Abstract
Targeted anticancer drugs such as imatinib, trastuzumab and erlotinib dramatically improved treatment outcomes in cancer patients, however, these innovative agents are often associated with unexpected side effects. The pathophysiological mechanisms underlying these side effects are not well understood. The availability of a comprehensive knowledge base of side effects associated with targeted anticancer drugs has the potential to illuminate complex pathways underlying toxicities induced by these innovative drugs. While side effect association knowledge for targeted drugs exists in multiple heterogeneous data sources, published full-text oncological articles represent an important source of pivotal, investigational, and even failed trials in a variety of patient populations. In this study, we present an automatic process to extract targeted anticancer drug-associated side effects (drug-SE pairs) from a large number of high profile full-text oncological articles.
We downloaded 13,855 full-text articles from the Journal of Oncology (JCO) published between 1983 and 2013. We developed text classification, relationship extraction, signaling filtering, and signal prioritization algorithms to extract drug-SE pairs from downloaded articles. We extracted a total of 26,264 drug-SE pairs with an average precision of 0.405, a recall of 0.899, and an F1 score of 0.465. We show that side effect knowledge from JCO articles is largely complementary to that from the US Food and Drug Administration (FDA) drug labels. Through integrative correlation analysis, we show that targeted drug-associated side effects positively correlate with their gene targets and disease indications. In conclusion, this unique database that we built from a large number of high-profile oncological articles could facilitate the development of computational models to understand toxic effects associated with targeted anticancer drugs.
Keywords: text mining, information extraction, targeted anticancer drugs, drug side effects, drug discovery, drug repositioning, drug toxicity prediction
Graphical abstract
1. Introduction
Targeted anticancer drugs control cancer cell growth by interfering with specific molecular targets involved in tumor growth and progression. Targeted cancer therapies have significantly (positively) impacted the survival and quality of life of cancer patients [1]. For instance, treatment of Philadelphia-positive chronic myeloid leukemia (CML) with tyrosine kinase inhibitor (TKI) imatinib confers a significant survival advantage and an overall 80–90% response rate [2]. Trastuzumab, a monoclonal antibody that binds to the extracellular domain of HER2, is used to treat patients with metastatic HER2-positive breast cancer and has decreased the cancer recurrence risk in treated patients by 52% and reduced relative risk of mortality by 33% [3]. Erlotinib, a TKI that induces cancer cell apoptosis by blocking the EGFR signaling pathway, has been associated with complete to partial response and improved overall survival in patients with non-small-cell lung cancer [4].
Targeted anticancer drugs promised new ways to personalize cancer treatments based on unique molecular targets expressed by tumor cells. However, recent studies have shown that these innovative drugs are often associated with unanticipated high toxicities [5]. Recent meta-analysis studies show that most newly-approved targeted anticancer drugs are more toxic than standard treatments and are associated with increased rates of toxic death, treatment discontinuation, and severe adverse events [6, 7]. Besides the overall toxicity levels, many targeted anticancer drugs are associated with unanticipated toxicities, such as cardiovascular events, that are idiosyncratic and their underlying molecular mechanisms remain largely unidentified [5, 8, 9, 10]. Unlike side effects induced by cytotoxic chemotherapeutics, which are similar among drugs, side effects associated with targeted anticancer drugs often differ among drugs of the same class such as erlotinib and gefitinib [11]. These toxicities may be caused by the receptor cross-reactivity, the presence of receptors on normal cells [12], or the multiplicity of affected off-target proteins [13, 14, 15]. In order to maintain the balance between tumor control and drug-induced toxicities, research is needed to improve our understanding of the molecular mechanisms of targeted anticancer drug-related toxicities [1]. Currently, approximately 500 novel targeted agents are under preclinical or clinical development for the treatment of specific types of cancers [16]. The availability of a comprehensive side effect knowledge base for targeted drugs and innovative computational approaches to predicting unexpected toxicities are important for the successful development of targeted anticancer agents in the near future.
Current systems approaches to studying phenotypic or side effect relationships among drugs rely exclusively on information extracted from the US Food and Drug Administration (FDA) drug labels [17, 18, 19, 20, 21]. It was recently demonstrated that 39% of serious events associated with targeted cancer drugs are never reported in clinical trials and 49% are not described in FDA drug labeling [22]. Therefore, in constructing a comprehensive knowledge base of drug-side effect (drug-SE) relationships for cancer drugs, it is important to extract knowledge from multiple sources, including FDA drug labels, the FDA post-market drug safety surveillance system (FAERS), patient electronic health records (EHRs), and the large body of published biomedical literature. Recently, we developed automatic signal prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from FAERS [23]. We developed an large-scale approach to combine signals from both biomedical literature and FAERS to improve post-marketing drug safety signal detection [24]. For drug-SE relationship extraction from biomedical literature abstracts, we developed an automatic approach to extract anticancer drug-specific side effects from MEDLINE by developing specific filtering and ranking schemes [25]. We also developed a pattern-based learning approach to accurately extract drug-SE pairs from MEDLINE sentences [26].
The Journal of Clinical Oncology (JCO) is the official journal of the American Society of Clinical Oncology and the leading journal in oncology. JCO articles include a variety of cancer-related research articles, including clinical trials reporting drug efficacy and toxicity in cancer patients, trial reports evaluating the effectiveness of biomarkers, clinical case reports, and meta-analysis studies, among other article types. JCO articles not only include pivotal clinical trials that have led to drug approval, but also trials that are still in investigational stages and even failed trials. Side effect knowledge for both commercial, investigational and failed drugs is crucial to our understanding of the molecular mechanisms underlying the observed toxicities. In one of our recently studies, we downloaded a total of 13,855 full-text JCO articles published between 1983 and 2013. We combined automatic table classification and relationship extraction approaches to extract anticancer drug-associated side effects from a total of 31,255 tables embedded in these JCO articles. We extracted a total of 26,918 drug-SE pairs from SE-related tables with a precision of 0.605, a recall of 0.460, and a F1 of 0.520 [27]. Complementary to our previous study, our current study presents an integrated system combining text classification, relationship extraction, signal filtering, and signal prioritization algorithms to extract targeted anticancer drug-associated side effects from the full-text part of JCO articles.
