Abstract
S100P signaling through the receptor for advanced glycation end-products (RAGE) contributes to colon cancer invasion and metastasis, but the mechanistic features of this process are obscure. Here, we investigate whether activation of S100P/RAGE signaling regulates oncogenic microRNA-21 (miR-21). We show that exogenous S100P up-regulates miR-21 levels in human colon cancer cells, whereas knockdown of S100P results in a decrease of miR-21. Furthermore, blockage of RAGE with anti-RAGE antibody suppresses S100P induction of miR-21. In addition, we found that S100P induction of miR-21 expression involves ERK and is suppressed by the MEK inhibitor U0126. Also, S100P treatment stimulates the enrichment of c-Fos, and AP-1 family members, at the miR-21 gene promoter.
INTRODUCTION
Approximately half of the patients diagnosed with colon cancer will develop liver metastasis1. Metastasis is the major cause of death in cancer patients and is largely considered incurable due to a lack of effective therapy other than hepatic resection2,3. Metastasis is a complex multi-factorial and multi-step process which promotes the detachment, migration, and proliferation of malignant lesions from the primary tumor site to distant site4,5. Defining the gene targets underlying the metastatic process is essential for the development of an effective targeted therapy6.
Inflammation plays a direct role in colorectal cancer progression. Several studies show that inflammation is associated with cancer progression and an increased infiltration of inflammatory cells and inflammatory molecules/factors are present in colon cancers during tumor progression (reviewed in Terzic et al.7). Recent studies by our group and others indicate that S100P is an important mediator of cancer related inflammation8–10. Extracellular S100P can act as a ligand for the receptor for advanced glycation endproducts (RAGE) and activate key signaling pathways such as extracellular regulated kinases (ERK1/2), NF-kB, and the JAK/STAT pathway10–12. S100P levels are increased in several cancers including colon cancers and are associated with metastasis13. Downstream target within the S100P/RAGE signaling pathway that contribute to cancer progression remain an active area of investigation.
Furthermore, the mechanistic linkage between inflammation and colon cancer progression remain to be elucidated. Recent studies indicate that microRNA (miRNAs) dysregulation represents a potential molecular mechanism for inflammatory pathways to mediate cancer development and progression14. Specifically, miR-21 has been shown to be over-expressed in many types of human cancers, including colon cancer15. Additional studies have demonstrated an association between elevated levels of miR-21 and down-regulation of several target genes such as programmed cell death 4 (PDCD4), tissue inhibitor of metalloproteinase 3 (TIMP3), phosphatase and tensin homolog (PTEN), Sprouty, and reversion-inducing cysteine-rich protein with Kazal motifs (RECK)16–18. Hence, these studies implicate miR-21 in the participation of several key biological processes important in the malignant phenotype. However, the factors that lead to the dysregulation of miR-21 expression have not been fully explored. In the present study, we investigate the effects of S100P/RAGE activation on the induction of miR-21 expression.
MATERIALS AND METHODS
Cell culture, S100P over-expression and stable lentiviral knock-down using shRNA
SW480 and LS174T human cancer cell lines were purchased from the ATCC and cultured in complete DMEM medium (DMEM 1X, 10% FBS and penicillin/streptomycin). The cells were incubated in humidified atmosphere of 5% CO2 at 37° C. We have previously described the generation of cells overexpressing S100P and knockdown of S100P in cells8,9. In regards to the generation of S100P overexpressing cells, one million SW480 or LS174T cells in 2 mL of OptiMEM medium were transfected according to instructions of Lipofectamine 2000 (Invitrogen). Cells were selected with 500µg/mL of G418 and S100P expression was confirmed by western blots. To knockdown S100P levels in colon cancer cells, lentiviral production for pLKO.1, pLKO.1/sh1-S100P and pLKO.1/sh2-S100P and infection were performed according to the RNAi Consortium protocol (http://www.broadinstitute.org/rnai/trc). Envelope (pCMV-dR8.2 dvpr) and packaging (pCMV-VSV-G) plasmids were obtained from ADDGENE Inc. The lentivirus particles were titrated by infecting one million LS174T cells with 15µL, 25µL, 50µL, 100µL, 250µL and 500µL particles. Cells were selected with 2µg/mL of puromycin. Confirmation of S100P knock-down expression was done by qRT-PCR and western blot analyses. Cells transduced with 100µL of viral particles were used for further experiments.
Expression and purification of S100P
The expression and purification of human recombinant S100P protein was performed as previously described by our group8. Briefly, full length human S100P cloned in pTRCHis2 vector was transformed in TOP10 E. coli (Invitrogen). His-S100P was purified using the Probond resin column (Invitrogen) as described by the manufacturer. Purified protein was concentrated with AMICON centrifugal filters. SDS-PAGE and Western blotting confirmed purity of the protein.
RNA isolation and qRT-PCR analysis
The procedures for RNA isolation and qRT-PCR have been previously described by our group8. Small RNAs were isolated with mirVana miRNA Isolation kit (Ambion) and total RNA was isolated with Qiagen RNA isolation kit (Qiagen). First strand cDNA for miR-21 and U6 were synthesized in the same reaction with Reverse transcriptase (Fermentas) using 20 pmoles of stem-loop RT primer (miR-21: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3’, U6: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATATG-3’) and 0.5µg of miRNA. QRT-PCR was performed with cDNA representing 50ng of miRNA/reaction in quadruplicates with forward primers only and the LightCycler 480 SYBR Green I Master (Roche) mix. The sequence of miR-21 forward primer was 5’-GCCCGCTAGCTTATCAGACTGATG-3’ and for U6 is 5’-GCGCGTGAAGCGTTC-3’. Data were normalized to U6 levels.
