Abstract
Objective
We evaluated the performance of the GS fourth-generation antigen/antibody assay and compared CDC’s proposed alternative algorithm (repeatedly reactive [RR] fourth-generation immunoassay [IA] followed by an HIV-1/HIV-2 differentiation IA and, if needed, nucleic acid testing [NAT]) with the current algorithm (RR third-generation IA followed by HIV-1 Western blot [WB]).
Design
A convenience sample of the following four specimen sets was acquired: 10,014 from insurance applicants, 493 known WB-positive, 20 known WB-indeterminate specimens, and 230 specimens from 26 HIV-1 seroconverters.
Methods
Specimens were tested with the GS third- and fourth-generation IAs, the Multispot HIV-1/HIV-2 differentiation IA, NAT, and WB. We applied the two algorithms using these results.
Results
Among insurance specimens, 13 (0.13%) specimens were IA RR: 2 were HIV-positive (RR by third- and fourth-generation IAs, and WB and Multispot positive); 2 third-generation RR and 9 fourth-generation RR specimens were false-positive. Third- and fourth-generation specificities were 99.98% (95%CI: 99.93%–100%) and 99.91% (95%CI: 99.84%–99.96%) respectively.
All HIV-1 WB-positive specimens were RR by third- and fourth-generation IAs. By Multispot, 491 (99.6%) were HIV-1 positive and 2 (0.4%) were HIV-2 positive.
Only eight (40%) WB-indeterminate specimens were fourth-generation RR: 6 were Multispot and NAT negative and 2 were Multispot HIV-1 positive but NAT negative.
The alternative algorithm correctly classified as positive 102 seroconverter specimens with the third-generation IA and 130 with the fourth-generation IA compared with 56 using the WB with either IA.
Conclusions
The alternative testing algorithm improved early infection sensitivity and identified HIV-2 infections. Two potential false-positive algorithm results occurred with WB-indeterminate specimens.
Keywords: Fourth-generation immunoassay, HIV testing algorithms, Specificity
INTRODUCTION
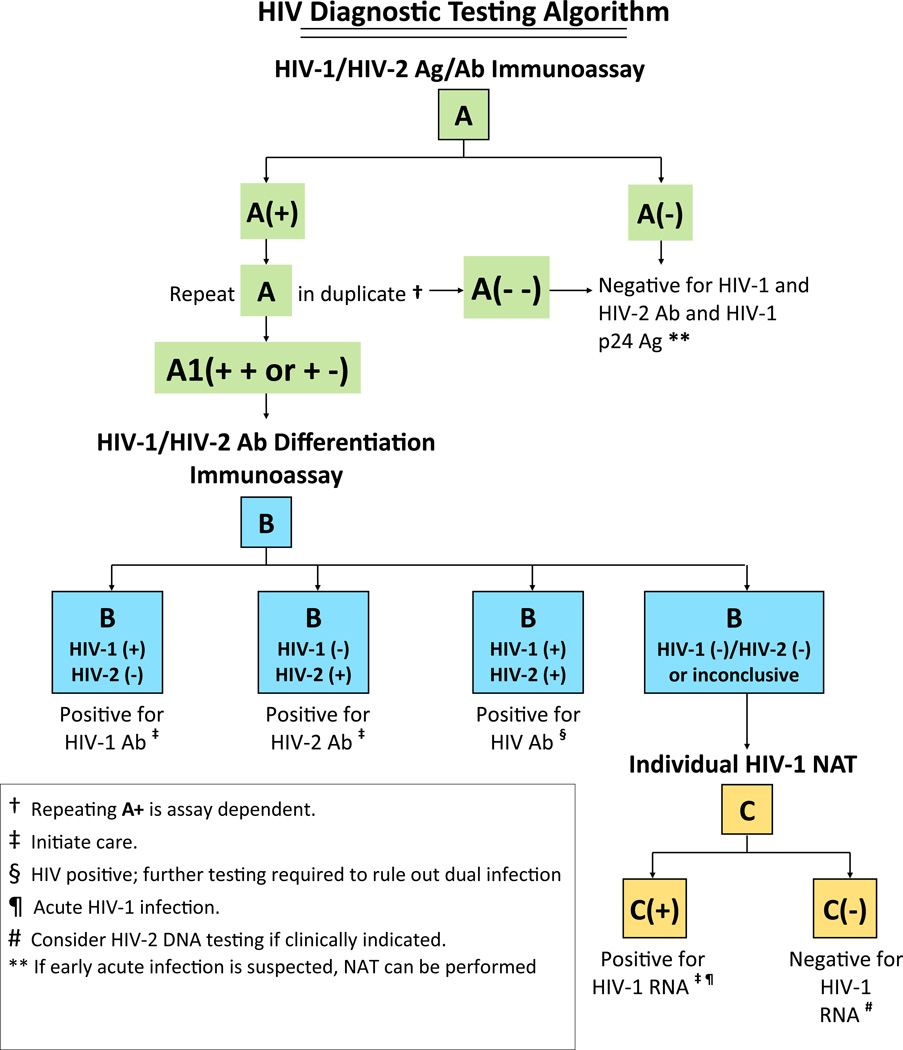
The current HIV testing algorithm, which was recommended by the Centers for Disease Control and Prevention (CDC) in 1989, indicates “no positive test results should be given to clients/patients until a screening immunoassay (IA) has been repeatedly reactive (RR) on the same specimen and a supplemental, more specific test such as the Western blot (WB) has been used to validate those results.”1 The WB detects anti-HIV antibody in a human serum sample infected with HIV; however, it cannot detect acute infections (period prior to detectable antibody) which have been associated with a higher probability of disease transmission compared with established infections.2–4 The HIV-1 WB also misclassifies many HIV-2 infections as HIV-1, which is problematic because HIV-2 infections do not respond to many first-line antiretroviral agents, including non-nucleoside reverse transcriptase inhibitors and