2. Approach
We first developed a support vector machine (SVM) classifier to classify downloaded articles into drug SE-related and -unrelated. We then extracted drug-SE co-ocurrence pairs from articles that were classified as SE-related. We developed a filtering approach to remove false positives (drug-disease treatment pairs) from the extracted drug-SE co-ocurrence pairs. We then developed ranking algorithms to further prioritize extracted drug-SE pairs based on their term and document frequencies. We investigated whether the drug side effect knowledge from JCO articles is complementary to that in FDA drug labeling by exhaustively curating all articles containing the drug sunitinib in their titles and comparing drug-SE pairs extracted from these JCO articles to those extracted from the FDA drug label. To show the potential of these targeted drug-associated side effects in developing systems approaches to understanding the molecular mechanisms underlying the observed drug phenotypes (side effects) and drug repositioning, we linked drugs to their corresponding gene targets and disease indications and systematically studied the correlations between targeted anticancer drug-associated side effects and their known gene targets and disease indications.
Our study is different from many literature-based drug-SE relationship extractions [24, 25, 26, 30] in at least two ways. First, most literature-based drug-SE relationship extraction tasks used only the abstracts of biomedical research articles, while we used full-text articles. While full-text articles contain richer drug-SE association knowledge compared to abstracts, they also contain much noise, which renders the extraction task more challenging. Second, while previous studies applied either machine learning approach[30, 26] or specific signal filtering and ranking approaches [23], we here combined both approaches in extracting drug-SE pairs from full-text articles. This study is complementary to our previous study in extracting drug-SE pairs from tables of JCO articles. In this study, we used the text part of JCO articles for drug-SE extraction. In addition, we focus on targeted anticancer drugs. Our main contribution is that we extracted a large number of targeted anti-cancer drug-associated side effects from high-profile oncological articles, the majority of which have not included in FDA drug labeling yet. In addition, we show that these extracted drug-SE pairs have the potential to illuminate complex pathways of targeted drug-induced side effects and to discover novel drug indications.
3. Methods
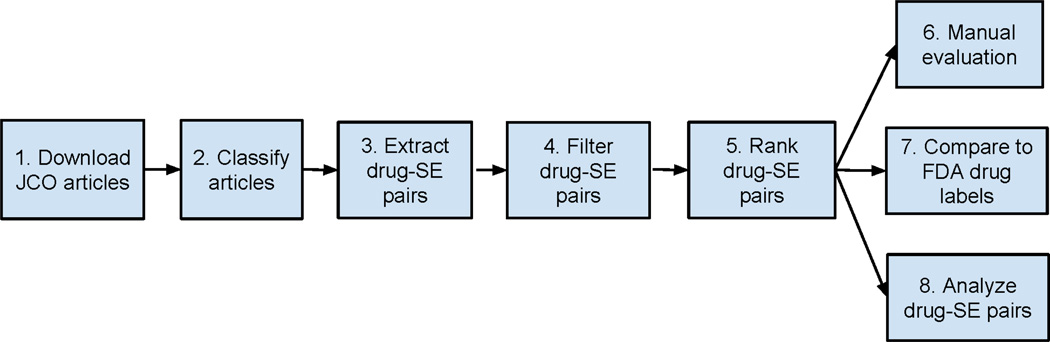
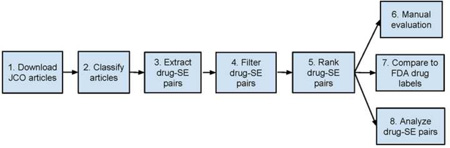
The overall experiment consists of the following steps: (1) download JCO full-text articles; (2) Classify JCO articles into drug SE-related and -unrelated; (3) Extract drug-SE pairs from articles classified as SE-related; (4) Filter out drug-disease treatment pairs; (5) Rank filtered pairs; (6) Manually evaluate the performance of drug-SE pair extraction; (7) Compare the drug-SE knowledge captured in JCO articles to that in FDA drug labels; and (8) Analyze the correlations between extracted drug-SE pairs and drug targets as well as drug disease indications (Figure 1).
Figure 1.
Experiment flowchart.
3.1. Download JCO full text articles
In our previous study, we downloaded a total of 13,855 JCO full text JCO articles published from 1983 through 2013 and extracted anticancer drug-SE pairs from the tables embedded in the articles [27]. In this study, we used the text part of these downloaded JCO articles for targeted anticancer drug-SE relationship extraction. We used the publicly available information retrieval library Lucene (http://lucene.apache.org) to create a search engine with indices created on article titles, abstracts, and all text. Each article was assigned a unique identification number.
3.2. Classify articles into drug SE-related and -unrelated
We randomly selected 500 articles from the 13,855 downloaded full-text JCO articles and manually classified them into drug SE-related and -unrelated. Among these 500 articles, 103 are SE-related and 393 are SE-unrelated. These articles were randomly split into the training dataset (60%) and testing dataset (40%). An SVM classifier [33] was trained on the training dataset and tested on the testing dataset. The SVM-based classifier used polynomial kernel, bag-of-words feature, TF-IDF weighting, stemming and stopwords-removal. The bag-of-words feature was used since it is often the case that the appearance of one specific word such as ‘toxicity’ or ‘adverse’ can be used to determine whether a sentence is drug-SE-related. The 10-fold cross validation was used in training the classifier. When evaluated on the testing dataset, the classifier achieved a precision of 0.862, a recall of 0.677, a F1 score of 0.759, a false positive rate of 0.029, and a false negative rate of 0.323.
3.3. Extract drug-SE pairs from classified JCO articles
The inputs to the drug-SE pair extraction algorithm were a list of targeted anticancer drugs, a list of SE terms, and JCO articles that were automatically classified as SE-related.
3.3.1. Targeted drug lexicon
A list of 45 targeted cancer drugs was obtained from the National Cancer Institute (NCI)1. The 45 targeted drugs are: alemtuzumab, litretinoin, anastrozole, bevacizumab, bexarotene, bortezomib, bosutinib, brentuximab, cabozantinib, carfilzomib, cetuximab, crizotinib, dasatinib, denileukin, erlotinib, everolimus, exemestane, fulvestrant, gefitinib, ibritumomab, imatinib, ipilimumab, lapatinib, letrozole, nilotinib, ofatumumab, panitumumab, pazopanib, pertuzumab, pralatrexate, regorafenib, rituximab, romidepsin, sorafenib, sunitinib, tamoxifen, temsirolimus, toremifene, tositumomab, trastuzumab, tretinoin, vandetanib, vemurafenib, vorinostat, and ziv-aibercept.