The binding of recombinant S100P to the RAGE was blocked with anti-RAGE monoclonal antibody (R&D Systems) as previously described by our group8. Two millions SW480 cells were seeded overnight in plates with complete medium. Cells were starved overnight in OptiMEM medium and incubated at 37° C, 5% CO2. Next day, 40µg/mL of anti-RAGE monoclonal antibody was added and after 2hrs 200nM of hr-S100P was added to the cells. Cell were pre-treated with blocking anti-RAGE antibody and then treated with recombinant S100P for different time periods (0, 20 min, 1hr, 2hrs, 24hrs) followed by qRT-PCR to measure miR-21 induction as described above.
Western Blot Analysis
To determine the activation of ERK1/2, AP-1 and NF-kB by S100P/RAGE signaling, two millions SW480 cells were plated overnight in culture plates in complete medium. Next day, cells were serum starved overnight in OptiMEM medium. The following day, cells were treated with 200nM of hr-S100P and retrieved at different time intervals (0, 15min, 30min, 1hr and 2hrs). Activation of pERK1/2, p c-fos, and pNF-kB-p-p65 was determined from protein extracts as previously described19.
Pharmacologic inhibition
Two millions SW480 cells were pre-treated with chemical inhibitors for AP-1 and NF-kB signaling. Since ERK1/2 transcribes c-fos and activates c-fos and c-jun (components of AP-1), we used U0126 to inhibit AP-1 transcription and activity by inhibiting ERK1/2, and CAY10512 to inhibit NF-kB20. One million cells were seeded overnight in culture plates. Next day, cells were pre-treated separately with 10µM of U0126 for 30min and with 0.5µM of CAY10512 for an hour. Cells were treated with 100nM of hr-S100P for 24hrs and control samples with DMSO. Cells were harvested to determine miR-21 expression levels by qRT-PCR as described above.
Luciferase assays
We obtained the luciferase wild-type (WT) and mutant constructs of the pri-miR-21 promoter from Dr. Heike Allgayer21. Briefly, mutations of AP-1 are at positions −59/−52 bp (AP-1-I), −166/−159 bp (AP-1-II), −225/−220 bp (AP-1-III), and at −656/−663 (AP-1-IV). We used two mutant constructs, one containing AP-1 mutations at AP-1/I, AP-1/II and AP-1/III, and the second containing all AP-1 mutations (AP-1/I – AP-1/IV) and one NF-kappa B mutation at −209/−211. Five hundred thousand cells/well were seeded in 12-well plates overnight at 37° C, 5% CO2 in complete medium. Next day, transfections were performed according to reagent, TranslT-LT1 (Mirus Bio LLC) instructions. Co-transfection of each Firefly luciferase construct with a Renilla Luciferase control reporter vector (Promega) was done in replicas of three. Plasmid control for the pri-miR-21 promoter constructs was pGL3-Basic. Next day after transfection, 200nM of hr-S100P in complete medium was added to the cells and thereof every 24hrs for 48hrs. Cell homogenization and luciferase assays were performed according to Dual-Luciferase Reporter Assay System kit’s instructions (Promega). Luciferase activity was measured with Sirius Luminometer. Experiments were performed in triplicates.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to EpiSeeker ChIP Kit-One Step (ABCAM) protocol using the c-fos and phospho-NF-kB-p65 antibodies. One million SW480 cells were seeded overnight in complete medium in culture dishes. Next day, cells were stimulated with 200nM hr-S100P for 24hrs. Cells were then rinsed with PBS, fixed with 10 % formaldehyde/PBS for 1hr and scraped into lysis buffer for chromatin immunoprecipitation as described previously19. Binding of c-fos and phospho-NF-kB-p65 to the pri-miR-21 promoter was measured by qRT-PCR with SYBR Green and respective primers for AP-1 and NF-kB binding. We designed two sets of primers (AP-1(1) and AP-1(2)) comprising the AP-1 regions mutated by Mudduluru21, AP-1 binding motifs, − 50/−45 bp, −59/−52 bp, −66/−159 bp, −656/−663 bp, −692/−698 bp, −701/−705 bp, and a motif after the TSS 26/33 bp. Primer set AP-1 (1) sequences are, FW-1: 5’-GCC TCC CAA GTT TGC TAA TG-3’ and REV-1: 5’-TGT ACT CTG GTA TGG CAC AAA GA-3’, and it includes AP-1 binding motifs at AP-1/I, AP-1-II, AP-1-III (see Luciferase Assays above), which are proximal to the transcription start site (TSS). Primer set AP-1 (2) (FW-2: 5’-GAG ATC AGG CCA TTG CAC TC-3’ and REV-2: 5’-GCA ACA CTG CCT AAT GCT TG-3’) includes the AP-1 binding motif AP-1-IV. Primers sequences for NF-kB (1) comprise regions outside the promoter and were obtained from literature22. NF-kB (2) primer sequences (FW: 5’-CTA TCC CAA TCA TCT CAG AAC AAG CTG TTA CTA-3’ and REV: 5’-GGA CAA TCT GTG CGT CAT CCT TAT CC-3’) comprise two NF-kB binding motifs at positions −300/−308 bp and −301/−292 bp from the TSS within the promoter region.
