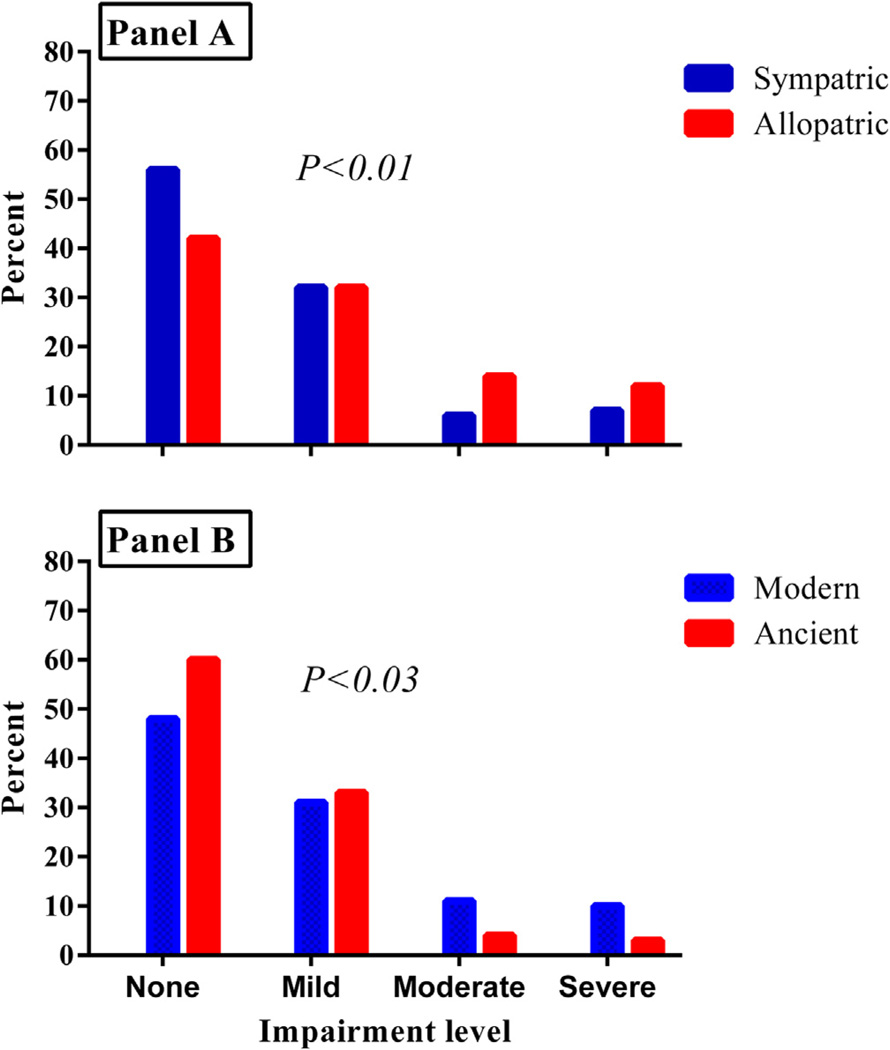
Overall, nearly half (154/311) of the patients had pulmonary impairment of varying levels of severity (Table 1; Fig. 1). There was no significant association between lineage and pulmonary impairment found in univariate (p = 0.13) or multivariate analysis (p = 0.73). As shown in Tables 2 and 3 the associations between host-pathogen relationships and PIAT were variable, but significantly associated among all enrolled subjects and within the East Asian, Euro-American and Indo-Oceanic lineages (Table 3). The adjusted OR for PIAT was 2.1 (95% CI 1.1, 4.1) higher for patients with disease resulting from allopatric versus sympatric infection among patients with Euro-America strains and 6.0 (95% CI 1.5, 23.9) among those with the East Asian lineage. Disease resulting from allopatric infection among patients with Indo-Oceanic strains were inversely associated with PIAT, adjusted OR 0.14 (95% CI 0.02, 0.91) (Table 3). A second multivariate logistic regression model that controlled for age and smoking volume and was restricted to patients with allopatric relationships; relative to the Indo-Oceanic lineage, the East Asian lineage was associated with PIAT and Euro-American lineage was not. The aOR was 5.4 (95% C.I. 1.1, 27.0). On the contrary, both lineages (i.e. East Asian and Euro-American) were not associated with PIAT using an exact multivariate model restricted to patients with sympatric relationships (Data not shown).
