Abstract
Insertion of green fluorescent protein (GFP) encoding-gene into virus genes has provided a valuable tool for flavivirus research. This study aimed to develop dengue virus (DENV) replicons expressing GFP reporter that would provide a fast in vitro system to analyze functional roles of specific DENV sequences in viral replication. Two classes of recombinant replicon constructs were generated; one was a RNA-launched replicon with a GFP gene directly inserted into a full-length DENV genome (FL-DENV/GFP), and the other consisted of 4 types of DNA-launched DENV subgenomic replicons with GFP replacement at various structural genes (Δ-DENV/GFP). The FL-DENV/GFP resulted in GFP expression in transfected cells with no viable DENV being recovered from the transfection. The Δ-DENV/GFP constructs with partial structural gene deletion (ΔC-, ΔCprM/M-, ΔprM/M-, or ΔE-) expressed bright and long lasting GFP. The GFP expression intensity in living cells correlated well with the level of RNA replication. Various mutations in the 5′noncoding region of DENV-2 previously shown to be important genetic determinants for virus replication and mouse virulence were incorporated into the 5 different replicon constructs. Characterizations of 29 mutants demonstrated that these replicons can provide a useful platform for a quick and powerful in vitro system to analyze genetic determinants of DENV replication. These constructs can also be useful for development of vectors expressing foreign genes for various researches.
Keywords: Dengue virus type 2, Subgenomic replicons, DENV/GFP plasmid, 5′ NCR mutation
1. Introduction
Dengue virus (DENV) is a positive strand RNA virus in the Flaviviridae family. This virus family member includes yellow fever virus (YFV), Japanese encephalitis virus (JEV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV). DENV is transmitted to humans by infected mosquitoes and causes diseases, ranging from mild, self-limiting dengue fever, to severe dengue hemorrhagic fever, and dengue shock syndrome. The global distribution of DENV results in more than 50 million cases of infection, and an estimated 2.5 billion people are at risk each year (Gubler, 2002). Based on distinct antigenic characteristics, DENVs are divided into four serotypes; DENV-1, -2, -3 and -4. Re-infection with a second serotype virus increases the risk of developing the more severe dengue hemorrhagic fever/shock syndrome due to induction of antibody-dependent enhancement of the infection and/or T-cell mediated immunopathogenesis (Halstead et al., 1984; Rothman and Ennis, 1999). Although recent advances in DENV vaccine development puts several potential candidates on pre-clinical and clinical trials, there is currently no DENV vaccine yet available, partly due to lack of understanding of viral and cellular factors during the viral infection.
The positive-strand RNA genome of DENV is organized into 5′ noncoding region (5′NCR)-capsid protein (C)-pre membrane/membrane (prM/M)-envelope protein (E)-non-structural proteins 1–5 (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5)-3′NCR. The C protein is essential in encapsidation of the viral genome. prM/M protein serves as a chaperon for E protein during protein processing in cells and is involved in maturation of the virions. The mature viral envelope contains the M and E transmembrane proteins. The E protein is responsible for cell receptor binding, induction of the major neutralizing antibody response, mediation of virus-specific membrane fusion in acid pH endosomes, and virus assembly in cells. The NS proteins are involved in host cellular immune response, helicase and protease enzyme activity, and viral RNA-dependent RNA replication. As with the other positive strand RNA viruses, the 5′NCR and 3′NCR of DENV are predicted to form secondary RNA structures and cyclization which interact with cellular and viral proteins for viral protein translation (Ali and Siddiqui, 1995) and RNA genome replication (Guesdon et al., 2001; Kuhn et al., 2002). Sequences of 5′NCR together with capsid protein gene and 3′NCR are required to structurally interact during virus replication (Alvarez et al., 2005; You et al., 2001). The functional significance of dengue 5′NCR is demonstrated by the restricted growth of DENV-4 deletion mutants (Cahour et al., 1995), attenuation effect of the DENV-2 PDK-53 vaccine candidate (Butrapet et al., 2000), and reduced virus replication of DENV-2 5′NCR mutants (Leardkamolkarn et al., 2010; Sirigulpanit et al., 2007).
Accumulating data from DENV studies at the molecular level are the basis for dengue vaccine development and anti-DENV drug design. Reporter genes, such as green fluorescent protein (GFP) and luciferase tagged to viruses have been used to study flavivirus biology. Most previous studies reported constructs with parts of the viral structural gene region removed resulting in replicon plasmids that can be replicated in cells. For example, subgenomic WNV replicon expressing reporter gene was developed to promote antiviral drug screening (Lo et al., 2003a; Rossi et al., 2005; Shi et al., 2002). DENV-2 New Guinea C replicons containing GFP or luciferase reporter stabilized in BHK-21 cells were established for antiviral screening and siRNA testing (Ng et al., 2007). DENV-2 16681 replicon containing luciferase reporter was created and used to study 3′NCR structure and an anti-viral compound targeting the 3′NCR (Alvarez et al., 2005; Holden et al., 2006). In this study, we reported various strategies in constructing DENV-2 replicons with GFP reporter that provided a convenient and fast system to investigate the effects of 5′NCR mutations in viral replication. We also compared 4 types of subgenomic DENV/GFP constructs to identify an optimal replicon expressing the GFP.
2. Materials and methods
2.1. Cells and DENV-2 cDNA plasmid
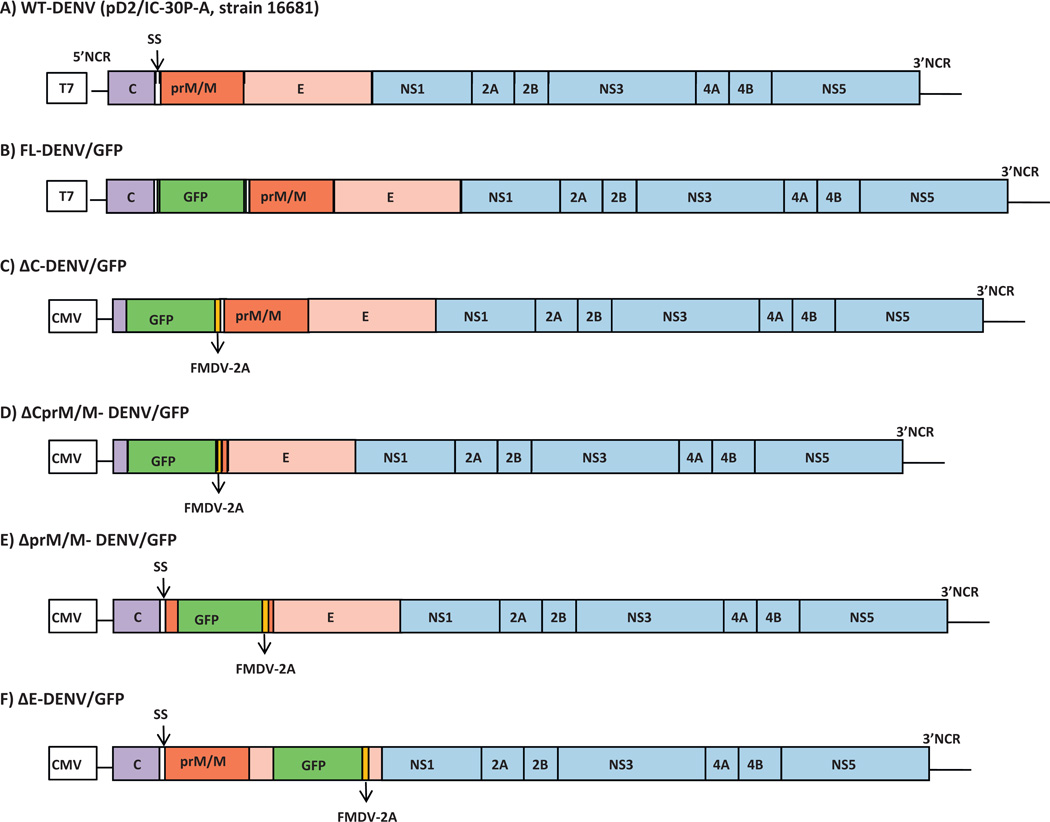
Vero cells (African green monkey kidney cells) and BHK-21 cells (baby hamster kidney cells), were cultured in Eagles minimum essential medium with Earle’s balanced salts (MEM/EBSS; HyClone®, Thermo Scientific, USA) containing 10% fetal bovine serum (FBS, Hyclone, USA), 100 U/ml penicillin and 100 µg/ml streptomycin at 37 °C in a 5% CO2 incubator. The plasmid, D2/IC-30P-A (Fig. 1A) (Kinney et al., 1997), containing full-length DENV-2 cDNA derived from wild type (wt) strain 16681, and its subclones containing different parts of DENV-2 cDNA were used for construction of the clones in this study. The pD2/IC-30P-A was also used to derive wt virus for this study.