The Cancer Genome Atlas Data Analysis (TCGA)
Exon expression analysis of S100P, RAGE, mir21 and RECK in The Cancer Genome Atlas dataset of colorectal adenocarcinoma (COADREAD) was performed by RNA sequencing (IlluminaHiSeq) of primary colorectal tumors (n=347) and normal solid tissues (n=47). Statistical analysis was performed using Student’s T-test with Benjamini-Hochberg correction with p<0.05 considered as statistically significant.
Statistical analysis
All experiments were performed at least in triplicates. Results are presented as means of ±S.E.M. Statistical comparisons between two groups were made using two-tailed unpaired Student's t-test. A p-value of <0.05 was considered statistically significant. Any p-value of <0.05 is denoted as *, p-value of <0.01 as **, and p-value of <0.001 denoted ***.
RESULTS
S100P/RAGE up-regulates the expression of miR-21
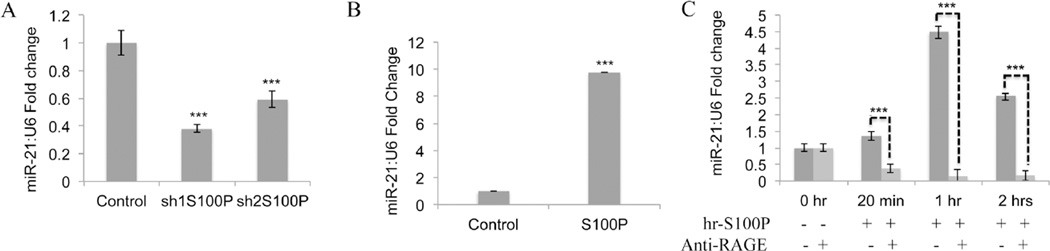
To first investigate the relationship between miR-21 and S100P expression, we accessed whether changes in S100P expression could alter miR-21 levels in colon cancer cells. Studies by our group have previously shown that LS174T cells express S100P protein and knockdown of S100P affects cell proliferation and motility9. Interestingly, knock-down of S100P in LS174T cells form smooth colonies lacking filopodia formation compared to the controls, whereas over-expression of S100P in LS174T cells have the opposite effect. S100P over-expression induces filopodia formation and invasion of cells into the surrounding collagen (Supplemental Figure S1A). S100P overexpressing in both LS174T and SW480 cells also display a more invasive phenotype (Supplemental Figure S1B). These data are in agreement with previous findings regarding the effects of genetic maninpulation of S100P levels in colon cancer cells9,10,23,24. The knockdown of S100P led to reduced miR-21 levels in the LS174T cells (Figure1A). We also found that the over expression of S100P resulted in significant elevation of miR-21 in LS174T cells (Figure1B). These data suggest that S100P may regulate miR-21 expression in human colon cancer cells.
Figure 1. S100P/RAGE signaling induces miR-21 expression.
A. S100P knock-down in LS174T decreases miR-21 mRNA levels. B. S100P over-expression increases significantly miR-21 expression in LS174T colon cancer cells. C. Induction of miR-21 expression at 20min, 1hr and 2 hrs by hr-S100P is inhibited by anti-RAGE blocking antibody in SW480 cells. Expression levels of miR-21 were normalized to U6 levels. Error bars represent mean ± SEM, Statistical significance was calculated using two tailed Student’s t-test, p< 0.05 =*, p< 0.01 = **, p< 0.001 = ***.
Because extracellular S100P can regulate microRNA expression via the RAGE8, we evaluated whether S100P control of miR-21 expression is RAGE mediated. Previous studies demonstrated that SW480 cells express the RAGE but not S100P protein10. SW480 cells were treated with purified human recombinant S100P for different time points and analyzed for changes in miR-21 expression using qRT-PCR. The levels of miR-21 began to increase within 20 min after exposure to recombinant S100P and were maximal at 1hr after S100P treatment. A similar rapid induction of pri-miR-21 and miR-21 expression has been observed in human hepatocellular carcinoma cells treated with Dehydroepiandrosterone (DHEA)25. Blocking anti-RAGE antibody attenuated recombinant S100P induction of miR-21 at the time points tested (Figure 1C). Collectively, these results indicate that the stimulation of miR-21 by S100P is RAGE dependent.