some protease inhibitors.5 In 2010 an alternative laboratory HIV diagnostic testing algorithm was proposed6 (Figure 1) that is designed to detect early infections, reduce indeterminate results, and identify HIV-2 infections.7–10 The alternative diagnostic algorithm involves screening with a sensitive fourth-generation antigen/antibody HIV-1/2 IA, or if unavailable, a third-generation HIV-1/2 IA. When the screening IA is repeatedly reactive, it is followed with an HIV-1/HIV-2 antibody differentiation test. If the differentiation test is reactive, the result is positive for either HIV-1 or 2 antibodies or both. However, when the HIV antibody differentiation test results are negative, an HIV-1 nucleic acid test (NAT) is used to resolve infection status. Persons with a positive NAT and a negative differentiation test are considered to have acute HIV-1 infection.
Figure 1.
Alternative laboratory HIV diagnostic testing algorithm
To date, HIV NAT has not been used widely for diagnosis due to its labor requirements, cost, and uncertainty about whether acute infections would be identified in certain populations.4, 11 Recently, the Food and Drug Administration (FDA) approved fourth-generation IAs that detect p24 antigen and HIV-1 and HIV-2 antibodies.12 These assays have the ability to detect more than 80% of acute HIV infections otherwise detectable only by NAT.13–15 The commercial availability of fourth-generation HIV-1/2 assays will make simultaneous screening for both acute and established HIV infections feasible for most clinical laboratories. However, the specificity of these screening tests must be evaluated in low prevalence settings because of cost implications associated with NAT to resolve false-positive fourth-generation IA screening test results. In this study we evaluated the performance of the FDA-approved fourth-generation assay, the GS HIV Combo Ag/Ab IA (Bio-Rad Laboratories, Redmond, WA),16 as part of the alternative laboratory HIV diagnostic testing algorithm compared to the current algorithm (RR third-generation IA/ HIV-1 WB). The evaluation was conducted using specimens from a low prevalence population, persons with established infections, and seroconverters.
METHODS
Specimens
Quest Diagnostics obtained three sets of de-identified residual serum/plasma specimens and processed them at their Lenexa, Kansas facility: (1) 10,014 specimens from life insurance applicants, a population that typically has low HIV prevalence (<0.1%)17; (2) 493 previously tested GS HIV-1 WB-positive specimens from life insurance applicants; (3) 20 previously tested GS HIV-1 WB-indeterminate specimens submitted for diagnostic testing. In addition, CDC obtained 230 serial plasma specimens from 26 U.S. donors early in the process of HIV seroconversion (presumably infected with subtype B) from BBI-SeraCare Diagnostics (West Bridgewater, MA) and Zeptometrix, Inc. (Buffalo, NY).