Fig. 1.
Genetic maps of DENV-2, FL-DENV/GFP and Δ-DENV/GFP clones (A) Original wt full-length DENV-2 16681 cDNA plasmid, pD2/IC-30P-A (Kinney et al., 1997). (B) Full-length DENV-2 16681 cDNA with GFP insertion. (C)–(F) 4 types of subgenomic Δ-DENV/GFP constructs. T7: T7 promoter, CMV: cytomegalovirus IE promoter. SS: signal sequences for prM at C-terminal of C gene (between C and prM). GFP: green fluorescent protein. FMDV-2A: foot-and-mouth disease virus 2A protease. Color blocks without any label indicated partial deletion or disruption.
2.2. Construction of a full-length dengue virus plasmid expressing GFP fusion protein (pFL-DENV/GFP)
Aequorea coerulescens green fluorescent protein (GFP) gene was amplified from pAcGFP1-N1 (Clontech, USA) by polymerase chain reaction (PCR) using forward (AcGFP-N1-F2: ACAGATGATCATTATGGTGAGCAAGGGCGCCGAGC) and reverse (AcGFP-BclI-R:GGCTGATCATCTTGTACAGCTCATCCATGCCGTGG) primers containing natural BclI restriction site (bold) and extra bases of the DENV-2 (underlined) to ensure in frame cloning of the GFP gene. Plasmid containing wt DENV-2 cDNA nt 1–1380 (Kinney et al., 1997) was used for insertion of the GFP fragment at the BclI site (DEN-2 cDNA nt-407 position). The DENV-2/GFP fusion fragment of the subclone was then cloned into the full-length DEN-2 cDNA clone, pD2/IC-30P-A by SstI and SphI restriction sites. The final FL-DENV/GFP (FL for “full-length”) plasmid containing the GFP gene inserted in the nt 407 position, which is located within the signal sequence (SS) between the capsid and prM genes (Fig. 1B) was confirmed by restriction enzyme analysis, PCR, and sequencing.
2.3. Construction of subgenomic DENV-2 replicon with GFP reporter gene (Δ-DENV/GFP)
Four different types of subgenomic DENV-2 replicons (ΔC, ΔCprM/M, ΔprM/M, and ΔE) with reporter GFP were made by replacing part of the DENV-2 structural gene region with the GFP gene (Fig. 1C–F). A foot-and-mouth-disease virus 2A protease (FMDV-2A) cleavage factor was included in the constructs at the 3′end of GFP to ensure C-terminal cleavage of the GFP. The GFP with FMDV-2A fragment was directly PCR amplified from pAcGFP1 plasmid (Clontech, USA) using pairs of forward and reverse primers as follows: SalI-AcGFP-F (5′TATAATGTCGACTATGGTGAGCAAGGGC GCCGAGC3′) and FMDV2-BclI-R (5′ATATGATCATGGGCCCGGGGTTGGACTCGACGTCT3′) for ΔC-DENV/GFP; SalI-AcGFP-F and FMDV2-BoxI-R (5′ATATATAGACAGCTGTCAGGGGCCCGGGGTTGGACTCGACGTCT3′) for ΔCprM/MDENV/GFP; MscI-AcGFP-F (5′TATAATTGGCCATGATGGTGACCAAGGGCGCCGAG3′) and FMDV2-BoxI-R for ΔprM/MDENV/GFP; and SphI-AcGFP-F (5′TATAATGCATGCAATGGTGAGCAAGGGCGCCGAGC3′) and FMDV2-BamHI-R (5′ATATATATAGGATCCGGGCCCGGGGTTGGACTCGACGTCT3′) for ΔE-DENV/GFP. Underlined letters are the DENV-2 nucleotide sequences before and after GFP gene to ensure in frame cloning, bold letters indicate the natural restriction sites in the DENV-2 clone used for cloning. The resulting Δ-DENV/GFP plasmids have the following partial DENV-2 structural gene deletions: partial C gene (nt 172–405) deletion in ΔC construct, partial C-prM/M (nt 172–909) deletion in the ΔCprM/M construct, partial prM/M deletion (nt 556–909) deletion in ΔprM/M plasmid, and partial E deletion (nt 1386–2203) in the ΔE construct.
The original full length pD2/IC-30PA clone contains the T7 promoter, and required in vitro transcription to derive viral RNA prior to mammalian cell transfection. In the Δ-DENV/GFP constructs, the original T7 promoter on the plasmid was replaced with a human cytomegalovirus immediate early (CMV-IE) promoter which allowed direct RNA transcription in the plasmid-transfected mammalian cells. The CMV-IE sequence was also amplified from pAcGFP1-N1 (Clontech, USA) and cloned upstream to the 5′-end of DENV-2 gene in each replicon plasmid.
2.4. Cell transfection of FL-DENV/GFP RNA or Δ-DENV/GFP plasmid
To derive RNA transcripts from the FL-DENV/GFP plasmid with T7 promoter, 200 ng of linearized plasmid was in vitro transcribed using m7GpppA cap analog (New England Biolabs, USA) and reagents from Ribomax™ Large Scale RNA Production system T7 (Promega, USA). Then, total volume of in vitro transcribed RNA was transfected into 4 × 106 BHK-21 or Vero cells using the Bio-Rad Genepulse electroporator (BioRad, USA). Two pulses of 1.5 kV, 25 µF and infinite ohms were applied to BHK-21 cells (in a 0.2 cm gene pulser cuvette) and one pulse of 225 V with 25 ms of pulse length was applied to Vero cells (in a 0.4 cm gene pulser cuvette).
For Δ-DENV/GFP plasmids containing CMV-IE promoter, 1 × 106 BHK-21 cells were transfected (285 V, 1100 µF, 500 ohms, 2 mm cuvette, 1 pulse) with 8 µg of the plasmid. The transfected cells were cultured in MEM containing 10% FBS and penicillin/streptomycin.
2.5. Analysis of viral protein expression and GFP expression
Viral protein expression in the FL-DENV/GFP RNA transfected cells was determined by immunofluorescence assay (IFA) at 24 and 48 h post transfection (pt), using mouse hyperimmune ascetic fluid polyclonal antibodies against DENV-2 New Guinea C strain and anti-mouse-IgG antibody conjugated with FITC. The cells were counterstained with Evan’s blue and were observed under the fluorescence microscope. The transfected cells were also observed for GFP expression in the living cells by periodical examination directly under an inverted fluorescence microscope (Olympus, USA). When intracellular GFP expression was observed, the transfected cells were harvested, fixed, and counterstained with DAPI.
The cells transfected with Δ-DENV/GFP plasmids were harvested by trypsinization at 24, 48, and 72 h pt and re-suspended in fresh media at 4 °C. The GFP mean fluorescent intensity (MFI) measured at 488 nm in each cell was analyzed by FACSCanto flow cytometer using FACSDiva software version 2.4 (BD Bioscience, USA). The cells harvested on day 7 pt were also analyzed for viral NS1 protein by Western blot. Briefly, the cells were lysed in m-RIPA buffer (150 mM NaCl, 50 mM Tris–HCl, pH 7.4, 1% NP-40, 50 mM EDTA and aprotinin) (Sigma, USA). Cell lysates were subjected to 10% SDS-PAGE, and proteins were transferred to nitrocellulose membranes and immunoblotted with 1:100 dilution of monoclonal antibody against dengue anti-NS1, Clone IPF5SC(2G6) (Puttikhunt et al., 2003), and 1:5000 secondary antibody conjugated to horseradish peroxidase (Abcam, USA). The protein bands were detected by chemiluminescence (Amersham, USA) on the ECL film.
2.6. Analysis of virion production
Supernatants of the transfected cells collected on days 1, 4, 6 and 8 pt were determined for infectious viral titer by standard plaque titration assay as previously described (Leardkamolkarn et al., 2010). Briefly, 200 µl of serially diluted supernatant was inoculated onto the confluent monolayer of BHK-21 cells. Then 4 ml of 1st agarose overlay medium was applied. The cultures were incubated at 37 °C, in 5% CO2 condition for 7 days, and 2 ml of 2nd overlay medium with 1% Neutral Red (Sigma, USA) was applied on top. Plaque formation was observed during the next few days. Viral titers were calculated as follows:
Titer (PFU/ml) = numbers of plaques × sDF × iDF, where PFU is plaque formation unit, sDF is the serial dilution factor; iDF is infection dilution factor (1 ml/0.2 ml = 5). Detection limit was 5 PFU/ml.