S100P/RAGE transcriptionally regulates miR-21 promoter through AP-1
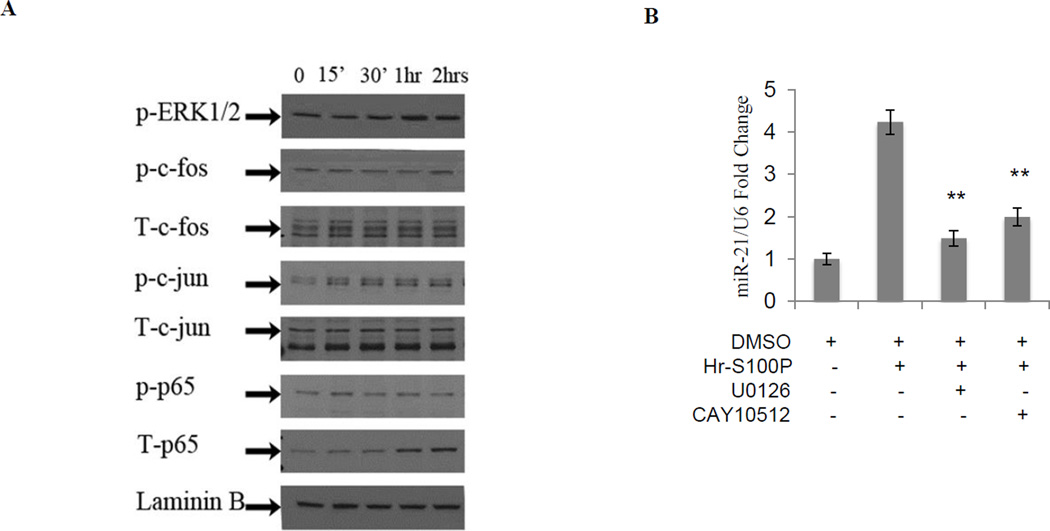
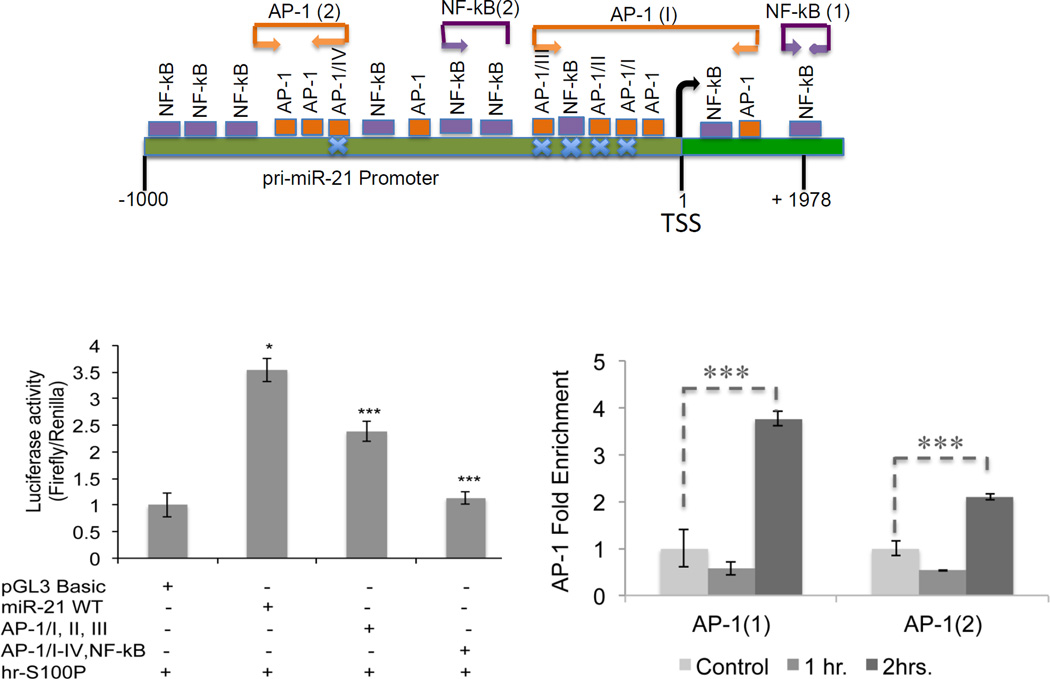
Using the ChIP bioinformatics, MAPPER (Multi-genome Analysis of Positions and Patterns of Elements of Regulation), which combines TRANSFAC and JASPAR matrices (http://mapper.chip.org/), we found ten AP-1 (four of them previously reported and mutated21) and seven NF-kB binding sites (data not shown). We confirmed our previous observation that extracellular S100P could stimulate both ERK and NF-kB signaling pathways (Figure 2A)8. Next, we analyzed the effect of pharmacologic blockage on S100P stimulation of miR-21 in colon cancer cells. We found that both U0126 and CAY10512 reduce miR-21 expression compared to controls (Figure 2B). To further elucidate the involvement of AP-1 and NF-kB in S100P-mediated induction of miR-21, we measured luciferase activity of S100P induced SW480 cells transfected with constructs containing either wild type (WT), or mutated AP-1 binding sequences (AP-1/I, AP-1/II, AP-1/III) or mutated AP-1 and NF-kB sequences of the pri-miR-21 promoter (AP-1/I, AP-1/II, AP-1/III, AP-1/IV and NF-kappa B) (Figure 3A). These results show that S100P decreases the activity of mutated pri-miR-21 promoter compared to WT construct at 24hrs (Figure 3B). We observed a similar induction of pri-miR-21 promoter induction in LS174T and SW480 cells over expressing S100P (Supplemental Figure S2). These data suggest that S100P/RAGE signaling activates the pri-miR-21 promoter via AP-1. To verify these observations, we performed Chip assays in SW480 stimulated with S100P at the indicated time points. The results showed that S100P treatment enhanced c-Fos occupancy of the pri-miR-21 promoter at 2hrs (Figure 3C). In contrast, p-p65 does not bind to the pri-miR-21 promoter (data not shown). Taken together, these data show that S100P stimulate expression of miR-21 via AP-1 signaling.
Figure 2. S100P activates ERK and NF-kB signaling.
A. Serum-starved SW480 cells were induced with hr-S100P. Analysis of protein levels by western blots shows activation of ERK1/2 after 30min of cells with S100P. AP-1 components c-fos and c-jun are activated after 2hrs and 15min of hr-S100P treatment, respectively. NF-kB (p65) activation was on and off at 15min and 2hrs, respectively. Hr-S100P treatment increased expression of total c-fos (T-c-fos) at 15min and total p-65 at 1hr. Laminin B is the sample loading control. B. qRT-PCR analysis of SW480 cells induced with hr-S100P and treated with U0126 and CAY10512 inhibit ERK1/2 and NF-kB respectively, showing about 50% decrease in miR-21 expression levels. Error bars represent mean ± SEM, statistical significance was calculated using two tailed Student’s t-test, p< 0.05 =*, p< 0.01 = **, p< 0.001 = ***.