Fig. 1.
Panel A shows the comparison of the frequency of host–pathogen relationships across all levels of pulmonary impairment in treated patients. Panel B shows the comparison of the frequency of ancient versus modern Mycobacterium tuberculosis genotypes across all levels of pulmonary impairment.
Table 2.
Factors associated with pulmonary impairment after tuberculosis (PIAT)
| Variable | n | Crude OR (95% CI) |
P-value | Adjusted OR (95% CI) |
P-value | |
|---|---|---|---|---|---|---|
| Lineage | East-Asian | 72 | 2.02 (0.98, 4.17) | 0.06 | 1.29 (0.59, 2.85) | 0.54 |
| Euro-American | 182 | 1.28 (0.69, 2.38) | 0.44 | 1.01 (0.52, 1.93) | 0.99 | |
| Indo-Oceanic | 51 | referent | 0.13 | referent | 0.73 | |
| East Africa-Indian | 6 | 0.34 (0.04, 3.27) | 0.35 | 0.41 (0.04, 4.10) | 0.45 | |
| Host-pathogen relationships | Sympatric | 184 | referent | -- | referent -- | -- |
| Allopatric | 127 | 1.72 (1.09, 2.91) | 0.02 | 1.71 (1.03, 2.84) | 0.04 | |
| Smoking volume: | <1 | 153 | referent | <0.01 | referent-- | 0.04 |
| (Pack-years) | 1 – 5 | 38 | 0.63 (0.30, 1.33) | 0.23 | 0.60 (0.28, 1.28) | 0.19 |
| 6 – 15 | 54 | 1.22 (0.65, 2.27) | 0.54 | 1.14 (0.58, 2.25) | 0.70 | |
| 16 – 29 | 26 | 1.65 (0.72, 3.85) | 0.24 | 1.51 (0.62, 3.69) | 0.36 | |
| >30 | 40 | 3.65 (1.70, 7.99) | <0.01 | 3.06 (1.30, 7.19) | 0.01 | |
| Age group | <30 | 56 | referent | 0.18 | referent-- | 0.82 |
| (years) | 30 – <40 | 50 | 0.97 (0.45, 2.09) | 0.93 | 0.81 (0.36, 1.81) | 0.61 |
| 40 – <50 | 65 | 1.07 (0.52, 2.21) | 0.85 | 0.84 (0.38, 1.83) | 0.65 | |
| 50 – <60 | 73 | 2.02 (1.00, 4.10) | 0.05 | 1.29 (0.59, 2.81) | 0.52 | |
| 60 – <70 | 37 | 1.96 (0.84, 4.54) | 0.12 | 1.17 (0.46, 2.95) | 0.75 | |
| >70 | 30 | 1.17 (0.48, 2.84) | 0.74 | 0.87 (0.33, 2.26) | 0.77 | |
| Sex* | Male | 216 | 1.36 (0.84, 2.21) | 0.22 | ||
| Female | 95 | referent | -- | |||
| BMI^ | 18.5 – <25 (Healthy weight) | 167 | referent | 0.08 | ||
| <18.5 Underweight | 41 | 2.08 (1.01, 4.29) | 0.05 | |||
| 25 – <30 (Over weight) | 62 | 0.74 (0.41, 1.34) | 0.32 | |||
| >30 (Obese) | 17 | 0.68 (0.25, 1.86) | 0.45 | |||
| Biomass Exposure | None | 185 | 1.17 (0.66, 2.05) | 0.59 | ||
| Some | 66 | referent | -- |
OR(95%CI) = Odds ratio (95% Confidence Interval [CI]);
data available for 293 (94%);
Not all variables tested in univariate were included in the initial multivariate model shown; Final results of the stepwise logistic multivariate regression modeling are given in the text.
These variables were not included in multivariate logistic regression.
Hence multivariate analysis model comprised of Age group, Smoking Volume, Lineage and Host-pathogen relationships.
BMI = Body Mass Index (kg/m2).
Figures in bold and italics are statistically significant, p < 0.05.
Table 3.