2.7. Analysis of intracellular replicon RNA in transfected cells
One million BHK-21 cells transfected with 8 µg of each Δ-DENV/GFP plasmid were harvested at 24, 48, and 72 h pt, and total intracellular RNA was extracted using the RNeasy kit (Qiagen, USA). Residual DNA was removed using RNase-Free DNase kit (Qiagen, USA). For FL-DENV/GFP RNA transfected cells, intracellular RNA was extracted from the transfected cells on days 4, 6, and 8 pt. First strand cDNA was synthesized using a RevertAid™M-MuLV Reverse Transcriptase (Fermentas, Canada) with 1 µl (2% of the total extracted RNA) and specific anti-sense primer binding to the 3′NCR (nt 10680–10700; 5′CATTCCATTTTCTGGCGTTCT3′). Real-time PCR was performed using SYBR Green I in the Bio-Rad iQ5 instrument and specific sense primer (nt 10549–10572; 5′AAGGTGAGATGAAGCTGTAGTCTC3′) and anti-sense primer (nt 10680-10700; 5′CATTCCATTTTGGCGTTCT3′). Briefly, sample was added in a 25 µl reaction mixture containing 1 µl (out of 20 µl RT reaction) of the cDNA, 2.5 µl of 10× High Fidelity Reaction buffer, 0.2 µl of 25 µM dNTPs, 1.25 µl of 10 µM of each primer, 0.75 µl of SYBR Green I stock solution (1:1000), 0.2 µl of High Fidelity PCR enzyme mix (Fermentas, Canada). The thermocycler profile consisted of 3 min of Taq DNA polymerase activation at 94 °C, followed by 45 cycles of PCR at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min. To verify the specificity of the amplified product, melting curve analysis was performed. The results were analyzed by BioRad iQ5 standard software version 2.0. A standard curve was established in every experiment using 10-fold serial diluted standards between 1.16 and 1.16 × 1010 copies of DENV-16681 infectious clone plasmid, pD2/IC-30P-A. The RNA copy numbers were calculated according to the standard curve generated in the real-time PCR for comparison. The total RNA copy number = the copy number per reaction × 20 (1/20 of the RT sample used in PCR) × 50 (2% of total RNA used in RT).
2.8. Construction and characterization of DENV-2 5′NCR mutants using FL-DENV/GFP and Δ-DENV/GFP replicons
Full-length DENV-2 cDNA plasmids containing various mutations in the 5′NCR (Table 1) were made previously (Sirigulpanit et al., 2007; Leardkamolkarn et al., 2010). A cDNA fragment with A69T substitution isolated from full-length DENV-2.5′-69M plasmid was cloned into the FL-DENV/GFP plasmid to replace the corresponding wt fragment. In vitro transcribed RNA derived from the resulting FL-DENV-2.5′-69M/GFP plasmid was transfected into BHK-21 cells as described above.
Table 1.
Wild-type (wt) and 5′NCR mutations.
| Name | Mutation | |
|---|---|---|
| nt position | Substitution | |
| DENV-2 16681 | None (wt) | |
| DENV-2.5′-69M | 69 | A → T |
| DENV-2.5′-14M | 14 | C → A |
| DENV-2.5′-15M | 15 | G → T |
| DENV-2.5′-55M | 55 | A → G |
| DENV-2.5′-57M | 57 | C → T |
| DENV-2.5′-60M | 60 | A → G |
| DENV-2.5′-D57 | 57 | Deletion |
| DENV-2.5′-D5758 | 57 and 58 | Deletion |
cDNA fragments from other full-length infectious cDNA 5′ NCR-mutant clones, DENV-2.5′-14M, DENV-2.5′-15M, DENV-2.5′-55M, DENV-2.5′-57M, DENV-2.5′-60M, DENV-2.5′-D57M, and DENV-2.5′-D5758M were also cloned into the corresponding region of the 4 types of subgenomic Δ-DENV/GFP constructs described above and the resulting 28 plasmids were each transfected into BHK-21 cells as described above.
The methods described above were used to evaluate GFP expression, viral RNA quantification, and viral NS1 expression of the mutant transfected cells. The DENV-2.5′-D57M and DENV-2.5′D5758M deletion mutants were included as negative control constructs, since our previous results demonstrated that these deletions were totally lethal for DENV-2 (Sirigulpanit et al., 2007).
2.9. Statistical analysis
The MFI and RNA copies analysis experiments were performed independently in triplicates. The MFI results were analyzed using a one-way ANOVA, followed by a Tukey’s post hoc test. The RNA copies were compared by a two-tailed, unpaired, and unequal variant Student’s t-test. The differences were considered significant between groups at P < 0.05.
3. Results
3.1. Characterization of the RNA-launched FL-DENV/GFP constructs
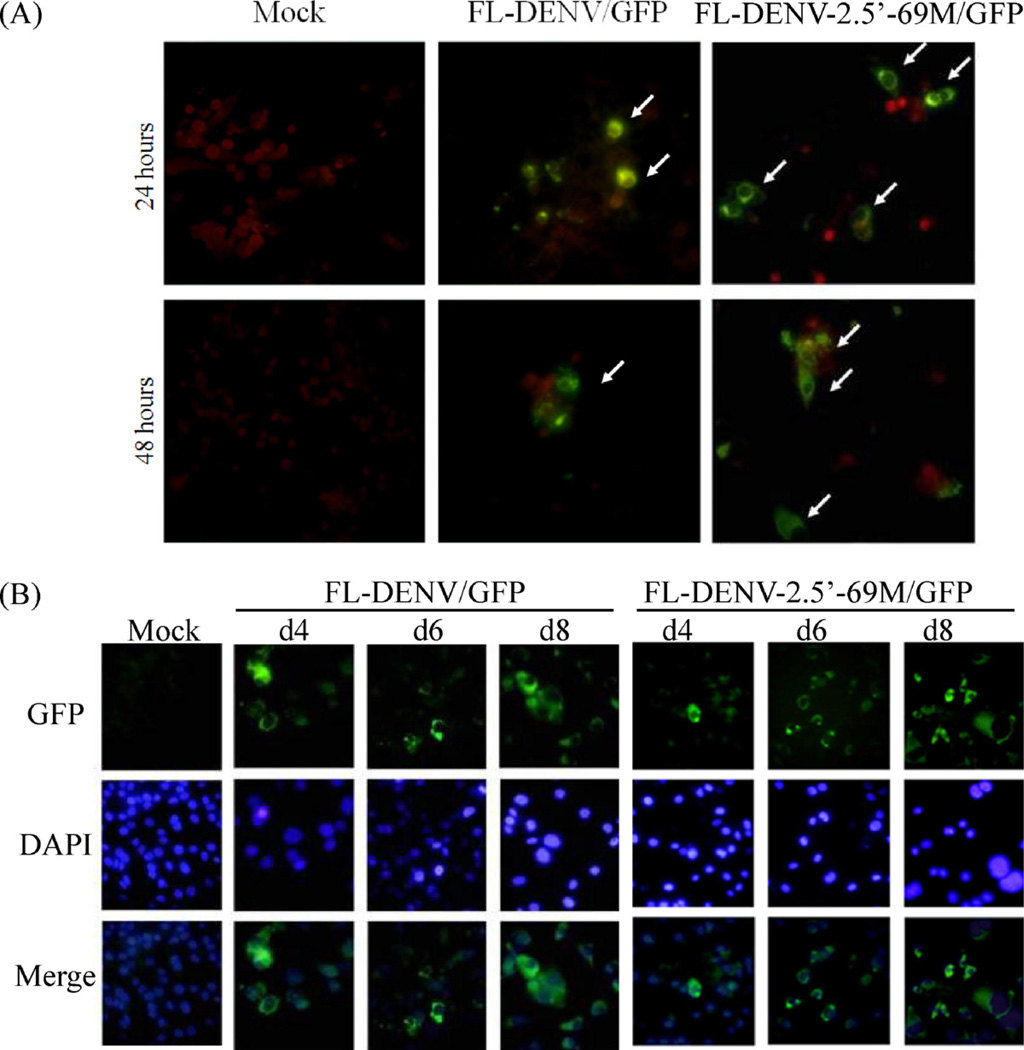
In vitro transcribed recombinant RNA from the wt FL-DENV/GFP or FL-DENV-2.5′-69M/GFP mutant (sometimes abbreviated as 5′-69M from this point) was transfected into Vero cells. Viral proteins were detected in the cytoplasm of both transfected cells at 24 and 48 h pt (Fig. 2A). In addition, GFP expression was detectable in living cells directly under an inverted fluorescent microscope. Cells were harvested, fixed, and stained with DAPI on day 4, 6 and 8 pt, and the results showed that the GFP expression was restricted to the cytoplasm, and morphology of the transfected cells appeared normal (Fig. 2B). No significant difference in protein expression was observed between the wt FL-DENV/GFP and the 5′-69M transfected cells during day 4–8 pt.