Assays
At Quest Diagnostics, all specimens (except those from the HIV seroconversion panels) were tested according to package insert instructions with both the GS HIV-1/HIV-2 Plus O third-generation IA18 and the GS HIV Combo Ag/Ab fourth-generation IA (Bio-Rad Laboratories, Redmond, WA) using the EVOLIS Automated Microplate System (Bio-Rad Laboratories). Specimens that were initially reactive with the third- or fourth-generation IA were tested in duplicate on the same assay, and specimens reactive on one or both of the repeat tests were considered to be repeatedly reactive. Specimens repeatedly reactive (RR) by third or fourth generation IA were tested with a rapid HIV-1/HIV-2 differentiation assay, Multispot HIV-1/HIV-2 Rapid test (Bio-Rad Laboratories, Redmond, WA),19 and if negative, by the APTIMA HIV-1 RNA Qualitative assay (Gen-Probe Incorporated, San Diego, CA).20 Specimens with HIV-2 positive results on the differentiation assay were also tested with HIV-2 IA (Bio-Rad Laboratories, Redmond, WA) and research use only (RUO) QualiCode™ HIV-1/2 Western Blot Kit (QualiCode, Immunetics, Boston, MA).
At the CDC, specimens from the HIV seroconversion panels were tested manually by GS third- and fourth-generation IAs in singlet, Multispot and APTIMA. Because all specimens used in this study were unlinked from personal identifiers, this study was determined by the CDC to be research not involving identifiable human subjects.
Data management and analysis
Test results generated at Quest Diagnostics were recorded in an Excel database that was subsequently provided to CDC via secure data network. For specimens from low HIV prevalence life insurance applicants, we calculated the specificities of third- and fourth-generation IAs and used the Mid-P Exact Test to calculate 95% confidence intervals (CI). For WB-positive and WB-indeterminate specimens, we compared the results of the current and alternative laboratory HIV diagnostic testing algorithms. Samples from all specimen sets were considered to be from HIV infected individuals if they were either WB or NAT positive. Analyses were performed using SAS 9.2 (SAS Institute, Inc., Cary, NC, USA) statistical software.
For seroconversion panels, the analyses were conducted as described previously.7 Differences in the number of HIV-1 infections detected by the current and alternative laboratory HIV diagnostic algorithms were statistically analyzed using McNemar’s test with one degree of freedom and continuity correction.
RESULTS
Specimens from a Low Prevalence Population
Thirteen (0.13%) of 10,014 specimens from life insurance applicants were RR on at least one third- or fourth-generation IA (Table 1). Of these, two specimens RR by both third- and fourth-generation IAs were HIV-1 positive by WB and Multispot (i.e., considered HIV-1 positive by both algorithms). Two third-generation IA and 9 fourth-generation IA results were false-positive based on available testing data (Table 1): one specimen RR only by third-generation IA and 8 RR only by fourth-generation IA were negative by WB, Multispot and NAT; one specimen RR only by third-generation IA and another RR only by fourth-generation IA were WB-indeterminate and negative by Multispot and NAT. Specificities of third-generation (10,010/10,012) and fourth-generation IAs (10,003/10,012) were 99.98% (95% CI: 99.93–100%) and 99.91% (95% CI: 99.84–99.96%), respectively. Specificity values for both assays were within the confidence bounds reported within the product package inserts for low risk populations. No acute HIV infections were identified among specimens from these life insurance applicants.
Table 1.
HIV-1 Western blot, Multispot, and nucleic acid test (NAT) results of specimens repeatedly reactive (RR) by third- or fourth-generation immunoassays in a low prevalence population (10,014 insurance applicants)
| Number of specimens |
Third-generation RR |
Fourth-generation RR |
Western blot | Multispot | Nucleic acid test |
|---|---|---|---|---|---|
| 2†† | Yes | Yes | Positive | Positive | Not Done |
| 1§ | Yes | No | Negative | Negative | Negative |
| 8† | No | Yes | Negative | Negative | Negative |
| 1§ | Yes | No | Indeterminate | Negative | Negative |
| 1† | No | Yes | Indeterminate | Negative | Negative |
HIV-1 uninfected, false-positive on third-generation IA;
HIV-1 uninfected, false-positive on fourth-generation IA;
HIV-1 positive
HIV-1 Western blot-Positive Specimens
All 493 HIV-1 WB-positive specimens were third- and fourth-generation RR (i.e., classified HIV-1 positive by current algorithm). With the alternative algorithm, 491 (99.6%) were HIV-1 positive by Multispot and 2 (0.4%) were HIV-2 positive by Multispot, HIV-2 IA, and HIV-2 WB.