Within lineage multivariate logistic regression analysis.
| Variable | Mycobacterium tuberculosis lineage | ||
|---|---|---|---|
| Euro-American (n = 182) AOR (95% CI) |
East-Asian (n = 72) AOR (95% CI) |
Indo-Oceanic (n = 51) AOR (95% CI) |
|
| Host–pathogen relationships | |||
| Sympatric | Referent | Referent | Referent |
| Allopatric | 2.09 (1.06, 4.11)a | 5.99 (1.48, 23.86)a | 0.14 (0.02, 0.91)a |
| Smoking volume (pack-years) | |||
| <1 | Referent | Referent | Referent |
| 1–5 | 0.69 (0.25, 1.77) | 0.28 (0.03, 3.09) | 0.61 (0.07, 5.02) |
| 6–15 | 2.13 (0.79, 5.73) | 0.50 (0.12, 2.12) | 0.78 (0.11, 5.56) |
| 16–29 | 2.48 (0.73, 8.44) | 0.26 (0.04, 1.81) | 9.14 (0.43, 195.77) |
| >30 | 4.63 (1.56, 13.74)a | 1.06 (0.17, 6.69) | 0.70 (0.03, 16.28) |
| Age groups (years) | |||
| <30 | Referent | Referent | Referent |
| 30–39 | 1.22 (0.40, 3.74) | 0.83 (0.11, 6.44) | 0.11 (0.01, 1.21) |
| 40–49 | 0.83 (0.29, 2.33) | 0.29 (0.03, 3.12) | 0.41 (0.03, 5.18) |
| 50–59 | 1.85 (0.67, 5.11) | 0.46 (0.06, 3.66) | 0.18 (0.01, 2.42) |
| 60–69 | 1.63 (0.47, 5.67) | 0.72 (0.07, 7.10) | 0.03 (0, 1.30) |
| >70 | 1.61 (0.44, 5.87) | 0.33 (0.04, 2.60) | 0.10 (0, 2.21) |
AOR = Adjusted Odds Ratio.
Figures in italics denotes statistically significance, p < 0.05.
Out of twelve self-identified Asians infected with allopatric Euro-American lineages, 58% (7/12) were impaired and the degree of impairment was moderate-to-severe in 5 (71%). Conversely, among the 27 Asians infected with sympatric East-Asian lineages 41% (11/ 27) were impaired (p < 0.02). Among those impaired only 1 (9%) had moderate-to-severe impairment (p < 0.02). Thus Asians with disease due to allopatric Euro-American lineages were more likely to have pulmonary impairment and it was more severe than subjects with disease due to the sympatric East Asian strains. Similar to Asians, European/Caucasians with disease from infection with allopatric lineages had more frequent and more severe PIAT. Among 19 self-identified European/Caucasians with disease from infection with allopatric East-Asian lineages, 84% (16/19) were impaired, and in 8 (50%) the impairment was moderate-to-severe. Among the 45 European/Caucasians infected with sympatric Euro-American lineages, 64% (29/45) were impaired (p < 0.05). Among those impaired, 11 (38%) had moderate-to-severe impairment (p < 0.05). Of the 14 European/Caucasian non-smokers, 60% (6/10) infected with sympatric versus 100% (4/4) with allopatric were impaired (p = 0.25). Small numbers and high smoking prevalence of lifetime exposure among European/Caucasians precluded attaining statistical significance in univariate analysis (Table 3). Nonetheless, combined (Tables 3 and 4) these results show that the variable distribution of PIAT may be due to host–pathogen relationships.
Table 4.
Distribution of pulmonary impairment among European/Caucasian and Asians infected with sympatric or allopatric lineages by smoking status.