Fig. 2.
Fluorescent images of cells transfected with the FL-DENV/GFP RNA or FL-DENV-2.5′-69M/GFP RNA. (A) IFA images showing expression of viral protein in transfected cells at 24 and 48 h pt. Green cells (arrows) indicate positive cells, and red cells represent negative cells counter stained with Evans blue. (B) Green fluorescence emitted by GFP in the transfected cells on days 4, 6 and 8 pt. The cells were nuclear stained with DAPI (blue color). The micrographs were taken at 400× magnification. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Release of infectious virus from the transfected cells was analyzed by plaque assay of the culture supernatant collected on days 1, 4, 6 and 8 pt. Unlike the cells transfected with the control wt DENV RNA (from pD2/IC-30P-A, without GFP insertion), which produced high titers of the infectious DENV-2, culture of the FL-DENV/GFP RNA transfected cells yielded undetectable or minimal levels of infectious virus (only one titration experiment resulted in 5 PFU/ml, but other repeated experiments showed no detectable virus plaques). Attempts to amplify the virus by cell passage of transfected culture supernatant failed to demonstrate viable virus, suggesting that the GFP insertion in the RNA genome caused significant defect in virus infectivity.
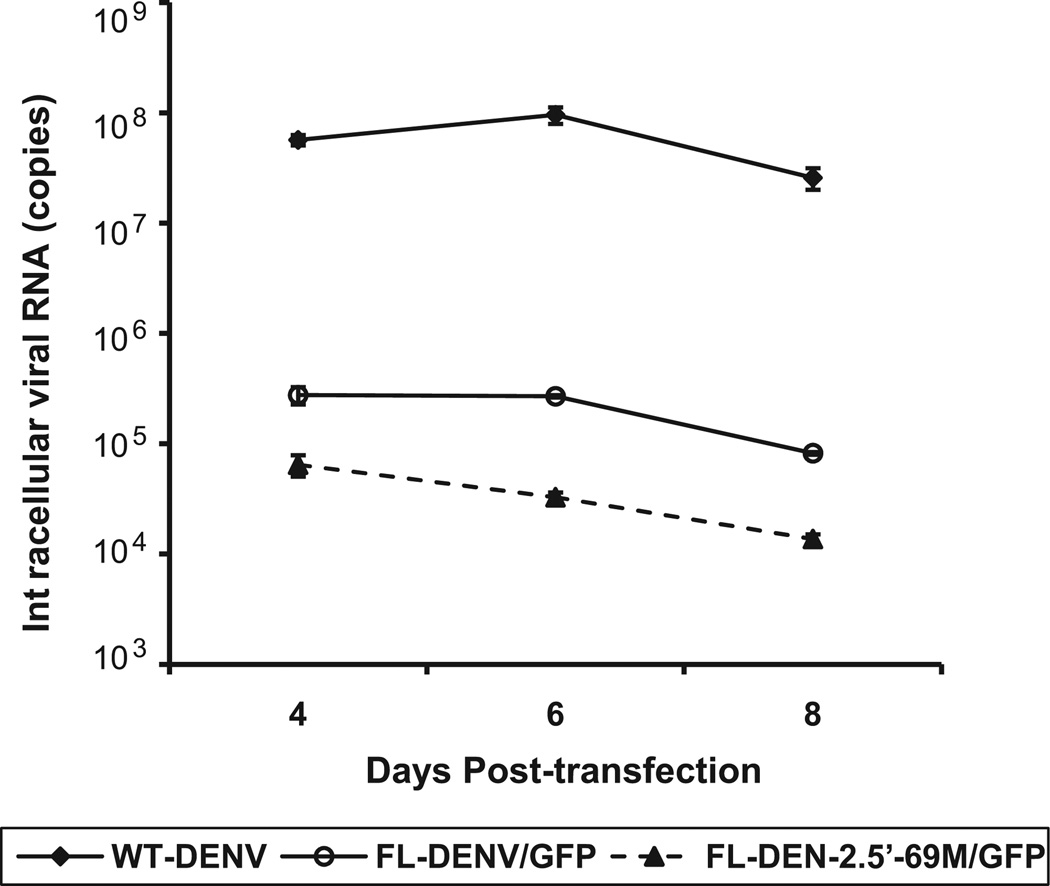
Quantification of the intracellular viral RNA in transfected BHK-21 cells was conducted on days 4, 6 and 8. RNA levels in cells transfected with either FL-DENV/GFP or FL-DENV-2.5′-69M/GFP were 2–3 log10 less than those in cells transfected with replicable D2/IC-30P-A infectious viral RNA (without GFP) (Fig. 3). The 5′-69M transfected cells exhibited substantially lower RNA levels than the FL-DENV/GFP transfected cells at all time points tested (P < 0.05).
Fig. 3.
Intracellular viral RNA copy numbers in wt DENV, FL-DENV/GFP and FL-DENV-2.5′-69M/GFP RNA transfected BHK-21 cells on days 4, 6 and 8 pt.
3.2. Characterization of the DNA-launched Δ-DENV/GFP replicons
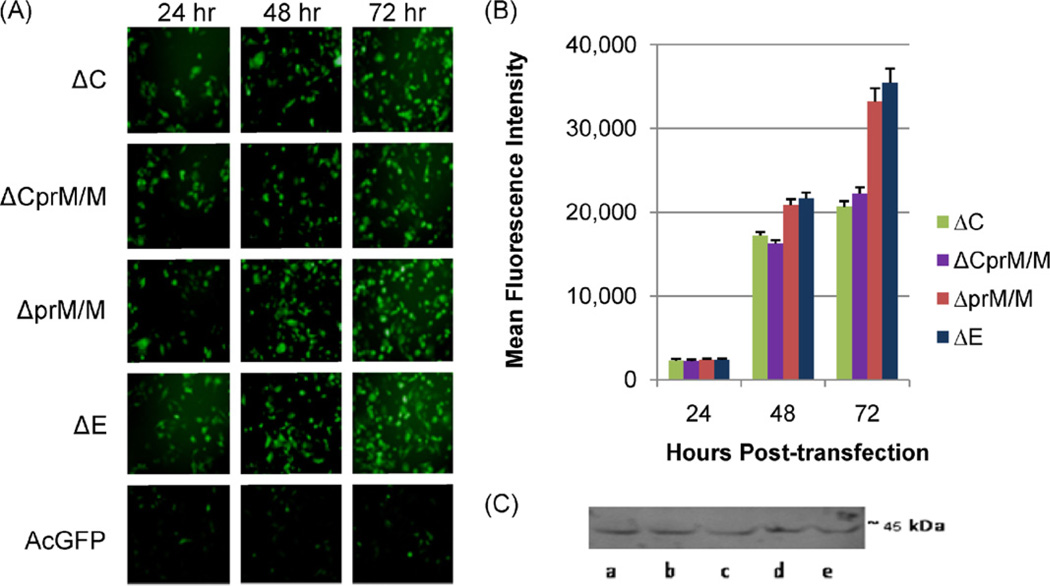
The GFP expression in transfected-BHK-21 cells was similar among all the 4 types of the subgenomic Δ-DENV/GFP plasmids (ΔC, ΔCprM/M, ΔprM/M and ΔE), and the GFP signals were very strong compared to that in the control transfected cells with commercial pAcGFP plasmid (Fig. 4A). Unlike the FL-DENV/GFP RNA transfected cells, the GFP expressed by the DNA-launched Δ-DENV/GFP plasmids was detected in both cytoplasm and cellular nuclei. The numbers of positive GFP cells were similar among the 4 types of Δ-DENV/GFP at 24 h pt, and continued to increase between 24 and 72 h (Fig. 4A). The mean fluorescent intensity (MFI) of each GFP-autofluorescing cell measured by flow cytometry (Fig. 4B) was also comparable among all 4 types of Δ-DENV/GFP transfected cells at 24 h pt, suggesting that they achieved similar reporter activity early in the cells. However, at 48–72 h pt both MFI and the numbers of GFP-positive cells from transfection of ΔprM/M- and ΔE-DENV/GFP plasmids became significantly higher (P < 0.05) than those from transfection of the ΔC- and ΔCprM/M-DENV/GFP plasmids. In addition, DENV-2 NS1 proteins were detectable by Western blot in all 4 types of Δ-DEN/GFP transfected cells at least up to day 7 pt (Fig. 4C).