3.3.2. Manually curated clean side effect (SE) lexicons
An accurate and comprehensive SE lexicon is critical for the task of drug-SE relationship extraction from free-text. We have built two clean SE (or disease) lexicons and demonstrated that these clean lexicons are important in improving precisions in biomedical relationship extraction tasks, including drug-SE relationship extraction [23, 24, 25, 26, 27] and disease-phenotype relationship extractions [28, 29]. The first SE lexicon was built based on the Medical Dictionary for Regulatory Activities (MedDRA) [31] by manually removing many non-SE terms such as medical procedures, lab tests, and protein names. After manual curation, the lexicon contained 49,625 terms. The second SE (or disease) lexicon was based on the Unified Medical Language System (UMLS) (2011AB version) [32] and was built by manually removing incorrectly classified disease terms, ambiguous terms, and overly general terms. The final UMLS-based clean lexicon consisted of 75,558 terms. In this study, we demonstrated that these clean lexicons considerably improved upon the overall precision of the subsequent drug-SE relationship extraction from full-text JCO articles.
3.3.3. Drug-SE pair extraction from automatically classified articles
We then trained a SVM classifier using these 500 annotated articles and used it to classify all 13,855 JCO articles. A total of 2602 articles are classified as SE-related. We used each targeted anticancer drug as a search query to the local search engine. If a drug term appeared in the title or text of an SE-related article, the term, its frequency, and the article ID was recorded. Similarly, we used each term from the clean SE lexicons as a search query to the local search engine. If a SE term appeared in the title or text of an article, the term, its frequency, and the article ID was recorded. Drug-SE pairs, along with their document frequency and term frequency, were extracted by joining the article IDs associated with drug terms and with SE terms.
3.4. Filter extracted drug-SE pairs by removing drug-disease (cancer) treatment pairs
Drug-associated side effects are often reported in the context of drug treatments in patients with cancers. Therefore, one of the main challenges in extracting drug-SE pairs from JCO articles is to differentiate drug-SE causal pairs from drug-disease treatment pairs. This task is made easier by the fact that we can in general classify the extracted pairs into causal or treatment relationship based on the medical condition entities (the SE terms) alone. If the SE term in a drug-SE pair is a cancer term, then this pair is more likely to be a drug-disease treatment pair than a drug-SE causal pair (though some drugs also cause cancers). In this study, we first removed many cancer terms from the SE lexicons by filtering out terms of the semantic type “Neoplastic Process” based on UMLS classification. We then extracted drug-SE pairs using these filtered SE lexicons. We showed that this filtering strategy removed many false positives and significantly improved the precision while keeping the high recall of the extracted drug-SE pairs.
3.5. Rank filtered drug-SE pairs based on term and document frequency
We developed two ranking algorithms to rank the filtered drug-SE pairs. The first one is to rank drug-SE pairs according to their total occurrences in the entire corpus, which is equivalent to the term frequency used in information retrieval. The second one is to rank drug-SE pairs according to their document frequencies (the number of documents where a pair appeared). The intuition is that if a drug-SE pair appears often in many different articles, then it is likely that there is a true semantic association between the drug and the SE entity. This semantic association can be “DRUG cause SE” or “DRUG treat DISEASE.” Since we have already filtered out the drug-disease treatment pairs, then the top-ranked pairs are more likely to be drug-SE causal pairs. We measured the ranking efficacy using 11-point interpolated average precision, which is commonly used to evaluate retrieved ranked lists for search engines [34]. For each ranked list, the interpolated precision was measured at the 11 recall levels of 0.0, 0.1, 0.2, ‥, 1.0. A composite precision-recall curve showing 11 points was then graphed and used to evaluate whether the ranking algorithms work effectively in prioritizing extracted drug-SE pairs.
3.6. Manual evaluation
Currently, there exists no gold standard that accurately represents drug-associated side effect knowledge captured in JCO articles. For example, as we will show later in this study, many of drug-SE pairs for FDA-approved drugs were reported in JCO articles but not included in FDA drug labels yet; therefore, drug-SE pairs derived from FDA drug labels can not serve as a gold standard to evaluate drug-SE relationship extraction from JCO articles. In addition, JCO articles also include many investigational drugs and failed drugs while FDA drug labels contain only commercial drugs. In this study, we first classified all articles into SE-related or -unrelated. We then automatically annotated all SE-related articles using drug and SE terms from the input lexicons as search queries. We then randomly selected 100 SE-related articles with titles containing one targeted anticancer drug term. We then manually extracted drug-SE pairs from these articles. Three curators with graduate degrees in biomedical sciences or clinical medicines independently performed the manual curation. It took approximate 24 hours for each annotator to curate these 100 articles. The inter-annotator agreement rate was 85%. For each article, only drug-SE pairs agreed upon by all three curators were used as the gold standard. We ran our algorithm on these articles and calculated precision, recall, and F1 for each article using the manually curated pairs from these articles as goldstandard. The final reported precision, recall, and F1 were averages of precisions, recalls and F1s across these 100 articles.
3.7. Compare side effect knowledge extracted from JCO articles to that from FDA drug label
We investigated whether the drug side effect knowledge from JCO articles is complementary to that in FDA drug labeling. We exhaustively curated all 49 articles that contain the targeted drug sunitinib in their titles. We then compared drug-SE pairs extracted from these 49 JCO articles to those extracted from its FDA drug label. Sunitinib is a multi-targeted receptor tyrosine kinase inhibitor approved for the treatment of renal cell carcinoma and gastrointestinal stromal tumor. Since sunitinib targets multiple receptors that are involved in both tumor growth and normal cell functions, it is associated with many different types of side effects. From the 49 JCO articles, we manually extracted 332 sunitinib-SE pairs. From the FDA drug label that we downloaded2, we manually extracted a total of 117 sunitinib-SE pairs. We compare]d the overlap of sunitinib-SE pairs between these two sources. In addition, we also compared the overlap of sunitinib-SE pairs between these two sources at difference frequency cutoffs in order to investigate whether more frequently reported pairs in JCO articles tend to be more likely captured in FDA drug labels.
3.8. Analyze drug-SE pairs
We investigated whether drug-drug pairs that shared side effects also tended to share gene targets and disease indications. We downloaded a total of 10,478 drug-gene pairs from DrugBank [35], a knowledge base for drugs, drug actions, and drug targets. These downloaded drug-gene pairs included a total of 24 targeted cancer drugs. For drug-drug pairs that shared different numbers of side effects, we calculated the average number of shared gene targets.