| Pulmonary impairment | |||||
|---|---|---|---|---|---|
| European/Caucasian (n = 64) | Asian (n = 39) | Total | |||
| Sympatric | Allopatric | Sympatric | Allopatric | ||
| Never-smokers | |||||
| None | 4 (40) | 0 | 3 (25) | 5 (45) | 12 (32) |
| Impaired | 6 (60) | 4 (100) | 9 (75) | 6 (55) | 25 (68) |
| Sub-total | 10 (100) | 4 (100) | 12 (100) | 11 (100) | 37 (100) |
| Ever-smokers | |||||
| None | 12 (34) | 3 (20) | 13 (87) | 0 | 28 (44) |
| Impaired | 23 (66) | 12 (80) | 2 (13) | 1 (100) | 38 (56) |
| Sub-total | 35 (100) | 15 (100) | 15 (100) | 1 (100) | 66 (100) |
Lastly, in backward stepwise regression modeling, only smoking and host–pathogen relationships where significantly associated with pulmonary impairment. A potential dose-response trend between smoking and pulmonary impairment was observed (p = 0.02); however, only those who smoked >30 pack-years significantly more likely to have PIAT (aOR: 2.6; 95% CI: 1.1, 6.2). Patients with allopatric host–pathogen relationships had nearly double the odds of PIAT (aOR: 1.8; 95% CI: 1.1, 2.9) compared to patients with sympatric host–pathogen relationships.
4. Discussion
Patients with tuberculosis often have very different clinical outcomes. There are a number of patient factors that influence the clinical presentation and outcome (Perez-Velez and Marais, 2012; Blumberg et al., 2003). However, factors specific to M. tuberculosis and the host pathogen relationship are unclear. We show, for the first time, that patients with pulmonary TB who had disease from organisms with allopatric host–pathogen relationships were more likely to have PIAT. Patients with TB disease from East Asian lineages, who also had allopatric host–pathogen relationships, had six-times greater odds of having PIAT when compared to patients who had sympatric host–pathogen relationships. Similarly among patients with TB disease from Euro-American lineages with allopatric host–pathogen relationships had approximately twice the odds of having PIAT when compared to patients with sympatric host–pathogen relationships. Overall patients with allopatric host–pathogen relations had greater PIAT; however, there was one exception. Patients infected with the Indo-Oceanic lineage, allopatric host–pathogen relationships were inversely associated with PIAT (Table 3). M. tuberculosis lineage alone was not significantly associated with PIAT. Thus, patients who had TB disease resulting from allopatric relationships were more likely to have PIAT than patients with disease from sympatric relationship.
Additionally, PIAT was more likely to be moderate-to-severe in patients with disease from allopatric host–pathogen relationships than sympatric. Patients with disease with a sympatric host– pathogen relationship were more likely to have no impairment. Previously, we showed that PIAT is more frequent and severe in certain risk groups and racial/ethnic populations and also demonstrated that PIAT could be life-long (Pasipanodya et al., 2010, 2012b; Vecino et al., 2011). In these studies, self-reported non-Hispanic white was associated with PIAT, while various measures of socio-economic status were not. Analysis of data of self-identified European/Caucasians and Asians infected with either Euro-American or East-Asian lineages demonstrated that an increased frequency of allopatric host–pathogen relationships explained the racial/ethnic differences in PIAT distribution that we reported earlier. Thus, the Whites were more likely to be infected with East-Asian genotypes which were allopatric and associated with severity of PIAT (Pasipanodya et al., 2012b).
Our finding on the effect of allopatric sympatric relationship and PIAT, rather than lineage per se, adds to the understanding of the disproportionate racial/ethnic distribution noted by others across the TB disease spectrum (Stead et al., 1990; Long and Jablon, 1955; Crowle and Elkins, 1990; Pasipanodya et al., 2012b). These data contribute to difference in pulmonary impairment among TB survivors. Previously described racial/ethnic differences were limited to: susceptibility to infection, progression to disease after infection, and mortality after TB disease (Stead et al., 1990; Long and Jablon, 1955; Crowle and Elkins, 1990). In a study of long-term care patients in the United States, Stead and others concluded that Blacks were twice as likely to become infected compared to White patients (Stead et al., 1990). These findings were consistent with an earlier study of U.S. soldiers, which revealed a higher incidence of progressive primary tuberculosis after infection among Blacks (4.2%) than among Non-Hispanic Whites (2.8%) under comparable environmental conditions (Long and Jablon, 1955). Crowle and Elkins demonstrated that macrophages inoculated with the same M. tuberculosis strain from Blacks were more ‘permissive’ of tuberculosis infection as compared to macrophages collected from Whites (Crowle and Elkins, 1990). In all three studies, pathogen genotypes were not identified; however, it is likely that Euro-American lineages were prevalent among the US population (Stead, 2001). Nonetheless, these data support the notion that under comparable conditions, TB outcomes after infection or treatment can be affected by host immune response genes; many of these genes can be segregated according continental ancestry (Baena et al., 2002; Zhivotovsky et al., 2003). We suspect that polymorphic gene variants responsible for TB immune function or those involved in xenobiotic metabolism, may differ significantly between racial/continental ancestries (Gumbo et al., 2007; Pasipanodya et al., 2012a;), and thus potentially contribute to PIAT. These findings suggest that difference in TB outcomes between ethnic/geographically defined populations groups might have a biological as well as evolutionary basis.