HIV-1 Western blot-Indeterminate Specimens
Twelve (60%) of the 20 HIV-1 WB-indeterminate specimens were RR by third-generation IA, but not by fourth-generation IA, and were negative by Multispot and NAT (i.e., classified as HIV-negative by the alternative diagnostic testing algorithm). Eight (40%) of the 20 WB-indeterminate specimens were RR by third- and fourth-generation IA. Of these, six were negative by Multispot and NAT (i.e., classified as negative by the alternative algorithm), and 2 were reactive by Multispot (i.e., classified as antibody-positive by the alternative algorithm) but NAT-negative. One of these two HIV-1 WB-indeterminate specimens had p24 and p55 WB bands present, and the other had a gp160 band.
Specimens from HIV-1 Seroconverters
The third -generation IA was reactive with 102 seroconverter specimens and the fourth -generation IA with 131 (Table 2a). The use of Multispot as a supplemental test improved the correct classification of HIV-1 infections compared with the WB regardless of the screening IA used. A total of 90 specimens were positive using Multispot as the supplemental test, whereas only 56 were confirmed positive by the WB (Table 2a). Two seroconverters with reactive NAT results at earlier bleeds had a total of four subsequent specimens that were negative by NAT but positive by Multispot and positive (n=3) or indeterminate (n=1) by WB. Multispot was negative in 12 seroconverter specimens that were reactive by third-generation IA and 41 that were reactive by fourth-generation IA. Use of the entire alternative algorithm including NAT further improved detection of HIV-1 infections compared with the WB, and correctly classified as positive 102 seroconverter specimens with the third-generation IA and 130 with the fourth-generation IA compared with 56 using the current algorithm with either IA (Table 2.b).
Table 2.
| a. Performance of third- and fourth-generation IA, and the current and alternative laboratory HIV testing algorithms among 230 specimens from HIV-1 seroconverters | ||||||
|---|---|---|---|---|---|---|
| Multispot | Western blot | |||||
| NR | R | negative | indeterminate | positive | ||
| Third-generation screening | ||||||
| R | n= 102 | 12 | 90 | 11 | 35 | 56 |
| NAT negative | 0 | 4† | 0 | 1† | 3† | |
| NAT positive | 12 | 86 | 11 | 34 | 53 | |
| NR | n= 128 | 128 | 0 | n/d | n/d | n/d |
| Fourth-generation screening | ||||||
| R | n=131 | 41 | 90 | 36 | 39 | 56 |
| NAT negative | 1 | 4† | 1 | 1† | 3† | |
| NAT positive | 40 | 86 | 35 | 38 | 53 | |
| NR | n=99 | 99 | 0 | n/d | n/d | n/d |
| b. Number of specimens detected by the current and alternative laboratory HIV testing algorithm among 26 HIV-1 seroconverters (230 specimens) | ||
|---|---|---|
| Screening IA | MS/NAT | WB |
| Third-generation | 102* ** | 56** |
| Fourth-generation | 130* ** | 56** |
Same specimens, and had been obtained subsequent to NAT-positive specimens from the same seroconverters
The table shows the number of specimens detected by the combination of different tests in the context of the current and alternative laboratory.
HIV testing algorithm including nucleic acid testing (NAT); n: number; n/d: not determined; NR: non-reactive; R: reactive; NAT: nucleic acid test
p< 0.001 (102 vs. 130);
p< 0.001 (102 vs. 56 and 130 vs. 56).
MS: Multispot; NAT: nucleic acid test; WB: Western blot*
DISCUSSION
The GS HIV Combo Ag/Ab fourth-generation IA reduced the diagnostic window compared with the third-generation IA,7, 15 enabling detection of acute HIV infections only otherwise detected by amplified RNA methods. This screening assay also performed with high specificity in a low HIV prevalence population.21 Similar performance characteristics have been observed in studies of the only other FDA-approved fourth-generation IA on the U.S. market, the Abbott ARCHITECT HIV Ag/Ab Combo.14 In the current study, the alternative laboratory HIV testing algorithm using the GS HIV Combo Ag/Ab fourth-generation IA not only correctly resolved the infection status of specimens from individuals with established HIV infections that had been detected using the HIV-1 WB assay, but also identified 2 HIV-2 infections in specimens misclassified as HIV-1 by WB.