Fig. 4.
(A) GFP expression of the ΔC-DENV/GFP, ΔCprM/M-DENV/GFP, ΔprM-DENV/GFP, ΔE-DENV/GFP, or the control AcGFP plasmid in BHK-21 cells at 24, 48 and 72 h pt. (B) MFI of GFP in Δ-DENV/GFP transfected BHK-21 cells at 24, 48 and 72 h pt. (C) Western blot analysis of DENV-2 NS1 protein in plasmid-transfected BHK-21 cells on day 7 pt. (a) wt DEN-2 16681 plasmid (pD2/IC-30P-A), and (b) ΔCprM/M-, (c) ΔprM/M-, (d) ΔC-, and (e) ΔE-DENV/GFP plasmids.
Despite the different types of construct, all the 4 Δ-DENV/GFP plasmid-transfected cells showed over 100–1000 fold increase in intracellular replicon RNA between 24 and 72 h pt. Intracellular RNA of ΔC and ΔCprM/M transfected cells increased from 5.9–6.0 log10 copies at 24 h pt to 8.8–9.2 log10 copies, while the increase for ΔprM/M and ΔE transfected cells were from 6.5–7.0 log10 copies to 9.4–9.7 log10 copies. Repeated experiments with the same amount of plasmid input for transfection resulted in similar RNA levels between ΔC and ΔCprM/M transfected cells, and between ΔprM/M and ΔE transfected cells. In general, the RNA levels of ΔC and ΔCprM/M transfections were approximately 10-fold lower than those of ΔprM/M and ΔE transfection at all 3 time points.
3.3. DENV-2 5′NCR mutagenesis analysis using DNA launched Δ-DENV/GFP replicons
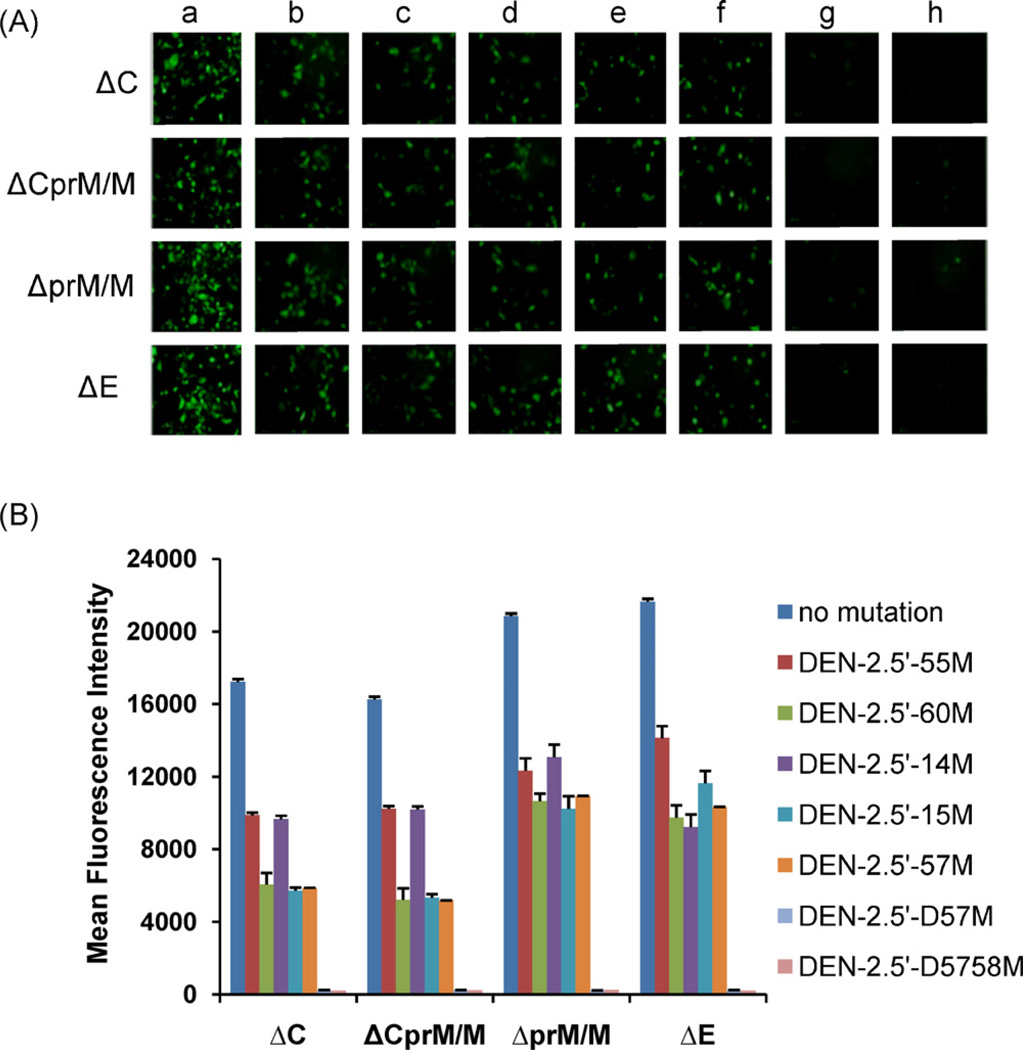
It is important to investigate the potential utility and accuracy of the Δ-DENV/GFP replicons in DENV research. For this purpose, we chose five 5′NCR mutations previously shown to have attenuation effects in mouse neurovirulence and 2 deletion mutants that were lethal for virus (Sirigulpanit et al., 2007; Leardkamolkarn et al., 2010) to be incorporated in the replicons (Table 1), and evaluated whether the mutation effects could be measured using the replicon reporter system in vitro. Positive GFP expressions were observed in cells transfected with all 4 types of Δ-DENV/GFP replicons containing 5′-55M, 5′-60M, 5′-14M, 5′-15M, 5′-57M, but the positive cell numbers were less than those of the Δ-DENV/GFP replicons with wt 16681 5′NCR at 48 h pt (Fig. 5A). The deletion mutants, 5′-D57M and 5′D5758M, on the other hand, only had a few cells that were GFP positive (Fig. 5A, panels g and h). Among the mutants, 5′-55M generally showed the highest numbers of GFP-positive cells (Fig. 5A, panel b). Except for samples transfected with deletion mutants (5′-D57M and 5′-D5758M), the numbers of the GFP positive cells transfected with the other 5 mutants increased between 24 and 72 h post-transfection (data not shown).
Fig. 5.
(A) Fluorescent images of Δ-DENV/GFP plasmid-transfected cells at 48 h pt. Δ-DENV/GFP replicons with (a) wt 5′NCR, (b) DENV-2.5′-55 M, (c) DENV-2.5′-60M, (d) DENV-2.5′-14M, (e) DENV-2.5′-15M, (f) DENV-2.5′-57M, (g) DENV-2.5′-D57M, and (h) DENV-2.5′-5758M. (B) MFI of GFP in BHK-21 cells transfected with Δ-DENV/GFP plasmids at 48 h pt. MFI in mutant-transfected cells were all significantly less (P < 0.001) than those in cells transfected with corresponding replicon containing wt 5′NCR.
The MFI levels of GFP in cells transfected with the deletion mutants were also very low at 48 h pt (Fig. 5B). Similar MFI were observed between ΔC and ΔCprM/M replicons, and between ΔprM/M and ΔE replicons carrying the same mutation (Fig. 5B). Similar to the comparison results among the 4 types of unmutated Δ-DENV/GFP replicons, the MFI levels produced from the mutant replicons were generally higher from ΔprM/M and ΔE types than those from the other 2 types of replicon. All the mutant replicons with 5′NCR mutation had significantly weaker (P < 0.001) MFI than their corresponding replicons containing wt 5′NCR. Except in the ΔE-DENV/GFP replicon, 5′-55M and 5′-14M mutants generally resulted in higher MFI levels than the other 3 substitution mutants within the same type of replicon. In the ΔE-DENV/GFP mutant group, the 5′-55M still produced higher MFI than the other mutants, but the MFI from 5′-14M mutant was the lowest one among the 5 substitution mutants.