Among subjects with East Asian TB lineages (Beijing strains), persons with allopatric host–pathogen relationships had six times greater odds of having PIAT compared to those with sympatric ones (Table 3). This finding could be partly explained in terms of immunogenicity and the particular lineage strain ability to evade host innate immune system. We hypothesize that both emergent properties are acquired over time in the fight for survival between host and pathogen. Interestingly, some East Asian lineage genotypes, such as HN878, W4 and W10 genotypes, have been shown to possess an intact polyketide synthase (pks 1/15) gene involved in the production of phenolic glycolipid (PGL) (Caws et al., 2008; Parwati et al., 2010). PGL which has been identified in other Euro-American strains, has been linked to hypervirulence phenotypes in laboratory studies that used macrophages and animal models (Coscolla and Gagneux, 2010; Malik and Godfrey-Faussett, 2005). Furthermore, East Asian isolates have been associated with more severe cavitary disease (Cowley et al., 2008; Parwati et al., 2010; van Crevel et al., 2009). Combined, our data provide evidence to demonstrate that ‘modern’ lineages particularly East Asian and Euro-America strains evolved non-lethal but aggressive/destructive properties in the lungs which enhanced intergenerational propagation. The fact that PIAT was more likely to be encountered and severe in allopatric rather than across all populations suggests more recent acquisition of this property.
An unexpected finding was the protective effect of allopatric relationship against PIAT among patients infected with “ancient” Indo-Oceanic lineages. These findings are consistent with several prior observations (Mitchison et al., 1960; Goren et al., 1974; Portevin et al., 2011; Krishnan et al., 2011). M. tuberculosis from southern India, the geographic area associated Indo-Oceanic lineages, were shown more than 50 years ago to be less virulent in guinea pigs than those collect in the United Kingdom strains (Mitchison et al., 1960). We speculate that aggressive/destructive lung damage with which we used PFT as a measure might not be an appropriate measure for virulent among “ancient” strains such as the Ino-Oceanic lineage. Regardless, our data adds to the growing evidence that virulence and host immune response vary considerably between hosts and among different M. tuberculosis lineages and should be considered in design of better TB vaccines (Adams et al., 2011; Brosch et al., 2002; Cowley et al., 2008; Domenech et al., 2005; Homolka et al., 2010; Krishnan et al., 2011; Lamichhane et al., 2005; Parwati et al., 2010; Portevin et al., 2011).
In the United States, spoligotyping and MIRU-VNTR are routinely performed as part of national surveillance (Ghosh et al., 2012). Our data suggest that mycobacterial genotype combined with patients’ continental ancestry may allow clinicians and TB programs to identify those at highest risk for PIAT. Indeed, there is an association between phylogenetic lineage and clinical presentation and time-to-culture conversion in patients receiving standard anti-TB therapy (Click et al., 2012b,a; Nahid et al., 2010). There are known inter-ethnic differences in the allele frequencies of enzymes responsible for metabolizing TB drugs that contributes to variable drug response (Pasipanodya et al., 2012a). Further study of these associations may identify appropriate treatments that can be tailored to specific lineages.
In our cohort, there were two variables independently associated with PIAT in multivariate analysis: cigarette smoking and allopatric host–pathogen relationships. Our data showed that patients smoking >30 pack-years were three times as likely to be associated with PIAT as compared to patients with smoking <1 pack-years after adjusting for lineage and age. This finding was expected. Smoking is well established to cause decreased pulmonary function (Boelaert et al., 2003; Davies et al., 2006; Kerstjens et al., 1997). Both biomass smoke and cigarette smoke are believed to induce functional and morphological changes in host alveolar macrophages thereby weakening macrophages through sustained alveolar inflammation, and facilitating M. tuberculosis persistence (Boelaert et al., 2003; Davies et al., 2006). Education about the role of cigarette smoking, its relationship to PIAT, and its potential for worsening PIAT should be part of TB treatment.