Figure 3. S100P/RAGE signaling activates binding of AP-1 to the pri-miR-21 promoter.
A. pri-miR-21 promoter (1 to −1000 bp) within the sequence of chromosome 17 showing the AP-1 and NF-kB binding sites and regions within the primer sets (AP-1(1), AP-1(2) and NF-kB(2). NF-kB (1) primer set for NF-kB binding site is within the sequence of the pri-miR-21 precursor (AY699265). B. pri-miR-21 promoter activity decreases significantly in SW480 cells transfected with pri-miR-21 promoter constructs mutated on AP-1 binding sites (AP-1/I, AP-1/II, AP-1/III and AP-1/IV) and on NF-kB binding site 24hrs after hr-S100P treatment compared to WT promoter. C. c-fos binding to the pri-miR-21 promoter within the regions of AP-1(1) and AP-1(2) primer sets increases 3.5-fold enrichment compared to the control after 2hrs of hr-S100P treatment. Blue crosses represent mutation sites. Firefly luciferase activity was normalized to Renilla activity of the pri-miR-21 promoter constructs. Error bars represent mean±SEM, Statistical significance was calculated using two tailed Student’s t-test, p< 0.05 =*, p< 0.01 = **.
S100P, RAGE, miR-21, and RECK expression in human colon cancers
MicroRNA-21 (miR-21) targets key genes such as RECK. Reversion-inducing cysteine-rich protein with Kazal motifs (RECK) is a tumor and metastasis suppressor gene, that is central for regulating invasive and metastatic tumor cell activity. RECK is an enzyme that regulates MMP-9 expression and is down-regulated in certain malignancies, and its expression is positively correlated with cancer patient survival26. We found that over-expression of S100P could lead to a decrease in RECK protein levels in colon cancer cells (Supplemental Figure S3).
TCGA expression data
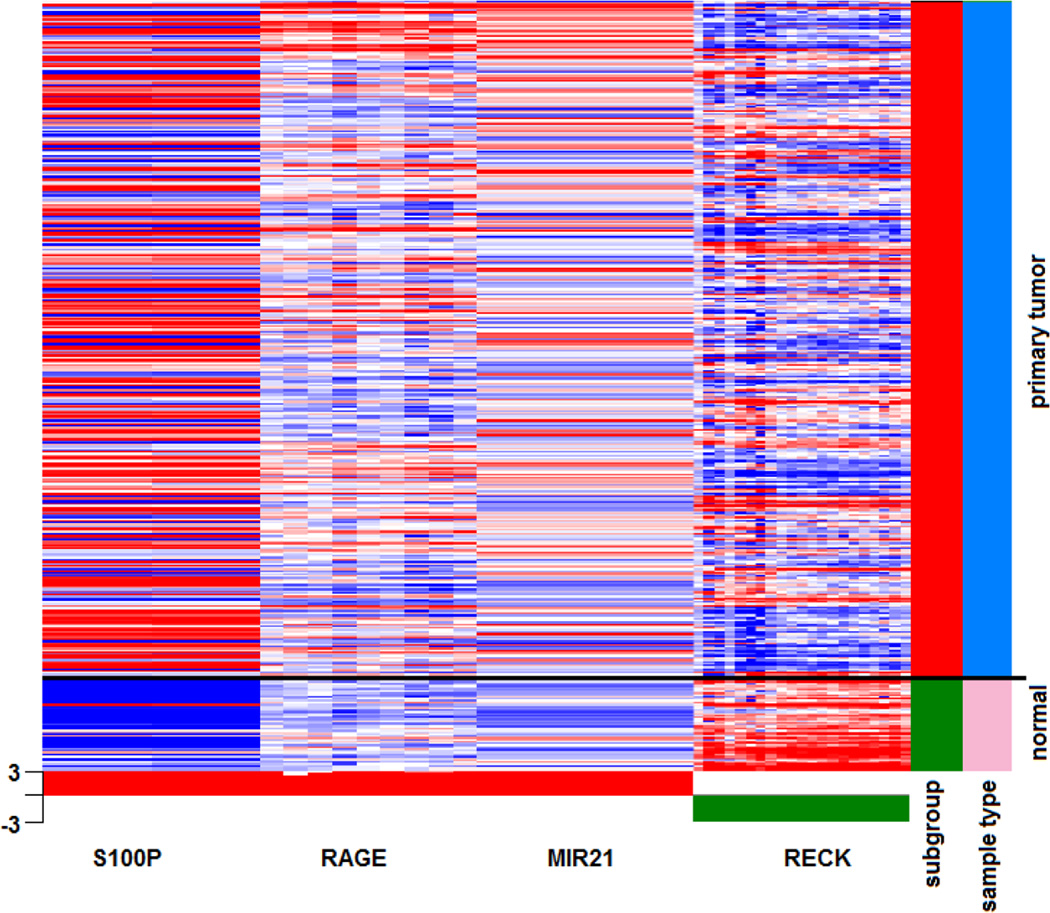
Given the role of the S100P/RAGE/miR-21 role in tumor progression and metastasis, we sought to determine the relationship of the S100P/RAGE/miR-21 and RECK in human colon cancer by mining The Cancer Genome Atlas (TCGA) database. Expression analysis revealed that S100P, RAGE (Advanced glycation endproduct receptor, gene encoding RAGE (AGER)), and miR-21 were significantly over-expressed in primary tumors (n=347) compared to normal control tissues (n=47). Whereas, the miR-21 target gene RECK was expressed in normal controls and decreased in cancer cases (Figure 4). This inference is consistent with our in vitro findings. Furthermore, previous studies have shown that high expression of S100P and miR-21 are associated with decreased survival13,27.