When used as part of the alternative algorithm in populations with persons at high risk of HIV during their early infection period, fourth-generation IAs will enable testing programs to detect HIV infections earlier. Even when a third-generation IA is used as the initial test in the alternative algorithm, more early infections will be detected than in the current algorithm when a third-generation IA is used with WB. Individuals in the acute stage of HIV infection are usually highly viremic, resulting in more virus shedding at mucosal sites, and are therefore more likely to transmit HIV infection through bodily secretions.4 Estimates of the proportion of total HIV transmissions attributable to early infections range widely from 6% to 49.4%.3, 22, 23 To use the alternate algorithm to its full advantage to detect early infections and reduce transmissions, laboratories must be prepared to conduct NAT testing with rapid turn-around and public health systems must be in place to conduct timely linkage to medical care and partner services.11 Detection of early HIV infection may also play a key role in the success of treatment as prevention by offering available antiretroviral drugs to high-risk people before or immediately after HIV exposure or as a means of secondary prevention.24
The Bio-Rad GS fourth-generation IA demonstrated high specificity in a very low-prevalence population. The specificity is similar to that obtained with third-generation IAs.25 However, with the WB-indeterminate specimens, more false-positive results appeared to occur with third-generation IA compared with the fourth-generation IA, potentially due to cross reaction with the p24 antigen present in the third-generation assay, but not the fourth. This study demonstrated that few NATs will be required to resolve specimens with false-positive screening assay results when the alternative HIV diagnostic testing algorithm is utilized. Further, in our study, the alternative algorithm correctly identified the HIV infection status among all HIV-1 WB-positives, highlighting its utility in a variety of settings in the United States. In addition, even though HIV-2 is rare in the United States,26 the alternative algorithm identified HIV-2 infections that would otherwise have been misclassified as HIV-1 infections by the HIV-1 WB, as has been noted in other studies.10, 27 With the availability of supplemental testing technologies that differentiate HIV-1 from HIV-2 and widespread use of the alternative algorithm, it is likely that more HIV-2 infections will be recognized.
The alternative HIV diagnostic testing algorithm has previously been shown to work well in high-risk individuals8 and in persons with established HIV-1 infection when used with a third-generation screening IA.9 In this study of over 10,000 specimens, excluding the seroconversion specimens, the alternative algorithm correctly resolved the infection status of all specimens except two. These two specimens were RR by third- and fourth-generation IA, and HIV-1 WB-indeterminate, Multispot HIV-1 reactive and NAT-negative, indicating discordance between serologic and virologic markers of infection. These specimens highlight the difficulty of resolving the true infection status without follow-up testing with a subsequent sample. It is possible that they represent false-positive results by the alternative algorithm. Alternatively, they might be similar to the four study specimens from seroconverters that were HIV-1 positive by serology but NAT-negative during follow-up. Other studies have shown that 3–5% of HIV-1 antibody-positive specimens28, 29 are negative by NAT. Persons classified as HIV antibody positive by the alternative algorithm (positive third- or fourth-generation IA and Multispot results) who have a negative NAT after entering care will need further diagnostic testing.30
There are several theoretical limitations associated with these analyses. First, factors beyond assay performance may have been responsible for the limited number of false-positive and false-negative results. False-positive results may have occurred due to contamination of samples or specimen mix up, and false-negative NAT results in stored, frozen specimens tested retrospectively may have happened due to degradation of viral RNA by microbes or multiple freeze-thaws.31–33 Second, for the HIV-1 WB-indeterminate specimens, we did not have the ability to obtain follow-up samples and thus the ultimate infection status is not known with certainty. Third, even though our study demonstrated the excellent performance of the alternative laboratory HIV diagnostic testing algorithm in selected specimen sets, we did not assess its performance with known HIV-2 positive specimens or in an actual population at high risk for acute infection. Finally, although the performance of the alternative diagnostic testing algorithm was assessed in this study using APTIMA, the only NAT approved by the FDA for diagnosis, it may be of additional benefit to assess the performance of quantitative HIV RNA assays, which are used widely in clinical settings for therapeutic monitoring.30
In conclusion, the Bio-Rad fourth-generation HIV-1/2 IA demonstrated high sensitivity for both early and established infections, and high specificity in a low HIV prevalence population. When coupled with the alternative supplemental tests, testing jurisdictions will gain the ability to corroborate reactive screening test results for early infections, identify HIV-2 infections, and reduce indeterminate results. These features make this algorithm a good alternative to the current HIV testing algorithm, which utilizes WB or immunofluorescence assay (IFA) as the definitive supplemental test. However, as with any diagnostic testing, low numbers of false-positive results may occur that will warrant additional diagnostic testing.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the technical staff of the Quest Diagnostics - Lenexa, KS facility for processing and testing these specimens.