The study has several limitations. First, race/ethnicity is a social construct that varies with time, geography, and has limited taxonomic significance in biology; despite several studies showing that genetic clusters correspond closely with groups that self-identify by race/ethnicity or continental ancestry (Lee et al., 2008; Lin and Kelsey, 2000; Mountain and Risch, 2004; Risch and Ziv, 2002; Rosenberg et al., 2002; Zhivotovsky et al., 2003). Indeed, several human population studies have shown that genetic differentiation is greatest when populations are defined based on continental ancestry, and that self-description of ancestry is adequately discriminatory (Lin and Kelsey, 2000; Rosenberg et al., 2002; Zhivotovsky et al., 2003). Second, admixture among racial/ethnic groups might add misclassification bias. Even though mixed race/ethnicity was rare among persons who self-identified European/Caucasian ancestry, Asian or Pacific Islanders, the Hispanic and African American population in the United States has a heterogeneous ethnic ancestry comprising of Native American Indians, European and African origins. Thus, these findings are important in generating hypotheses for additional genome-wide association studies that examine human immunologic markers more strongly associated with area of geographic origin. Third, spirometry is a simple, inexpensive and noninvasive tool used widely to measure pulmonary functional impairment; however, it is not disease specific, difficult to interpret and subject to misclassification bias. Moreover, TB has been known to cause both obstructive and restrictive pulmonary impairment. The distribution of either impairment pattern between populations is affected by comorbid conditions, environmental conditions as well as several other factors not examined in this study. As such, we may have underestimated mildly to moderately restrictive disease among patients, as spirometry does not assess total lung capacity (Miller et al., 2005; Cocchiarella and Andersson, 2000). Finally, even though we have demonstrated significant variation of pulmonary impairment between allopatric versus sympatric host–pathogen groups among treated TB patients, it remains to be determined how much of this variation can be attributed to initial racial/ethnic population difference prior to TB. Nevertheless, as pulmonary function has been shown to predict premature death, these findings can be used in future genome-wide studies to identify actual host and pathogen genes associated with PIAT.
5. Conclusion
Lineages per se were not associated with PIAT; however, the sample size was not sufficient to state this for all lineages and patient groups. Allopatric host–pathogen relationships and certain life-style risk factors were associated with variable odds for pulmonary impairment among geographically defined ethnic populations. We concluded that allopatric host–pathogen relationships explained the disproportionate distribution of PIAT by race/ethnicity that we previously reported. The cross-sectional nature of the analysis did not make it possible to determine proportion of variation attributable to population difference in pulmonary function which is known to segregate by continental ancestry. Further study of this association may identify ways that treatment and preventive efforts can be tailored to specific lineages and race/ethnic populations to decrease the risk of PIAT.
Acknowledgements
This work was made possible by the cooperation of the Tarrant County Public Health Department and the University of North Texas Health Science Center at Fort Worth, as well as both the U.S. Centers for Disease Control and Prevention. This research did not receive direct funding from these or other sources; however, the Tuberculosis Epidemiologic Studies Consortium and the Tuberculosis Trials Consortium of the CDC provided salary support for Drs. Fernandez, Miller, Pasipanodya, Vecino, and Weis, and Ms. Drewyer, although neither consortium directly funded this study nor had any role in study design, data collection, data analysis, data interpretation, nor writing this report. We thank Drs. Juliana Grant, Michael Iademarco, Phil LoBue, Eugene McCray, and Tom Navin for their thoughtful review and helpful comments to the manuscript.
Footnotes
The authors have no significant conflicts of interest with any companies or organizations whose products or services maybe discussed in this article.