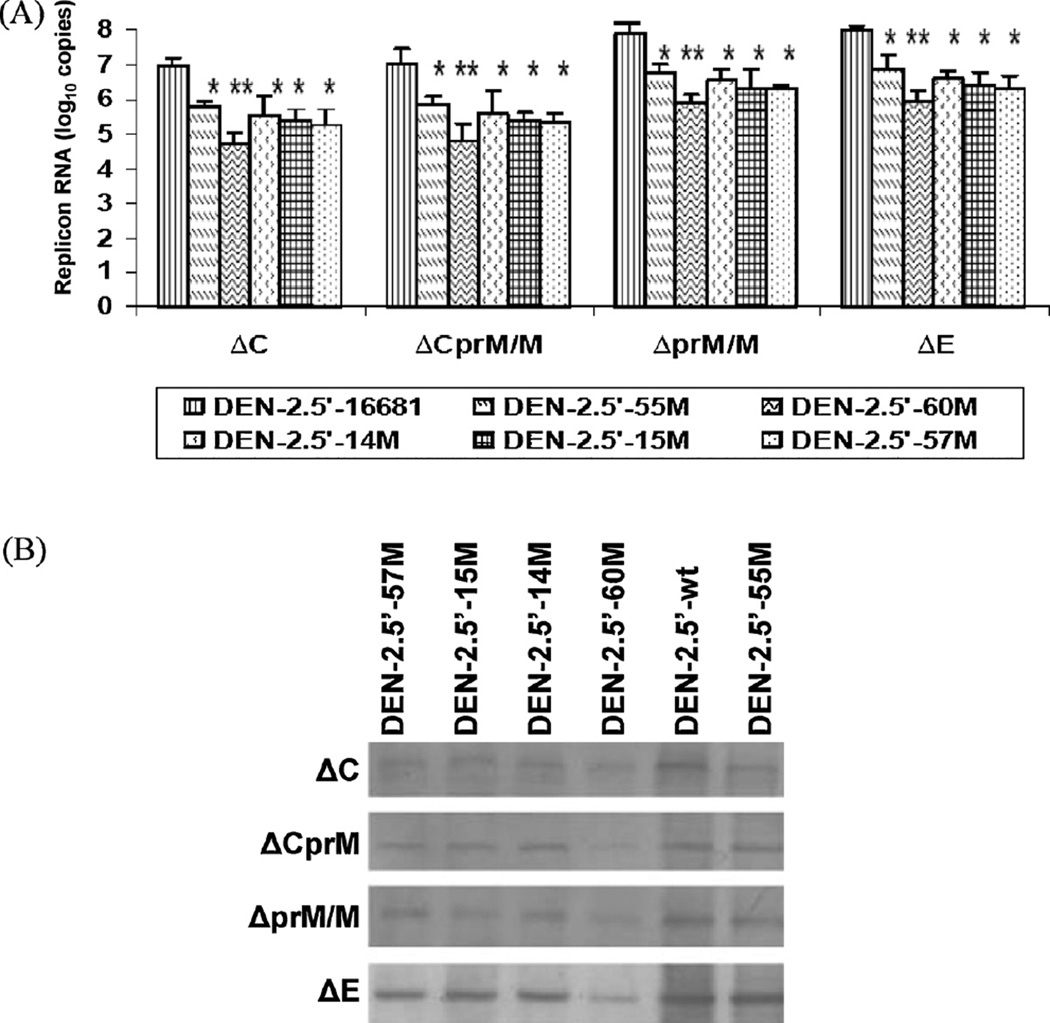
Production of replicon RNA in transfected BHK-21 cells was evaluated by 2-step RT/real-time PCR at 48 h pt. RNA levels of the deletion mutants (5′-D57M and 5′-D5758M) were under the detectable limit (data not shown), indicating that the initial RNA transcripts produced from the deletion mutant plasmids were not self-replicable, and they could be used as non-replicable controls for the replicon system. All the non-deletion mutants, on the other hand, produced significant amounts of RNA (Fig. 6A). Similar to the MFI results, RNA levels were comparable between ΔC and ΔCprM/M replicons, and between ΔprM/M and ΔE replicons. In general, the RNA levels of ΔC and ΔCprM/M replicons were approximately 10-fold lower than that of ΔprM/M and ΔE replicons. Compared to their corresponding ΔDENV/GFP replicon containing wt 5′NCR, all of the mutants had significantly less (P < 0.001) intracellular viral RNA levels, and the 5′-60M produced the least amount of RNA (P < 0.0001) at 24, 48, and 72 h pt (showed 48 h pt as a representative, Fig. 6A).
Fig. 6.
(A) Intracellular replicon RNA copy numbers of cells transfected with Δ-DENV/GFP plasmids at 48 h pt. *P < 0.001; **P < 0.0001(B) DENV-2 NS1 protein (Western blot) in BHK-21 cells transfected with Δ-DENV/GFP plasmids at day 7 pt.
The expression of DENV-2 NS1 protein also varied among different types of replicon (Fig. 6B). No NS1 protein was detected in cells transfected with any of the 5′-D57M and 5′-D5758M mutants on day 7 pt (data not shown). Among all of the non-deletion mutants, similar to the GFP MFI and intracellular RNA results, 5′-60M expressed the least amount of the NS1 protein. Interestingly, all mutants made in the ΔE replicon expressed highest amount of NS1 protein when compared to other replicons carrying the same mutation, suggesting it may be the optimal type of the Δ-DENV/GFP replicon. Infection of fresh cells with supernatant of transfected cells and plaque titration assay confirmed that no infectious virus was produced by any of the cells transfected with these subgenomic replicons.
4. Discussion
Successful construction of a full length DENV-2 cDNA clone expressing GFP was demonstrated in this study by direct insertion of GFP gene within the signal sequence, which is a hydrophobic domain located at the C-terminus of C and upstream of the N-terminus of prM (Lindenbach and Rice, 2003). The viral proteins and GFP expression were demonstrated in the cytoplasm of the FL-DENV/GFP RNA transfected BHK-21 cells. Since GFP gene was incorporated in monocistronic manner, the expression of GFP reporter can be used to indicate the viral RNA replication and protein translation efficiency of the recombinant plasmid. The FL-DENV/GFP RNA transfected cells failed to generate infectious DENV-2, and that was likely due to the insertion of GFP in the signal sequence. The GFP insertion at the fifth amino acid of the signal sequence, a 14-amino acid peptide in front of the DENV-2 prM, may impair the specific dynamic requirement for the correct cleavages of C and prM (Amberg et al., 1994; Amberg and Rice, 1999; Stocks and Lobigs, 1998; Lobigs and Lee, 2004). The signal sequence is critical for proper translocation of the prM into ER lumen during polyprotein processing. Results of the intracellular RNA also indicated that no second round of virus infection occurred in cells transfected with FL-DENV/GFP RNA. Their RNA levels were over 100-fold less than the cells transfected with infectious wt DENV-2 RNA on day 4–8 pt when new rounds of DENV-2 infection should have already happened. GFP reporter gene has been inserted between E and NS1 gene of a full-length YFV cDNA, resulting in recombinant YFV expressing GFP (Bonaldo et al., 2007). Pierson et al. (2005) also reported a recombinant infectious WNV expressing the GFP reporter that was cloned with an internal ribosome entry site (IRES) gene after the stop codon of the WNV polyprotein. However, GFP gene in this bicistronic WNV construct trended to be deleted after several replication cycles. Recently, construction of WNV was made successful by insertion of GFP in the capsid sequences, with some modification, resulting in viable virus that benefits to study infectivity, dissemination and transmission of the virus in mosquito vector as well as viral tracking in mice (McGee et al., 2010). Although our FL-DEN/GFP construct did not yield infectious virus, our main goal for this study was to generate a replicon system with reporter GFP to facilitate DENV-2 research. In our system, the GFP expression level would reflect the efficiency of initial viral polyprotein translation and the viral RNA self-replication within the single-round of the transfected cells. Previously, we have shown the 5′-69 mutation of DENV-2 virus is partially attenuated for neurovirulence in newborn mice (Sirigulpanit et al., 2007). This A69U substitution was conserved among the wt DENV-2 strains of the American genotype causing dengue fever, but not dengue hemorrhagic fever, in humans (Leitmeyer et al., 1999). It is located at the leading end of the first stem situated at the base of the predicted SLA structure at the 5′-end of the genome. The SLA is recognized by viral RNA polymerase (NS5) and participates in cyclization of the viral genome through the interaction with the 3′-end sequence, thereby promoting minus-strand viral RNA synthesis (Filomatori et al., 2006). Using the single-round FL-DENV/GFP RNA-launched system, we confirmed that the 5′-69M construct resulted in significantly less (P < 0.05) intracellular viral RNA levels than the FL-DENV/GFP containing wt 5′NCR construct (Fig. 3).