We extracted a total of 52,000 drug-disease pairs from ClinicalTrials.gov, a registry of federally- and privately-supported clinical trials conducted in the United States and around the world3. For drug-drug pairs that shared SEs at different cutoffs, we calculated the average number of shared disease indications.
4. Results
4.1. Performance of drug-SE relationship extraction from JCO full text articles
To measure the performance of the relationship extraction algorithm, we applied the algorithm to the 100 evaluation articles. For each article, we calculated the precision, recall, and F1 using the manually-extracted pairs from the same article as the gold standard. We then calculated the average precision, recall, and F1 of the algorithm across these 100 articles. We compared the performance of the algorithm using different SE lexicons (clean vs. original, separate vs. combined, cancer-filtered vs. unfiltered). These SE lexicons included (1) three MedDRA-based SE lexicons: original, clean, and clean lexicon with all cancer terms removed (“Clean minus cancer terms”); (2) three UMLS-based SE lexicon: original, clean, and clean lexicon with all cancer terms removed (“Clean minus cancer terms”); and (3) a combined clean lexicon consisted of terms from both MedDRA and UMLS, also with cancer terms removed. The drug lexicon was consisted of 45 targeted cancer drug terms obtained from NCI. The overall recalls of the algorithm were high, ranging from 0.708 to 0.899, meaning that the SE lexicons covered the majority of the SE concepts used in JCO articles (Table 1). However, the precisions varied greatly from 0.075 to 0.405.
Table 1.
Table classification performance.
| SE Lexicon | Lexicon Processing | Precision | Recall | F1 |
|---|---|---|---|---|
| MedDRA | Original | 0.112 | 0.891 | 0.176 |
| Clean | 0.230 | 0.886 | 0.303 | |
| Clean minus cancer terms | 0.405 | 0.899 | 0.465 | |
| UMLS | Original | 0.075 | 0.708 | 0.118 |
| Clean | 0.165 | 0.714 | 0.218 | |
| Clean minus cancer terms | 0.310 | 0.712 | 0.337 | |
| Combined | Clean minus cancer terms | 0.310 | 0.878 | 0.380 |
Comparing the clean SE lexicons with the original lexicons, we show that the precision significantly increased from 0.112 to 0.230 for the MedDRA-based SE lexicon, and from 0.075 to 0.165 for the UMLS-based SE lexicon. The MedDRA-based SE lexicon also had a better precision than the UMLS-based SE lexicon. Comparing the clean SE lexicons with the same lexicons removed of cancer terms, we showed that the precisions further significantly increased from 0.230 to 0.405 for the MedDRA-based SE lexicon, as well as increasing from 0.165 to 0.310 for the UMLS-based SE lexicon. By combining terms from two clean lexicons, we did not observe improvements in precision or recall.
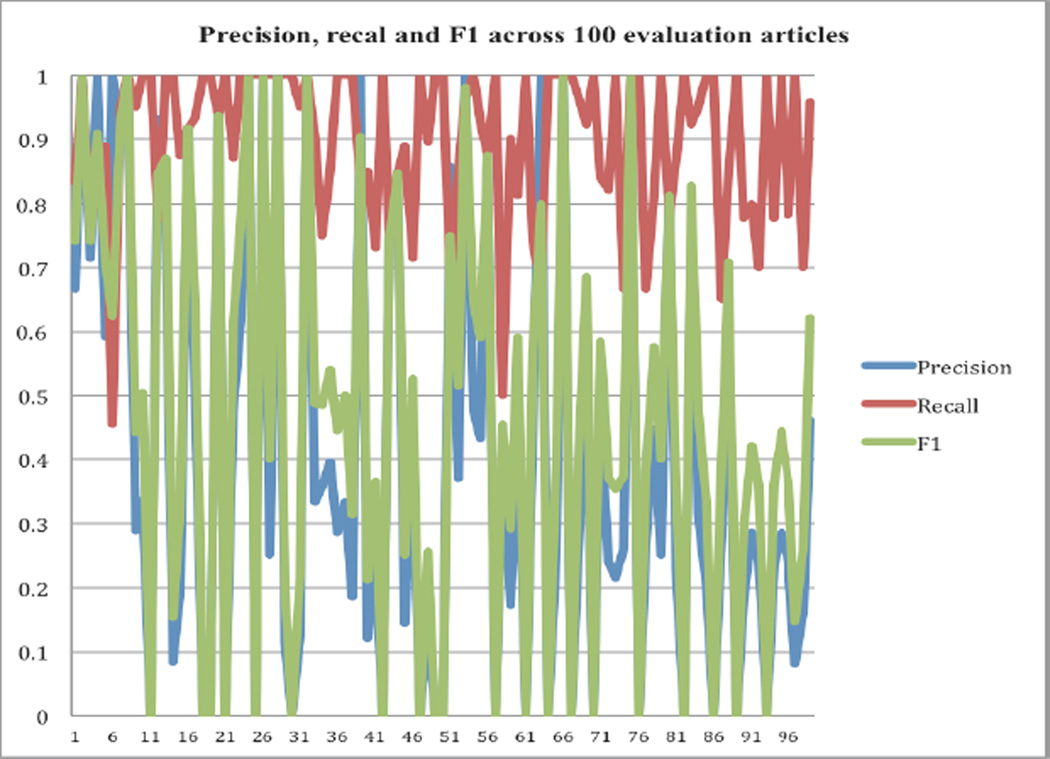
The precisions, recalls, and F1 values varied greatly across different articles. As shown the Figure 2, the recalls were consistently high, ranging from 0.67 to 1.0. However, the precisions and F1 values varied greatly from 0.0 to 1.0. Several factors may have contributed to the varying precisions. First, the text classifier (precision: 0.862, recall: 0.677, F1: 0.759) is not perfect in classifying articles into SE-related and -unrelated. Many SE-unrelated JCO articles such as those evaluating biomarkers in predicting the treatment outcomes contain both drug and disease terms (e.g. describing patient co-morbidities, outcomes measures, for instance); however, these disease terms are not drug-associated side effects. Second, even in SE-related clinical trial articles that report drug efficacy and toxicities, disease terms are often contained in the patient inclusion and exclusion criteria sections. For example, some studies may exclude patients with renal insufficiency or cardiovascular diseases. A potential way to avoid extracting pairs from these sections is to develop a nested classifier to further categorize sections or sentences in SE-related articles into toxicity-related or -unrelated.
Figure 2.