Figure 4. Expression analysis of S100P, RAGE, miR-21, and RECK in primary colorectal tumors using the TCGA database.
Exon expression analysis of S100P, RAGE, miR-21 and RECK in the TCGA dataset of colorectal adenocarcinoma (COADREAD) was performed by HiSeq Illumina sequencing of RNA samples obtained from fresh frozen tissues of primary colorectal tumors (in red, n=347) and normal solid tissues (in green, n=47). Red and blue in gene expression dataset represent over- and under-expression, respectively. The heatmap indicates increased expression of S100P, RAGE, and miR-21 and decreased expression of RECK in primary tumors compared to normal tissue. Statistical analysis was performed using Student’s t-test with Benjamini-Hochberg correction with p<0.05 considered as statistically significant.
DISCUSSION
A key characteristic of malignant colon cancer includes the ability to invade and metastasize. The molecular mechanisms and pathologic process involved in metastasis are beginning to be elucidated. Examples include the inflammatory microenvironment which appears to be a critical factor in the dysregulation of microRNA expression14.
In the present study, we demonstrate that activation of the S100P/RAGE signaling can lead to the up-regulation of oncogenic miR-21 via AP-1 signaling pathway. Our previous studies show that the expression of S100P can be stimulated by inflammatory prostaglandin E2 (PGE2)9. These data are consistent with previous findings demonstrating a functional role of miR-21 in colon cancer progression and important role of inflammation in enhancing miR-21 expression14,20,28. Results presented in this article indicate for the first time that S100P/RAGE signaling may be upstream factor that can influence the expression of oncogenic miR-21.
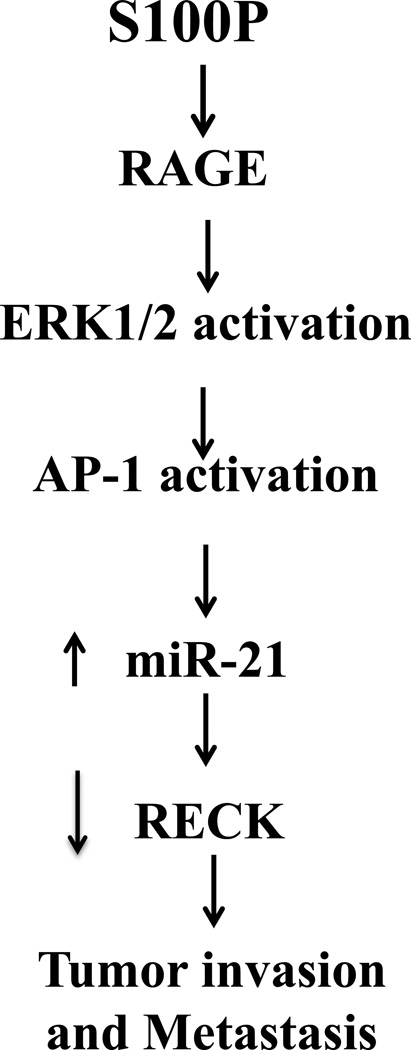
Increased expression levels of miR-21 have been found in many types of human cancers including colon cancer15. Additional studies have demonstrated an association between elevated levels of miR-21 and down-regulation of several target genes, such as PDCD4, TIMP3, PTEN, Sprouty, and RECK16–18. Our in vitro studies, bioinformatic analysis of the TCGA database, and western blot analysis demonstrate that the activation of the S100P/RAGE/miR-21 pathway can lead to the downregulation of RECK. Since RECK serves as an inhibitor of metalloproteinase activities of MT1-MMP, MMP-2 and MMP-929–31, the repression of RECK could lead to increased metalloproteinase activities in cells allowing the remodeling of ECM and consequently facilitating the onset of invasion and metastasis. Our studies are the first to show that S100P/RAGE signaling up-regulates miR-21 expression which could potentially lead to RECK repression, suggesting a role in the rearrangement or dissociation of cell-ECM interactions (Figure 5). However, further more detailed studies are required.
Figure 5. Proposed model of how S100P/RAGE signaling can promote colorectal cancer progression by linking inflammation to RECK down-regulation via the induction of miR-21.
Although extracellular S100P/RAGE signaling has been implicated in metastasis, our results further indicate that S100P/RAGE signaling can regulate expression of oncogenic miR-21. In addition, the regulation of miR-21 expression by S100P/RAGE signaling appears to be AP-1 dependent. Furthermore, our results support the notion that S100P could be a biomarker of early metastatic processes and functional blocking of extracellular S100P/RAGE signaling might be efficient for the treatment of metastatic disease.