In addition, we have successfully developed 4 types of subgenomic DENV-2/GFP DNA-launched-replicons. The deleted DENV-2 structural protein in the subgenomic plasmid was replaced by the GFP reporter gene with a FMDV-2A cleavage site to ensure C terminal cleavage of GFP (Varnavski et al., 2000). A human cytomegalovirus immediate early (CMV IE) promoter was applied upstream of the 5′NCR of DENV genome to initiate RNA transcription in the transfected cells (Akrigg et al., 1985; Varnavski et al., 2000). As in the FL-DENV/GFP construct, the AcGFP with optimized human codons was chosen as the reporter to enhance the GFP expression (Haas et al., 1996). To ensure the Δ-DENV/GFP replicons were capable of RNA self-replication, we included all DENV-2 genome parts essential for viral RNA-dependent RNA replication with deletion in the structural gene. For ΔC construct, most of the C gene except for the first 25 residues containing the cyclization motif for NS5 dependent RNA replication (Lo et al., 2003b; You and Padmanabhan, 1999) and the last 8 residues containing the signalase site was deleted (Chambers et al., 1990). In the ΔCprM/M, the cyclization motif of C and the last 5 residues of the M protein containing a signalase cleavage site (Chambers et al., 1990), were retained. In ΔprM/M and ΔE constructs, majority parts of the prM/M or E gene, except the N and C-terminal regions, were removed. These designs ensured correct protein processing of all NS proteins that are largely involved in viral replication, as well as intact 5′ and 3′NCRs of the DENV-2 genome, thereby retaining essential structures/elements for initial viral protein translation and viral RNA replication. Successful establishment of subgenomic flavivirus replicons including WNV, YFV and DENV-2 have been demonstrated (Shi et al., 2002; Jones et al., 2005; Ng et al., 2007), but most of them are RNA-launched system. DNA-launched subgenomic DENV-2 replicons based on NGC strain has been achieved before (Pang et al., 2001), but it did not include any reporter gene.
The GFP signals expressed by our constructs were readily detectable at 24 h pt, increased significantly within 24–72 h pt, and lasted stably for 9 days (data not shown). We were also able to detect NS1 at 7 days pt. The exponential increase of the RNA levels, the increase of positive GFP cells, and the prolonged expression of GFP and NS1, suggested that the subgenomic RNAs transcribed from the plasmid inside of the transfected cells were capable of protein translation, and self-replication through RNA-dependent RNA replication. The non-replicable control constructs; 5′-D57M and 5′-D5758M (deletion mutants), generated only minimal GFP and undetectable viral RNA levels (Figs. 5A and 6A). These results further demonstrated that all 4 types of the wt DNA-launched Δ-DENV-2/GFP replicons were able to generate self-replicative RNA in cells.
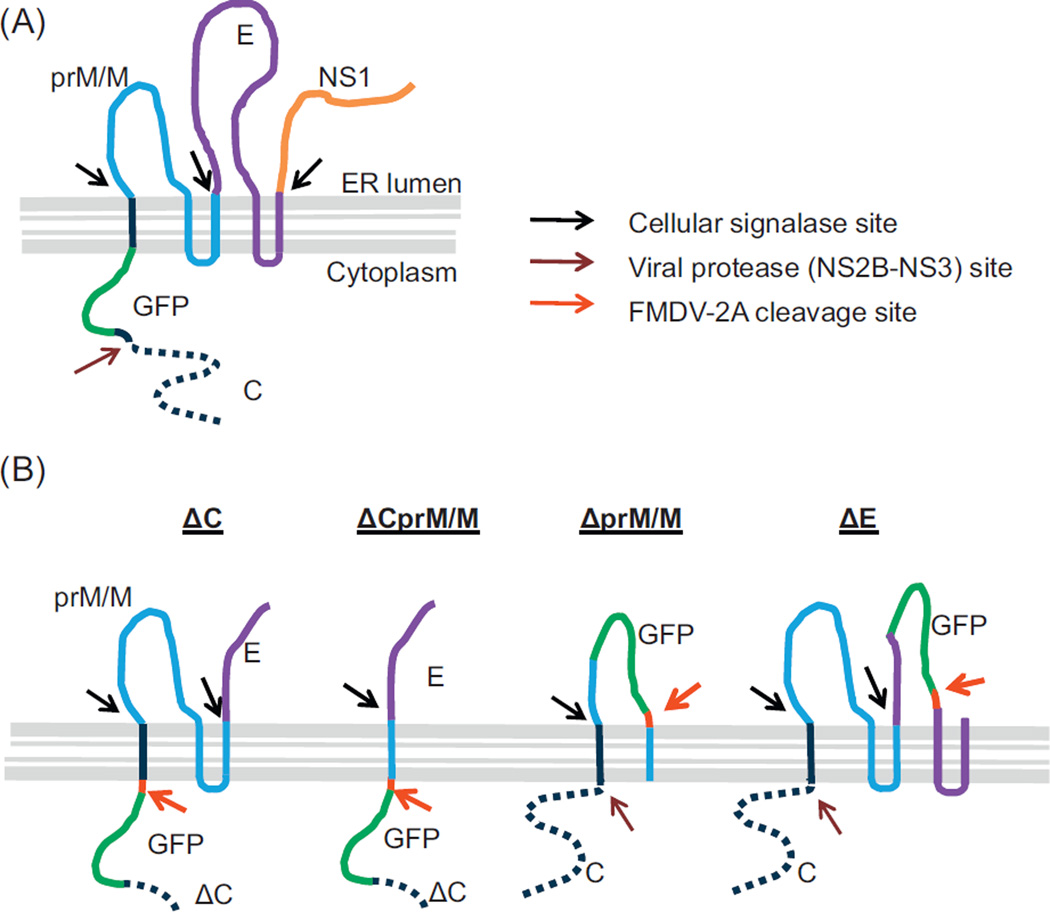
Unlike the FL-DENV/GFP RNA-launched construct expressing GFP only in cytoplasm, the GFP was observed in both cytoplasm and nuclei in cells transfected with the Δ-DENV/GFP DNA-launched constructs. It is not surprising that GFP signal was only observed in the cytoplasm of the FL-DENV/GFP RNA transfected cells, because it was fused with the hydrophobic signal sequence and likely was anchored in membranes of the cellular compartments (e.g. ER) (Fig. 7A). On the other hand, a FMDV-2A cleavage site was included at the C-terminal end of the GFP in all of the Δ-DENV/GFP constructs, resulting in small GFP fusion proteins (fused with only partial structural proteins) without any transmembrane domain to block its diffusion into nuclei (Fig. 7B). Because of its small size, GFP (239 amino acids) can penetrate into nuclei (Hanson and Kohler, 2001).
Fig. 7.
Predicted locations and protein processing of GFP within DENV-2 polyprotein. (A) FL-DENV/GFP and (B) Δ-DENV/GFP replicons.
When comparing the 4 types of the Δ-DENV/GFP replicons, the MFI levels of GFP and the replicon RNA were higher from cells transfected with ΔprM/M and ΔE than those with ΔC and ΔCprM/M, suggesting that some area within the deleted C gene of ΔC and ΔCprM/M replicons may still be involved in up-regulation of the viral RNA replication. This observation agrees with earlier reports (Mandl, 2004; Zhu et al., 2007) demonstrating that replicons with partial C deletion had adverse effect in flavivirus RNA replication. However, as depicted in Fig. 7B, the GFP expressed by ΔC or ΔCprM/M is expected to be translocated in cytoplasm, while the GFP expressed by ΔprM/M and ΔE would be in ER lumen. Therefore, it is also possible that efficiency of the viral polyprotein processing was affected by the alternate GFP sites, and consequently resulted in various efficiencies in RNA replication and GFP expression. Nevertheless, despite the different initial translocation sites of the GFP expressed by the various subgenomic replicons, we observed similar diffuse patterns of GFP signal throughout the cytosol and nuclei among all 4 types of the subgenomic replicontransfected cells (Fig. 4A). Although the GFP expressed by ΔprM/M and ΔE could be located in the ER lumen during polyprotein processing (Fig. 7B), without fusing with any signal sequence or transmembrane peptide, it could not secrete out of the cells through the ER-Golgi pathway (Laukkanen et al., 1996). Instead, the soluble GFP in the ER cleaved from the FMDV-2A site was capable of translocation across the ER membrane to the cytosol (Thomas et al., 2001; Tanudji et al., 2002) and further diffusing into nuclei.