Precisions, recalls and F1 values across the evaluation dataset of 100 randomly selected and manually curated articles.
4.2. Ranking by both term frequency and document frequency further improve the precisions
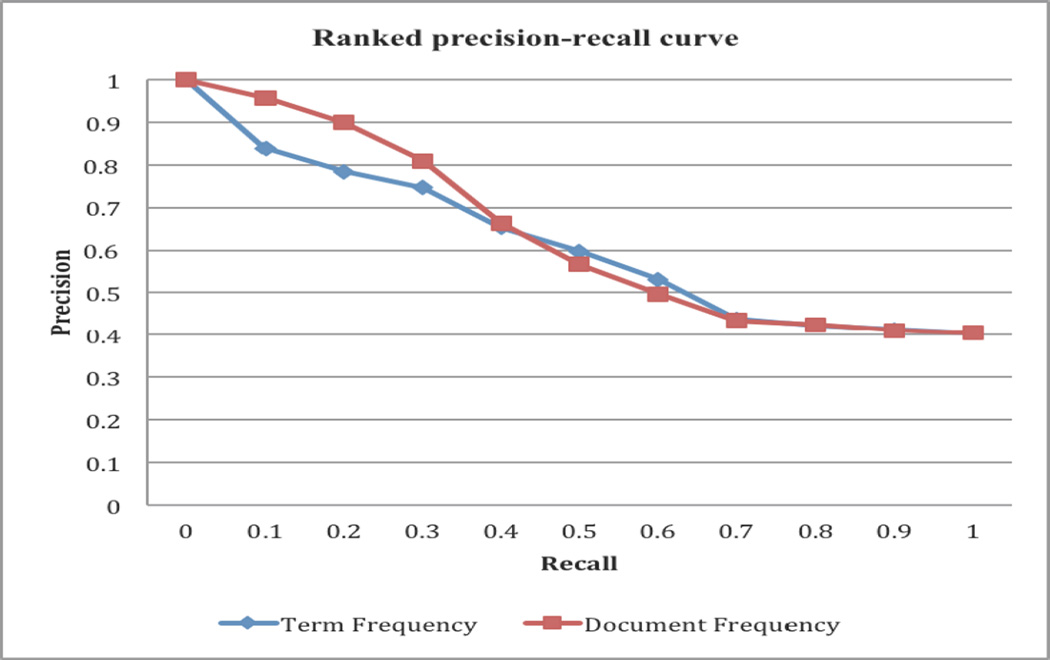
In the previous section, we show that we significantly improved the precision of drug-SE extraction from 0.112 to 0.405 by using manually-curated SE lexicons and by filtering out cancer terms from the clean SE lexicons. In this section, we developed a ranking algorithm to further prioritize the extracted drug-SE pairs. We ranked the filtered drug-SE pairs by term frequency and by document frequency. As shown in Figure 3, ranking by both term frequency and document frequency are effective in ranking true positives highly. For example, for pairs ranked by document frequencies, the top-ranked pairs at a recall of 0.1 had a precision of 0.957, representing a significant elevation in precision as compared to the overall precision of 0.405 for the whole list (at a recall of 1.0). Ranking by term frequency and by document frequency had similar results, even though the top-ranked pairs by document frequency had slightly higher precisions.
Figure 3.
Precision-recall curves for drug-SE pairs ranked by term frequency and by document frequency.
4.3. Comparison of toxicity knowledge contained in JCO articles to that in FDA drug labels
In this section, we investigated whether the drug side effect knowledge contained in JCO articles is complementary to that in FDA drug labeling. Side effect information from FDA drug labels is mainly derived from pivotal clinical trials or post-marketing experience of patients with the same diseases. Notably, the side effect information reported in JCO articles includes not only pivotal clinical trials, but also investigational and even failed trials in patients with the same or different cancers. For example, between April 2009 and May 2011, Pfizer has reported unsuccessful late-stage trials in using sunitinib in the treatment of breast cancer, metastatic colorectal cancer, advanced non-small-cell lung cancer, and castration-resistant prostate cancer.
From the 49 JCO articles (unclassified) containing drug term sunitinib, we manually extracted a total of 332 sunitinib-SE pairs, with each pair assigned a frequency count (number of times a pair appeared in these 49 articles). From the FDA drug label for sunitinib, we manually extracted a total of 117 side effects. Among these pairs, only 53 pairs, representing 15.8% of pairs extracted from JCO articles and 44.8% of pairs from FDA drug labels, appeared in both sources. This indicates that the drug side effect knowledge from these two resources has some overlap but is largely complementary.
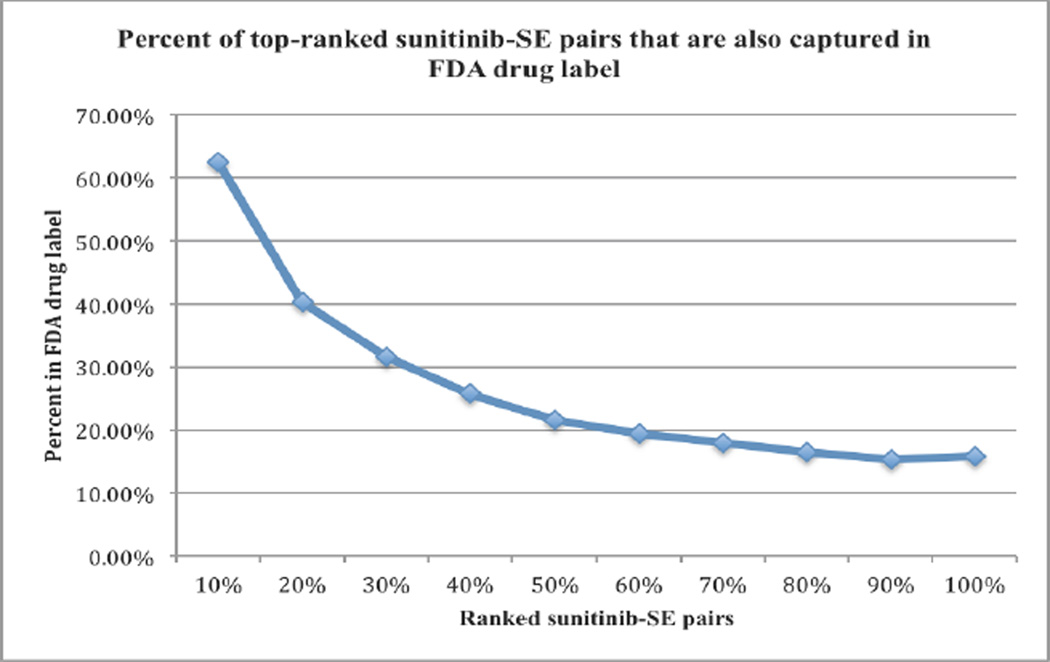
We then ranked the drug-SE pairs extracted from JCO articles by their term frequencies and investigated where frequent pairs were more likely to be included in both sources. We calculated the percentages of the top-ranked pairs extracted from JCO articles that were included in FDA drug labels. As shown in Figure 4, top-ranked (frequent pairs) drug-SE pair were more likely to be included in FDA drug labeling than less frequent pairs. For example, among the top 10% of ranked pairs extracted from JCO articles, 62.5% of them were also included in FDA drug labeling. The number steadily deceased to 40.3% for top the 20% of ranked pairs and to 25% for the top 40% of ranked pairs.