References
- Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena A, Leung JY, Sullivan AD, et al. TNF-alpha promoter single nucleotide polymorphisms are markers of human ancestry. Gene. Immun. 2002;3(8):482–487. doi: 10.1038/sj.gene.6363898. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449(7164):843–849. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- Blumberg HM, Burman WJ, Chaisson RE, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 2003;167(4):603–662. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- Boelaert JR, Gomes MS, Gordeuk VR. Smoking, iron, and tuberculosis. Lancet. 2003;362(9391):1243–1244. doi: 10.1016/S0140-6736(03)14529-1. [DOI] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caws M, Thwaites G, Dunstan S, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4(3):e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Diseases Control and Prevention (CDC) New CDC program for rapid genotyping of mycobacterium tuberculosis isolates. MMWR Morb. Mortal. Wkly. Rep. 2005;54(2):47. [Google Scholar]
- Click ES, Winston CA, Oeltman JE, Moonan PK, Cowan KS, Mac Kenzie WR. Association between Mycobacterium tuberculosis lineage and time to sputum culture conversion. Am. J. Respir. Crit. Care Med. 2012a;185:A3311. doi: 10.5588/ijtld.12.0732. [DOI] [PubMed] [Google Scholar]
- Click ES, Moonan PK, Winston CA, Cowan LS, Oeltmann JE. Relationship between Mycobacterium tuberculosis phylogenetic lineage and clinical site of tuberculosis. Clin. Infect. Dis. 2012b;54(2):211–219. doi: 10.1093/cid/cir788. [DOI] [PubMed] [Google Scholar]
- Cocchiarella L, Andersson GBJ. The Respiratory System. In: Cocchiarella L, Andersson GBJ, editors. Guides to the Evaluation of Permanent Impairment. 5th ed. Chicago: AMA Press; 2000. [Google Scholar]
- Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov. Today Dis. Mech. 2010;7(1):e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D, Govender D, February B, et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin. Infect. Dis. 2008;47(10):1252–1259. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- Crowle AJ, Elkins N. Relative permissiveness of macrophages from black and white people for virulent tubercle bacilli. Infect. Immun. 1990;58(3):632–638. doi: 10.1128/iai.58.3.632-638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PD, Yew WW, Ganguly D, et al. Smoking and tuberculosis: the epidemiological association and immunopathogenesis. Trans. R. Soc. Trop. Med. Hyg. 2006;100(4):291–298. doi: 10.1016/j.trstmh.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Barry CE., 3rd Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S. Host-pathogen coevolution in human tuberculosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367(1590):850. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Moonan PK, Cowan L, Grant J, Kammerer S, Navin TR. Tuberculosis genotyping information management system: enhancing tuberculosis surveillance in the United States. Infect. Genet. Evol. 2012;12(4):782–788. doi: 10.1016/j.meegid.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: phthiocerol dimycocerosate and the attenuation indicator lipid. Infect. Immun. 1974;9:150–158. doi: 10.1128/iai.9.1.150-158.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbo T, Louie A, Lui W, Brown D, Ambrose PG, Bhavnani SM, Drusano GL. Isoniazid bacterial activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics ro predict efficacy in different ethnic populations. Antimcrob. Agents Chemother. 2007;51(7):2329–2336. doi: 10.1128/AAC.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Maeda M, Bifani PJ, Kreiswirth BN, Small PM. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J. Clin. Invest. 2001;107(5):533–537. doi: 10.1172/JCI11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstjens HA, Rijcken B, Schouten JP, Postma DS. Decline of FEV1 by age and smoking status: facts, figures, and fallacies. Thorax. 1997;52(9):820–827. doi: 10.1136/thx.52.9.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Malaga W, Constant P, Caws M, Tran TH, et al. Mycobacterium tuberculosis lineage influences innate immune response and virulence and is associated with distinct cell envelope lipid profiles. PLoS One. 2011;6:e23870. doi: 10.1371/journal.pone.0023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun. 2005;73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Mountain J, Koenig B, et al. The ethics of characterizing difference. guiding principles on using racial categories in human genetics. Genome Biol. 2008;9(7):404. doi: 10.1186/gb-2008-9-7-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Kelsey JL. Use of race and ethnicity in epidemiologic research: concepts, methodological issues, and suggestions for research. Epidemiol. Rev. 2000;22(2):187–202. doi: 10.1093/oxfordjournals.epirev.a018032. [DOI] [PubMed] [Google Scholar]