Supplementary Material
Figure S1, A) S100P knock-down in LS174T cells leads to decrease motility in 3D collagen assays (top); whereas S100P over-expression in SW480 cells confers a migratory phenotype in 3D collagen assay. LS174T cells were stably transfected with empty pLKO1 vector (control) or two independent shRNA constructs against S100P (sh1-S100P and sh2-S100P) as previously described9. SW480 cells were stably transfected with empty vector pcDNA3.1 or pcDNA.1/S100P vector. The S100P proteins status of tumor cells lines was confirmed by by western blotting using S100P-specific antibody (1:1,000). Equal loading was confirmed by probing against α-tubulin or GAPDH. The collagen gel assay for EMT was performed essentially as described by with modification32. One thousand cells mixed with 300 µL of 1mg/mL of collagen (Invitrogen 100) in 10X M199/2.2% NaHCO3 were seeded in 24-well plates. The gels were allowed to polymerize for 4 hours at 37° C, 5% CO2. Six hundreds µL of complete medium were added on the polymerized gels and incubated for 12 days under the same conditions. Complete medium was added when needed. Pictures were taken under the microscope. B) Over-expression of S100P in LS174T and SW480 cells increases cell invasion. One hundred thousand control vector cells (LS147T-pcDNA 3.1 and SW480-pcDNA 3.1) or S100P over expressing cells (LS174T-pcDNA 3.1-S100P and SW480-pcDNA 3.1-S100P) were seeded on Transwell containing Matrigel. OpiMEM medium was added to the bottom chamber and cells were cells were incubated at 37° C, 5% CO2 for 24 hrs. The outer surfaces of the filters were stained with 50 µL of 0.5% of crystal violet/20% methanol for 1 min. Inserts were washed with distilled water, cleaned with cotton swabs and air dried overnight. The number of invaded cells was enumerated under the microscope. Experiments were repeated three times. The number of invaded cells are reported as mean ± SEM (*p<0.05).
Figure S2 A) S100P over-expression in LS174T cells (A) and SW480 cells (B) increase pri-miR-21 promoter activity. Transient transfection with wild-type and mutant pri-miR-21 promoter constructs was performed as described in Materials and Methods. Luciferase activity was assayed using the Dual Luciferase Reporter™ Assay (Promega) according to manufacturer's directions and luminescence was measured using the Sirius Luminometer (Berthold Detections Systems). Experiments were repeated at least three times. The results are reported as mean ± SEM (*p<0.05).
Figure S3. Reduced expression of RECK in LS174T cells over-expressing S100P. Protein isolation and Western analysis were performed as described in Materials and Methods. Western blots were reacted with 1:250 RECK antibody (BD Biosciences)/TBS dilution and 1:10,000 dilution of mouse IgG-HRP antibody. GAPDH was used as the loading controls.
Highlights.
In this paper we show the following:
Activation of S100P/RAGE signaling induces expression of the oncomiR- miR-21
The induction of miR-21 by S100P/RAGE signaling is AP-1 dependent
S100P, RAGE receptor, and miR-21 expression are inversely correlated with the miR-21 target gene RECK in human colorectal cancers
Acknowledgements
We would like to thank Dr. Thiru Arumugan and Dr. Craig D. Logsdon from M.D. Anderson Cancer Center for providing us the human recombinant S100P construct, and Dr. Giridhard Mudduluru and Dr. Heike Allgayer from Ruprecht-Karls-University of Heidelberg in Germany for supplying the luciferase wild type and AP-1 mutant pri-miR-21 promoter constructs.
Funding Support
This work was supported by CA 95060 (MEMP and MAN), ACS/IRG 74-001-34 (MEMP), and the Michele and Grant Senner Endowment (MAN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
Experimental design: MEMP, MAN, BCO
Execution of experiments: MEMP, BCO, ACP, QL
Data Analysis: MEMP, JJ
Preparation of manuscript: MEMP
Critical review: MAN, JJ
References
- 1.Sheth KR, Clary BM. Management of hepatic metastases from colorectal cancer. Clinics in colon and rectal surgery. 2005;18:215–223. doi: 10.1055/s-2005-916282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Elias D, Liberale G, Vernerey D, et al. Hepatic and extrahepatic colorectal metastases: when resectable, their localization does not matter, but their total number has a prognostic effect. Annals of surgical oncology. 2005;12:900–909. doi: 10.1245/ASO.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Gout S, Huot J. Role of cancer microenvironment in metastasis: focus on colon cancer. Cancer microenvironment : official journal of the International Cancer Microenvironment Society. 2008;1:69–83. doi: 10.1007/s12307-008-0007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse JC, Kalluri R. Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. Journal of cellular biochemistry. 2007;101:816–829. doi: 10.1002/jcb.21215. [DOI] [PubMed] [Google Scholar]
- 6.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114. doi: 10.1053/j.gastro.2010.01.058. e5. [DOI] [PubMed] [Google Scholar]
- 8.Onyeagucha BC, Mercado-Pimentel ME, Hutchison J, Flemington EK, Nelson MA. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Experimental cell research. 2013;319:2081–2090. doi: 10.1016/j.yexcr.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandramouli A, Mercado-Pimentel ME, Hutchinson A, et al. The induction of S100p expression by the Prostaglandin E(2) (PGE(2))/EP4 receptor signaling pathway in colon cancer cells. Cancer biology & therapy. 2010;10:1056–1066. doi: 10.4161/cbt.10.10.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuentes MK, Nigavekar SS, Arumugam T, et al. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Diseases of the colon and rectum. 2007;50:1230–1240. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 11.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) The Journal of biological chemistry. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 12.Rehbein G, Simm A, Hofmann HS, Silber RE, Bartling B. Molecular regulation of S100P in human lung adenocarcinomas. International journal of molecular medicine. 2008;22:69–77. [PubMed] [Google Scholar]
- 13.Wang Q, Zhang YN, Lin GL, et al. S100P, a potential novel prognostic marker in colorectal cancer. Oncology reports. 2012;28:303–310. doi: 10.3892/or.2012.1794. [DOI] [PubMed] [Google Scholar]