Figure 4.
Percentages of top ranked sunitinib-SE pairs extracted from JCO articles that are included in FDA drug label.
Many rare and severe adverse events associated with targeted drugs in cancer patients were reported in JCO articles but not included in FDA drug labeling. For example, in a case report article published in 2012 entitled “Takotsubo Syndrome in a Patient Treated With Sunitinib for Renal Cancer” reported that takotsubo syndrome was associated with sunitinib in patients with renal cancer. However, this association has not included in FDA drug labeling for sunitinib yet. In another article published in 2010 entitled “Recurrent Scrotal Hemangiomas During Treatment With Sunitinib” reported an instance of recurrent scrotal cutaneous capillary hemangiomas developed during therapy with sunitinib in a patient with renal cell carcinoma, and discussed a possible histopathogenetic mechanism of sunitinib. This adverse event is not included in drug labeling for sunitinib yet. Two other examples are acute myeloblastic leukemia and thyrotoxicosis that are reported to be associated with sunitinib in articles entitled “Phase II Study of Sunitinib Administered in a Continuous Once-Daily Dosing Regimen in Patients With Cytokine-Refractory Metastatic Renal Cell Carcinoma” and “Thyrotoxicosis during sunitinib treatment for renal cell carcinoma,” respectively.
4.4. Targeted cancer drug-associated side effects correlate positively with their target genes and disease indications
Drug-associated side effects may be caused by both drug ‘on-target’ and ‘off-target’ effects. In this section, we investigated the degree of targeted drug-associated side effects being correlated to their known drug-associated gene targets. We also investigated whether the observed drug side effects are correlated with drug indications, which may have implications in drug repositioning.
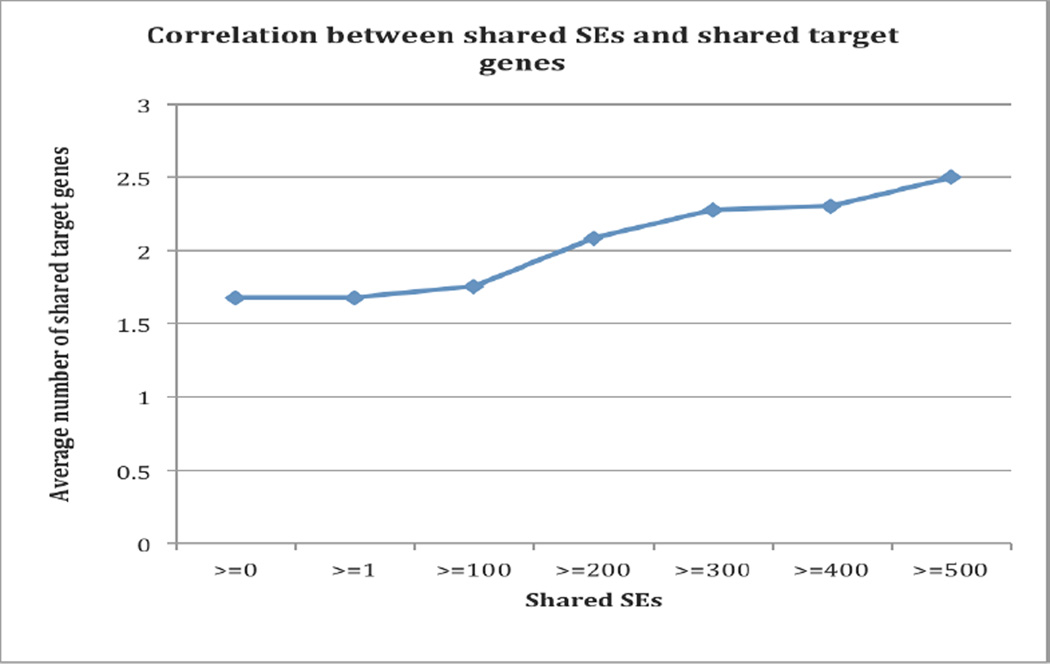
Among the 45 targeted drugs from the extracted 26,264 drug-SE pairs, 24 drugs have known associated target genes based on drug-gene association data from DrugBank. For all 276 drug-drug combinations for these 24 drugs (shared SEs ≥0), the average number of shared gene targets is 1.678. The number increases as drug-drug pairs sharing more SEs. The average number of shared gene targets is 2.081 for the 186 drug-drug pairs that shared at least 200 SEs, and the average number of shared gene targets is 2.278 for 115 pairs that shared at least 300 SEs (Figure 5). This demonstrates that some shared SEs among targeted anticancer drugs belongs to on-target effects and caused by their effects on normal cells. However, the modest positive correlation between shared SEs and shared ‘on-target’ genes indicates that many targeted drug-associated side effects may be caused by factors other than drug ‘on-targets’, such as unknown ‘off-targets, ’ drug metabolism, patient-specific characteristics including co-morbidities and performance status, and drug combinations or co-occurrent drugs.
Figure 5.
The correlation between drug side effects and drug targets.