- Long ER, Jablon S. Veterans Administration Medical Monograph. Washington, DC.: Veternas Administration; 1955. Tuberculosis in the army of the United States in World War II: An Epidemiological Study with an Evaluation of X-ray Screening. [Google Scholar]
- Malik AN, Godfrey-Faussett P. Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect. Dis. 2005;5(3):174–183. doi: 10.1016/S1473-3099(05)01310-1. [DOI] [PubMed] [Google Scholar]
- Miller MR, Crapo R, Hankinson J, et al. General considerations for lung function testing. Eur. Respir J. 2005;26(1):153–161. doi: 10.1183/09031936.05.00034505. [DOI] [PubMed] [Google Scholar]
- Mitchison DA, Wallace JG, Bhatia AL, Selkon JB, Subbaiah TV, et al. A comparison of the virulence in guinea-pigs of South Indian and British tubercle bacilli. Tubercle. 1960;41:1–22. doi: 10.1016/s0041-3879(60)80019-0. [DOI] [PubMed] [Google Scholar]
- Mountain JL, Risch N. Assessing genetic contributions to phenotypic differences among ‘racial’ and ‘ethnic’ groups. Nat. Genet. 2004;36(Suppl. 11):S48–S53. doi: 10.1038/ng1456. [DOI] [PubMed] [Google Scholar]
- Nahid P, Bliven EE, Kim EY, MacKenzie WR, Stout JE, et al. Influence of M. tuberculosis lineage variability within a clinical trial for pulmonary tuberculosis. PLoS One. 2010;5(5):e10753. doi: 10.1371/journal.pone.0010753. http://dx.doi.org/10.1371/journal.pone.0010753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwati I, Van CR, Van SD. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- Pasipanodya JG, Miller TL, Vecino M, et al. Pulmonary impairment after tuberculosis. Chest. 2007;131(6):1817–1824. doi: 10.1378/chest.06-2949. [DOI] [PubMed] [Google Scholar]
- Pasipanodya JG, McNabb SJ, Hilsenrath P, et al. Pulmonary impairment after tuberculosis and its contribution to TB burden. BMC Public Health. 2010;10:259. doi: 10.1186/1471-2458-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasipanodya J, Vecino E, Miller TL, et al. Non-Hispanic Whites have higher risk for pulmonary impairment from pulmonary tuberculosis. BMC Public Health. 2012a;12(119) doi: 10.1186/1471-2458-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasipanodya JG, Shashikant S, Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of anti-tuberculosis therapy. Clin. Infect. Dis. 2012b;55(2):169–177. doi: 10.1093/cid/cis353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Velez CM, Marais BJ. Tuberculosis in children. NEJM. 2012;367(4):348–361. doi: 10.1056/NEJMra1008049. [DOI] [PubMed] [Google Scholar]
- Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-comment2007. comment 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Shabbeer A, Cowan LS, Ozcaglar C, et al. TB-Lineage: an online tool for classification and analysis of strains of Mycobacterium tuberculosis complex. Infect. Genet. Evol. 2012;12(4):789–797. doi: 10.1016/j.meegid.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Stead WW. Variation in vulnerability to tuberculosis in America today: random, or legacies of different ancestral epidemics? Int. J. Tuberc. Lung Dis. 2001;5(9):807–814. [PubMed] [Google Scholar]
- Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N. Engl. J. Med. 1990;322(7):422–427. doi: 10.1056/NEJM199002153220702. [DOI] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44(12):4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant County Public Health Department. Division of Tuberculosis Elimination, Tarrant County Public Health, Client Services. 2010 [Google Scholar]
- van Crevel R, Parwati I, Sahiratmadja E, et al. Infection with Mycobacterium tuberculosis Beijing genotype strains is associated with polymorphisms in SLC11A1/NRAMP1 in Indonesian patients with tuberculosis. J. Infect. Dis. 2009;200(11):1671–1674. doi: 10.1086/648477. [DOI] [PubMed] [Google Scholar]
- Vecino M, Pasipanodya JG, Slocum P, et al. Evidence for chronic lung impairment in patients treated for pulmonary tuberculosis. J. Infect. Public Health. 2011;4(5–6):244–252. doi: 10.1016/j.jiph.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Weis SE, Slocum PC, Blais FX, et al. The effect of directly observed therapy on the rates of drug resistance and relapse in tuberculosis. N. Engl. J. Med. 1994;330(17):1179–1184. doi: 10.1056/NEJM199404283301702. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Global Tuberculosis Control: WHO, Report. Geneva, Switzerland: 2010. [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am. J. Hum. Genet. 2003;72(5):1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]