Footnotes
Disclaimer: The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services
Conflict of interest statement: No financial disclosures were reported by the authors of this paper.
REFERENCES
- 1.Centers for Disease Control and Prevention. Interpretation and use of the western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb Mortal Wkly Rep. 1989;38(Suppl 7):1–7. [PubMed] [Google Scholar]
- 2.Centers for Disease Control Prevention. Acute HIV infection - New York City, 2008. MMWR Morb Mortal Wkly Rep. 2009;58(46):1296–1299. [PubMed] [Google Scholar]
- 3.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195(7):951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 4.Pilcher CD, Eron JJ, Jr, Galvin S, Gay C, Cohen MS. Acute HIV revisited: new opportunities for treatment and prevention. J Clin Invest. 2004;113(7):937–945. doi: 10.1172/JCI21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ntemgwa ML, d'Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob Agents Chemother. 2009;53(9):3611–3619. doi: 10.1128/AAC.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branson BM. The future of HIV testing. J Acquir Immune Defic Syndr. 2010;55(S2):102–105. doi: 10.1097/QAI.0b013e3181fbca44. [DOI] [PubMed] [Google Scholar]
- 7.Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(Suppl 1):S17–S22. doi: 10.1016/j.jcv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Delaney KP, Heffelfinger JD, Wesolowski LG, Owen SM, Meyer WA, 3rd, Kennedy S, et al. Performance of an alternative laboratory-based algorithm for HIV diagnosis in a high-risk population. J Clin Virol. 2011;52(Suppl 1):S5–S10. doi: 10.1016/j.jcv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Wesolowski LG, Delaney KP, Hart C, Dawson C, Owen SM, Candal D, et al. Performance of an alternative laboratory-based algorithm for diagnosis of HIV infection utilizing a third generation immunoassay, a rapid HIV-1/HIV-2 differentiation test and a DNA or RNA-based nucleic acid amplification test in persons with established HIV-1 infection and blood donors. J Clin Virol. 2011;52(Suppl 1):S45–S49. doi: 10.1016/j.jcv.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Nasrullah M, Ethridge SF, Delaney KP, Wesolowski LG, Granade TC, Schwendemann J, et al. Comparison of alternative interpretive criteria for the HIV-1 Western blot and results of the Multispot HIV-1/HIV-2 Rapid Test for classifying HIV-1 and HIV-2 infections. J Clin Virol. 2011;52(Suppl 1):S23–S27. doi: 10.1016/j.jcv.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 11.Kelly JA, Morin SF, Remien RH, Steward WT, Higgins JA, Seal DW, et al. Lessons learned about behavioral science and acute/early HIV infection. The NIMH Multisite Acute HIV Infection Study: V. AIDS Behav. 2009;13(6):1068–1074. doi: 10.1007/s10461-009-9579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ly TD, Ebel A, Faucher V, Fihman V, Laperche S. Could the new HIV combined p24 antigen and antibody assays replace p24 antigen specific assays? J Virol Methods. 2007;143(1):86–94. doi: 10.1016/j.jviromet.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Pandori MW, Hackett J, Jr, Louie B, Vallari A, Dowling T, Liska S, et al. Assessment of the ability of a fourth-generation immunoassay for human immunodeficiency virus (HIV) antibody and p24 antigen to detect both acute and recent HIV infections in a high-risk setting. J Clin Microbiol. 2009;47(8):2639–2642. doi: 10.1128/JCM.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez P, Wesolowski L, Patel P, Delaney K, Owen SM. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab Combo Assay. J Clin Virol. 2011;52(Suppl 1):S51–S55. doi: 10.1016/j.jcv.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Bentsen C, McLaughlin L, Mitchell E, Ferrera C, Liska S, Myers R, et al. Performance evaluation of the Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA, a 4th generation HIV assay for the simultaneous detection of HIV p24 antigen and antibodies to HIV-1 (groups M and O) and HIV-2 in human serum or plasma. J Clin Virol. 2011;52(Suppl 1):S57–S61. doi: 10.1016/j.jcv.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Bio-Rad Laboratories. Product insert, GS HIV Combo Ag/Ab EIA. Redmond, WA, USA: Bio-Rad; 2011. [Google Scholar]