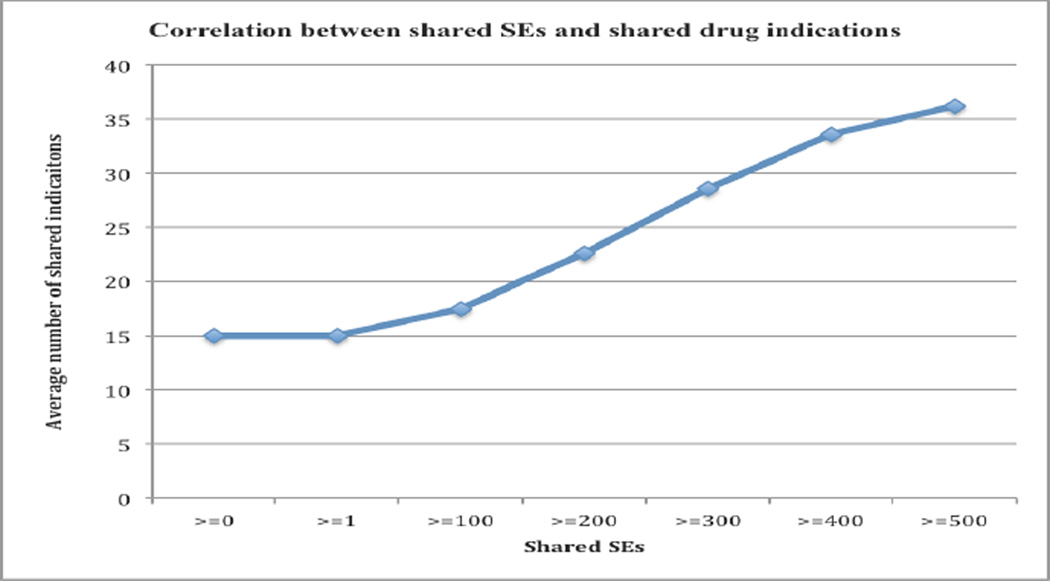
We investigated whether the observed drug side effects correlate with drug disease indications. A positive correlation implies that we may use the observed drug phenotype information in drug repositioning tasks. A total of 36 out of 45 targeted cancer drugs appeared in ClinicalTrial.gov. Note that many studies registered in ClinicalTrial.gov are still in investigational stages. As shown in Figure 6, there is a strong positive correlation between shared SEs and shared disease indications. The average number of shared disease indications for all 630 drug-drug combinations was 15.07. The number significantly increased to 22.61 for 333 drug-drug pairs that shared at least 200 SEs and to 33.59 for 114 drug-drug pairs that shared at least 400 SEs. The correlation of side effects with drug indications (Figure 6) is stronger than that with drug targets (Figure 5). These results indicate that we can leverage upon targeted drug-associated side effects for drug repositioning, even though we do not understand the pathophysiology underlying many these observed drug clinical phenotypes.
Figure 6.
The correlation between drug side effects and drug disease indications.
5. Discussion
In this study, we developed automatic relationship extraction, signal filtering, and ranking approaches to constructing a large scale drug-SE relationship knowledge base for targeted anticancer drugs from 13,855 full-text articles from the leading oncologic journal, JCO. Since our current goal is to build a comprehensive database of anticancer drug associated side effects, we biased our approach toward achieving high recall. Our algorithm achieved an overall precision of 0.405, a recall of 0.899, and an F1 score of 0.465. However, our approaches have limitations and can be further improved upon.
First, while the recall (0.899) of our relationship approach was high, the precision (0.465) could be further improved. We have manually created two large-scale SE lexicons, which significantly improved the precision from 0.112 to 0.230. We then filtered out many false positives due to drug-disease treatment pairs and further improved the precision from 0.230 to 0.405. The still modest precision is mainly caused by the inclusion of drug-disease co-occurrence pairs where the diseases are actually patient exclusion criteria. In the future, we will develop text classification approaches to categorize paragraphs (or sections) in JCO articles into toxicity-related or -unrelated before drug-SE relationship extraction. In addition, the current database as well as the automatically annotated or tagged text can serve as a pre-processing step for manual drug safely annotation.
Second, our knowledge base consisted of drug-SE pairs for individual drugs. In reality, cancer drugs, including targeted cancer drugs, are often used in combination with other drugs. Certain side effects may only occur for specific drug-drug combinations. Currently, the work on extracting side effects associated with drug-drug combinations from free-text is scant. A recent study led by Altman mined the FDA post-marketing FAERS database and found four pairs of drugs that seemed to cause symptoms only in combinations [36]. However, extracting side effects associated with drug combinations from free text will be different from mining patterns from the FAERS database.
Currently, we are integrating higher-level phenotypical drug side effect data with lower-level drug-related datasets such as drug targets, chemical structures, and gene expression as well as disease-related data such as disease-associated genes and disease phenotype data in order to develop systems approaches to drug target discovery, drug toxicity prediction and drug repositioning.
6. Conclusions
We presented an automatic process in combining text classification, relationship extraction, signal filtering and signal ranking approaches to extract side effects associated with targeted anticancer drugs from a large number of high profile full-text oncologic articles. Our extraction and filtering algorithms achieved a precision of 0.405, a recall of 0.899, and an F1 score of 0.465. This targeted drug-specific toxicity knowledge base consisted of 26,264 drug-SE pairs with drugs linked to their known “on-targets” and disease indications. We have shown that the toxicity knowledge in this knowledge base is largely complementary to that contained in FDA drug labeling. This unique toxicity knowledge base for targeted cancer drugs could facilitate the development of computational models to illuminate the complex pathways of drug-induced toxicities that up until now have remained obscure.
Highlights.
-
➢
Innovative targeted anticancer drugs are often associated with unexpected toxicities
-
➢
There exists no comprehensive toxicity knowledge base for targeted anticancer drugs.
-
➢
Systematic studies of targeted anticancer drug-associated toxicities can facilitate drug discovery and toxicity prediction.
-
➢
We developed an integrated approach to extract drug-SE pairs from full-text oncological articles.
Acknowledgement
Xu and Wang have jointly conceived the idea, designed and implemented the algorithms and prepared the manuscript. All authors read and approved the final manuscript. We would like to thank the three curators from ThinTek for the manual curation.