- 14.Peacock O, Lee AC, Cameron F, et al. Inflammation and MiR-21 Pathways Functionally Interact to Downregulate PDCD4 in Colorectal Cancer. PloS one. 2014;9:e110267. doi: 10.1371/journal.pone.0110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faltejskova P, Besse A, Sevcikova S, et al. Clinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cells. International journal of colorectal disease. 2012 doi: 10.1007/s00384-012-1461-3. [DOI] [PubMed] [Google Scholar]
- 16.Mudduluru G, George-William JN, Muppala S, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Bioscience reports. 2011;31:185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- 17.Sayed D, Rane S, Lypowy J, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Molecular biology of the cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clinica chimica acta; international journal of clinical chemistry. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 19.Chandramouli A, Onyeagucha BC, Mercado-Pimentel ME, et al. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer biology & therapy. 2012;13:175–183. doi: 10.4161/cbt.13.3.18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncia JV, Santella JB, 3rd, Higley CA, et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorganic & medicinal chemistry letters. 1998;8:2839–2844. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- 21.Mudduluru G, George-William JN, Muppala S, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Bioscience reports. 2011;31:185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- 22.Shin VY, Jin H, Ng EK, et al. NF-kappaB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011;32:240–245. doi: 10.1093/carcin/bgq240. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Lai YK, Zhang J, et al. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Molecular medicine. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Z, Deng H, Fang Y, et al. Identification of the interplay between SOX9 and S100P in the metastasis and invasion of colon carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y, Radde BN, Litchfield LM, et al. Dehydroepiandrosterone Activation of G-protein-coupled Estrogen Receptor Rapidly Stimulates MicroRNA-21 Transcription in Human Hepatocellular Carcinoma Cells. The Journal of biological chemistry. 2015;290:15799–15811. doi: 10.1074/jbc.M115.641167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Tseng SH. The potential of RECK inducers as antitumor agents for glioma. Anticancer research. 2012;32:2991–2998. [PubMed] [Google Scholar]
- 27.Kjaer-Frifeldt S, Hansen TF, Nielsen BS, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. British journal of cancer. 2012;107:1169–1174. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, Byrnes K, Han C, Wang Y, Wu T. miR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Molecular cancer research : MCR. 2014;12:890–900. doi: 10.1158/1541-7786.MCR-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noda M, Oh J, Takahashi R, Kondo S, Kitayama H, Takahashi C. RECK: a novel suppressor of malignancy linking oncogenic signaling to extracellular matrix remodeling. Cancer metastasis reviews. 2003;22:167–175. doi: 10.1023/a:1023043315031. [DOI] [PubMed] [Google Scholar]
- 30.Oh J, Takahashi R, Kondo S, et al. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell. 2001;107:789–800. doi: 10.1016/s0092-8674(01)00597-9. [DOI] [PubMed] [Google Scholar]
- 31.Shi Z, Zhang J, Qian X, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer research. 2013;73:5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 32.Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelial-mesenchymal transformation during cardiac valve formation. Developmental biology. 2007;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, A) S100P knock-down in LS174T cells leads to decrease motility in 3D collagen assays (top); whereas S100P over-expression in SW480 cells confers a migratory phenotype in 3D collagen assay. LS174T cells were stably transfected with empty pLKO1 vector (control) or two independent shRNA constructs against S100P (sh1-S100P and sh2-S100P) as previously described9. SW480 cells were stably transfected with empty vector pcDNA3.1 or pcDNA.1/S100P vector. The S100P proteins status of tumor cells lines was confirmed by by western blotting using S100P-specific antibody (1:1,000). Equal loading was confirmed by probing against α-tubulin or GAPDH. The collagen gel assay for EMT was performed essentially as described by with modification32. One thousand cells mixed with 300 µL of 1mg/mL of collagen (Invitrogen 100) in 10X M199/2.2% NaHCO3 were seeded in 24-well plates. The gels were allowed to polymerize for 4 hours at 37° C, 5% CO2. Six hundreds µL of complete medium were added on the polymerized gels and incubated for 12 days under the same conditions. Complete medium was added when needed. Pictures were taken under the microscope. B) Over-expression of S100P in LS174T and SW480 cells increases cell invasion. One hundred thousand control vector cells (LS147T-pcDNA 3.1 and SW480-pcDNA 3.1) or S100P over expressing cells (LS174T-pcDNA 3.1-S100P and SW480-pcDNA 3.1-S100P) were seeded on Transwell containing Matrigel. OpiMEM medium was added to the bottom chamber and cells were cells were incubated at 37° C, 5% CO2 for 24 hrs. The outer surfaces of the filters were stained with 50 µL of 0.5% of crystal violet/20% methanol for 1 min. Inserts were washed with distilled water, cleaned with cotton swabs and air dried overnight. The number of invaded cells was enumerated under the microscope. Experiments were repeated three times. The number of invaded cells are reported as mean ± SEM (*p<0.05).
Figure S2 A) S100P over-expression in LS174T cells (A) and SW480 cells (B) increase pri-miR-21 promoter activity. Transient transfection with wild-type and mutant pri-miR-21 promoter constructs was performed as described in Materials and Methods. Luciferase activity was assayed using the Dual Luciferase Reporter™ Assay (Promega) according to manufacturer's directions and luminescence was measured using the Sirius Luminometer (Berthold Detections Systems). Experiments were repeated at least three times. The results are reported as mean ± SEM (*p<0.05).
Figure S3. Reduced expression of RECK in LS174T cells over-expressing S100P. Protein isolation and Western analysis were performed as described in Materials and Methods. Western blots were reacted with 1:250 RECK antibody (BD Biosciences)/TBS dilution and 1:10,000 dilution of mouse IgG-HRP antibody. GAPDH was used as the loading controls.