- 17.Stout RL, Fulks M, Dolan VF. Trends in mortality of insurance applicants with HIV infection. J Insur Med. 2012;43(2):67–75. [PubMed] [Google Scholar]
- 18.Bio-Rad Laboratories. Product insert, GS HIV-1/HIV-2 Plus O EIA. Redmond, WA, USA: Bio-Rad; 2005. [Google Scholar]
- 19.Bio-Rad Laboratories. Product insert, Multispot HIV-1/HIV-2 Rapid Test. Redmond, WA, USA: Bio-Rad; 2004. [Google Scholar]
- 20.Gen-Probe Incorporated. Product insert, APTIMA HIV-1 RNA Qualitative assay. San Diego, CA, USA: Gen-Probe Incorporated; 2006. [Google Scholar]
- 21.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 22.Pinkerton SD. How many sexually-acquired HIV infections in the USA are due to acute-phase HIV transmission? AIDS. 2007;21(12):1625–1629. doi: 10.1097/QAD.0b013e32826fb6a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiridou M, Geskus R, de Wit J, Coutinho R, Kretzschmar M. Primary HIV infection as source of HIV transmission within steady and casual partnerships among homosexual men. AIDS. 2004;18(9):1311–1320. doi: 10.1097/00002030-200406180-00010. [DOI] [PubMed] [Google Scholar]
- 24.Cohen MS, Shaw GM, McMichael AJ, Haynes BF. Acute HIV-1 Infection. N Engl J Med. 2011;364(20):1943–1954. doi: 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wians FH, Jr, Briscoe D, Anderson KM, Hicks PS, Smith DL, Clark TA, et al. Evaluation of Four Qualitative Third-generation HIV Antibody Assays and the Fourth-generation Abbott HIV Ag/Ab Combo Test. Laboratory Medicine. 2011;42(9):523–535. [Google Scholar]
- 26.Centers for Disease Control and Prevention. HIV-2 Infection Surveillance --- United States, 1987--2009. MMWR Morb Mortal Wkly Rep. 2011;60:985–988. [PubMed] [Google Scholar]
- 27.Torian LV, Forgione LA, Punsalang AE, Pirillo RE, Oleszko WR. Comparison of Multispot EIA with Western blot for confirmatory serodiagnosis of HIV. J Clin Virol. 2011;52(Suppl 1):S41–S44. doi: 10.1016/j.jcv.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, et al. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol. 2008;46(5):1588–1595. doi: 10.1128/JCM.02196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel P, Mackellar D, Simmons P, Uniyal A, Gallagher K, Bennett B, et al. Detecting acute human immunodeficiency virus infection using 3 different screening immunoassays and nucleic acid amplification testing for human immunodeficiency virus RNA, 2006–2008. Arch Intern Med. 2010;170(1):66–74. doi: 10.1001/archinternmed.2009.445. [DOI] [PubMed] [Google Scholar]
- 30.AIDS info: Clinical guidelines portal. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents: Baseline Evaluation. [cited 2012 April 24];2012 Available from: http://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-treatment-guidelines/36/baseline-evaluation.
- 31.Ginocchio CC, Wang XP, Kaplan MH, Mulligan G, Witt D, Romano JW, et al. Effects of specimen collection, processing, and storage conditions on stability of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1997;35(11):2886–2893. doi: 10.1128/jcm.35.11.2886-2893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claassen M, van Zyl GU, Preiser W. Extraction buffer contaminated bacterially as a cause of invalid HIV-1 viral load results on the NucliSens EasyQ system. J Virol Methods. 2008;150(1–2):80–81. doi: 10.1016/j.jviromet.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Valentine-Thon E. Quality control in nucleic acid testing--where do we stand? J Clin Virol. 2002;25(Suppl 3):S13–S21. doi: 10.1016/s1386-6532(02)00196-8. [DOI] [PubMed] [Google Scholar]