Funding
RX was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under the NIH Directors New Innovator Award number DP2HD084068, the Training grant in Computational Genomic Epidemiology of Cancer (CoGEC) (R25 CA094186-06), and the Grant #IRG-91-022-18 to the Case Comprehensive Cancer Center from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The data is publicly available at: http://nlp.case.edu/public/data/TargetedToxicity_JCOFullText
References
- 1.Keefe DM, Bateman EH. Tumor control versus adverse events with targeted anticancer therapies. Nature Reviews Clinical Oncology. 2011;9(2):98–109. doi: 10.1038/nrclinonc.2011.192. [DOI] [PubMed] [Google Scholar]
- 2.Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, Druker B. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N. Engl. J. Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 3.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 4.Reguart N, Cardona AF, Rosell R. Role of erlotinib in first-line and maintenance treatment of advanced non-small-cell lung cancer. Cancer Manag. Res. 2010;2:143–156. doi: 10.2147/cmar.s5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleeland CS, Allen JD, Roberts SA, Brell JM, Giralt SA, Khakoo AY, Skillings J. Reducing the toxicity of cancer therapy: recognizing needs, taking action. Nature Reviews Clinical Oncology. 2012;9:471–478. doi: 10.1038/nrclinonc.2012.99. [DOI] [PubMed] [Google Scholar]
- 6.Kirk R. Targeted therapies: The toxic reality of new drugs. Nature Reviews Clinical Oncology. 2012;9(9):488–488. doi: 10.1038/nrclinonc.2012.134. [DOI] [PubMed] [Google Scholar]
- 7.Niraula S, Seruga B, Ocana A, Shao T, Goldstein R, Tannock IF, Amir E. The price we pay for progress: a meta-analysis of harms of newly approved anticancer drugs. Journal of Clinical Oncology. 2012;30(24):3012–3019. doi: 10.1200/JCO.2011.40.3824. [DOI] [PubMed] [Google Scholar]
- 8.Eschenhagen T, Force T, Ewer MS, de Keulenaer GW, Suter TM, Anker SD, Shah AM. Cardiovascular side effects of cancer therapies: a position statement from the Heart Failure Association of the European Society of Cardiology. European journal of heart failure. 2011;13(1):1–10. doi: 10.1093/eurjhf/hfq213. [DOI] [PubMed] [Google Scholar]
- 9.Ewer MS, Ewer SM. Cardiotoxicity of anticancer treatments: what the cardiologist needs to know. Nature Reviews Cardiology. 2010;7(10):564–575. doi: 10.1038/nrcardio.2010.121. [DOI] [PubMed] [Google Scholar]
- 10.Mellor HR, Bell AR, Valentin JP, Roberts RR. Cardiotoxicity associated with targeting kinase pathways in cancer. Toxicological Sciences. 2011;120(1):14–32. doi: 10.1093/toxsci/kfq378. [DOI] [PubMed] [Google Scholar]
- 11.Bonura F, Di Lisi D, Novo S, D’Alessandro N. Timely Recognition of Cardiovascular Toxicity by Anticancer Agents: A Common Objective of the Pharmacologist, Oncologist and Cardiologist. Cardiovascular toxicology. 2012;12(2):93–107. doi: 10.1007/s12012-011-9141-z. [DOI] [PubMed] [Google Scholar]
- 12.Ravaud A. How to optimise treatment compliance in metastatic renal cell carcinoma with targeted agents. Annals of oncology. 2009;20(suppl 1):i7–i12. doi: 10.1093/annonc/mdp073. [DOI] [PubMed] [Google Scholar]
- 13.Davies MA, Fox PS, Papadopoulos NE, Bedikian AY, Hwu WJ, Lazar AJ, Kim KB. Phase I study of the combination of sorafenib and temsirolimus in patients with metastatic melanoma. Clinical Cancer Research. 2012;18(4):1120–1128. doi: 10.1158/1078-0432.CCR-11-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molina AM, Feldman DR, Voss MH, Ginsberg MS, Baum MS, Brocks DR, Motzer RJ. Phase 1 trial of everolimus plus sunitinib in patients with metastatic renal cell carcinoma. Cancer. 2012;118(7):1868–1876. doi: 10.1002/cncr.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, Patnaik A. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clinical Cancer Research. 2012;18(8):2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 16.Chen MH, Kerkela R, Force T. Mechanisms of cardiac dysfunction associated with tyrosine kinase inhibitor cancer therapeutics. Circulation. 2008;118:84–95. doi: 10.1161/CIRCULATIONAHA.108.776831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3(114):114–127. doi: 10.1126/scitranslmed.3002774. [DOI] [PubMed] [Google Scholar]
- 18.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 19.Duran-Frigola M, Aloy P. Recycling side-effects into clinical markers for drug repositioning. Genome Med. 2012;4(3) doi: 10.1186/gm302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: From data to therapeutics. Clinical Pharmacology and Therapeutics. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 21.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seruga B, Sterling L, Wang L, Tannock IF. Reporting of serious adverse drug reactions of targeted anticancer agents in pivotal phase III clinical trials. J Clin Oncol. 29:174–185. doi: 10.1200/JCO.2010.31.9624. [DOI] [PubMed] [Google Scholar]
- 23.Xu R, Wang Q. Automatic signal prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS) J Biomed Inform. 2014:171–177. doi: 10.1016/j.jbi.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu R, Wang Q. Large-scale combining signals from both biomedical literature and the FDA Adverse Event Reporting System (FAERS) to improve post-marketing drug safety signal detection. BMC Bioinformatics. 2014;15:17. doi: 10.1186/1471-2105-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, Wang Q. Toward creation of a cancer drug toxicity knowledge base: automatically extracting cancer drug side effect relationships from literature. J Am Med Inform Assoc. 2014 Jan 1;21(1):90–96. doi: 10.1136/amiajnl-2012-001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu R, Wang Q. Automatic construction of a large-scale and accurate drug-side-effect association knowledge base from biomedical literature. J Biomed Inform. 2014 Jun 10; doi: 10.1016/j.jbi.2014.05.013. pii: S1532-0464(14)00138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, Wang Q. Combining automatic table classification and relationship extraction in extracting anticancer drug-side effect pairs from full-text articles. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R, Li L, Wang Q. Towards building a disease-phenotype knowledge base: extracting disease-manifestation relationship from literature. Bioinformatics. 29(17):2186–2194. doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R, Li L, Wang Q. dRiskKB: a large-scale disease-disease risk (causal) relationship knowledge base constructed from biomedical text. BMC Bioinformatics. 2014;15:105. doi: 10.1186/1471-2105-15-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurulingappa H, Mateen Rajput A, Toldo L. Extraction of potential adverse drug events from medical case reports. Journal of biomedical semantics. 2012;3(1):15–24. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Safety. 1999;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 32.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10–18. [Google Scholar]
- 34.Manning CD, Raghavan P, Schutze H. Introduction to information retrieval. Vol. 1. Cambridge: Cambridge University Press; [Google Scholar]
- 35.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research. 2008;36(suppl 1):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatonetti NP, Patrick PY, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Science translational medicine. 2012;4(125):125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]