Using the Δ-DENV/GFP replicons, we were able to dissect the mutation effects of seven 5′NCR mutations that we have previously analyzed using the infectious clone system (Leardkamolkarn et al., 2010; Sirigulpanit et al., 2007). Our results suggested that deletion of nt 57 or 57–58 severely diminished the GFP expression and RNA replication. It is very likely that essential RNA structure required for DENV RNA replication was severely destroyed with the deletions. All the 5 non-deletion mutants had less GFP intensity and RNA levels than the wt in all 4 types of replicons at 48 h pt. With few exceptions, results of the same mutation among different types of replicon were generally consistent and in most cases their GFP intensity correlated very well with RNA levels. For example, the 5′-55M expressing the strongest intensity of GFP had the highest RNA numbers among all the non-deletion mutants in all 4 types of replicon. The 5′-60M expressing GFP at much lower intensity than others also had least amount of RNA levels. Therefore, the GFP intensity expressed by these replicon systems can be used as a fast indicator to determine the RNA replication efficiency.
Previous studies using infectious clone demonstrated that each of the five 5′NCR substitutions caused partial attenuation of mouse neurovirulence (Leardkamolkarn et al., 2010; Sirigulpanit et al., 2007). Since the replicon systems reported here were measuring events within a single-round transfection, we further demonstrated that the attenuation effect of these 5′NCR mutations was due to less RNA replication efficiency. GFP expression intensity provided clear evidence of replication defects by the mutations when compared them to the wt replicons. Most interestingly, we have observed that different subgenomic constructs resulted in different levels of RNA self-replication and GFP expression, and constructs with deletions outside of C were generally better than the constructs with partial C deletion. Using both the FL-DENV/GFP and Δ-DENV/GFP constructs reported in this study, we have demonstrated that these reporter systems can provide a fast and powerful platform to study critical molecular determinants involved in viral protein translation and viral RNA replication. For example, mutant replicons with mutations at suspected molecular determinants can be first screened quickly by comparing their GFP intensity within 24–48 h pt. Mutations producing significantly lower GFP intensity can then be selected for further study using other platforms, such as the infectious clone system that is much more time and labor consuming. In addition, the single-round replication system can complement studies using whole replicable viruses. The multiple replicon constructs can also be very useful for screening antiviral molecules that target different stages of virus replication. In fact, we and others recently have successfully established cells lines that stably express subgenomic DENV-2 replicons with GFP tagging (Leardkamolkarn and Sirigulpanit, 2011; Massé et al., 2010), and both systems have provided a rapid and efficient cell-based fluorescent assay for anti-DENV compounds screening.
Acknowledgements
This study was funded by Thailand National Science and Technology Development Agency (NSTDA/BIOTEC), the Office of the Higher Education Commission and Mahidol University under the National Research Universities Initiative, Thailand Research Fund (TRF) and Faculty of Science, Mahidol University. We thank Dr. Richard M. Kinney, Division of Vector-Borne Diseases, Centersfor Disease Control and Prevention, Fort Collins, CO, USA for his valuable suggestions in preparation of this manuscript.
Contributor Information
Vijittra Leardkamolkarn, Email: scvlk@mahidol.ac.th.
Wipawan Sirigulpanit, Email: ws8007@yahoo.com.
Nunya Chotiwan, Email: nunya.chotiwan@gmail.com.
Supeecha Kumkate, Email: scskk@mahidol.ac.th.
Claire Y.-H. Huang, Email: chuang1@cdc.gov.
References
- Akrigg A, Wilkinson GW, Oram JD. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985;2:107–121. doi: 10.1016/0168-1702(85)90242-4. [DOI] [PubMed] [Google Scholar]
- Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirementin internal initiation of translation. J. Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation. RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Amberg SM, Nestorowicz A, McCourt DW, Rice CM. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 1994;68:3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg SM, Rice CM. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 1999;73:8083–8094. doi: 10.1128/jvi.73.10.8083-8094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo MC, Mello SM, Trindade GF, Rangel AA, Duarte AS, Oliveira PJ, Freire MS, Kubelka CF, Galler R. Construction and characterization of recombinant flaviviruses bearing insertions between E and NS1 genes. Virol. J. 2007;4:115. doi: 10.1186/1743-422X-4-115. doi:1.10.1186/1743-422X-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butrapet S, Huang CY, Pierro DJ, Bhamarapravati N, Gubler DJ, Kinney RM. Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5′ noncoding region and non-structural proteins 1 and 3. J. Virol. 2000;74:3011–3019. doi: 10.1128/jvi.74.7.3011-3019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 1995;207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- Guesdon FM, Greatorex J, Rhee SR, Fisher R, Hunter E, Lever AM. Sequences in the 5′ leader of Mason-Pfizer monkey virus which affect viral particle production and genomic RNA packaging: development of MPMV packaging cell lines. Virology. 2001;288:81–88. doi: 10.1006/viro.2001.1061. [DOI] [PubMed] [Google Scholar]
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Venkateshan CN, Gentry MK, Larsen LK. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J. Immunol. 1984;132:1529–1532. [PubMed] [Google Scholar]
- Hanson MR, Kohler RH. GFP imaging: methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 2001;52(356):529–539. [PubMed] [Google Scholar]
- Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology. 2006;344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Jones CT, Patkar CG, Kuhn RJ. Construction and applications of yellow fever virus replicons. Virology. 2005;331:247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, Gubler DJ. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukkanen M, Oker-Blom C, Keinanen K. Secretion of green fluorescent protein from recombinant baculovirus-infected insect cells. Biochem. Biophys. Res. Commun. 1996;226:755–761. doi: 10.1006/bbrc.1996.1425. [DOI] [PubMed] [Google Scholar]
- Leardkamolkarn V, Sirigulpanit W, Kinney RM. Characterization of recombinant dengue-2 virus derived from a single nucleotide substitution in the 5′ noncoding region. J. Biomed. Biotechnol. 2010 doi: 10.1155/2010/934694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leardkamolkarn V, Sirigulpanit W. Establishment of a stable cell line coexpressing dengue virus-2 and green fluorescent protein for screening of antiviral compounds. J. Biomol. Screen. 2011;7 doi: 10.1177/1087057111426903. doi: 0.1177/1087057111426903. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquitoborne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003a;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MK, Tilgner M, Shi PY. Potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 2003b;77:12901–12906. doi: 10.1128/JVI.77.23.12901-12906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M, Lee E. Inefficient signalase cleavage promotes efficient nucleocapsid incorporation into budding flavivirus membranes. J. Virol. 2004;78:178–186. doi: 10.1128/JVI.78.1.178-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW. Flavivirus immunization with capsid-deletion mutants: basics, benefits, and barriers. Viral Immunol. 2004;17(4):461–472. doi: 10.1089/vim.2004.17.461. [DOI] [PubMed] [Google Scholar]
- Massé N, Davidson A, Ferron F, Alvarez K, Jacobs M, Romette JL, Canard B, Guillemot JC. Dengue virus replicons: production of an interserotypic chimera and cell lines from different species, and establishment of a cell-based fluorescent assay to screen inhibitors, validated by the elevation of ribavirin’s activity. Antiviral Res. 2010;86:296–305. doi: 10.1016/j.antiviral.2010.03.010. [DOI] [PubMed] [Google Scholar]
- McGee CE, Shustov AV, Tsetsarkin K, Frolov IV, Mason PW, Vanlandingham DL, Higgs S. Infection, dissemination, and transmission of a west nile virus green fluorescent protein infectious clone by culex pipiens quinquefasciatus mosquitoes. Vector-Borne Zoonotic Dis. 2010;10:267–274. doi: 10.1089/vbz.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CY, Gu F, Phong WY, Chen YL, Lim SP, Davidson A, Vasudevan SG. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res. 2007;76:222–231. doi: 10.1016/j.antiviral.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Pang X, Zhang M, Dayton AI. Development of Dengue virus type 2 replicons capable of prolonged expression in host cells. BMC Microbiol. 2001;1:18. doi: 10.1186/1471-2180-1-18. (Epub 2001 Aug 24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS, Ahmed AA, Valentine LE, Davis CW, Samuel MA, Hanna SL, Puffer BA, Doms RW. An infectious West Nile virus that expresses a GFP reporter gene. Virology. 2005;334:28–40. doi: 10.1016/j.virol.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Puttikhunt C, Kasinrerk W, Srisa-ad S, Duangchinda T, Silakate W, Moonsom S, Sittisombut N, Malasit P. Production of anti-dengue NS1 monoclonal antibodies by DNA immunization. J. Virol Methods. 2003;109:55–61. doi: 10.1016/s0166-0934(03)00045-4. [DOI] [PubMed] [Google Scholar]
- Rossi SL, Zhao Q, O’Donnell VK, Mason PW. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology. 2005;331:457–470. doi: 10.1016/j.virol.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Rothman AL, Ennis FA. Immunopathogenesis of dengue hemorrhagic fever. Virology. 1999;257:1–6. doi: 10.1006/viro.1999.9656. [DOI] [PubMed] [Google Scholar]
- Shi PY, Tilgner M, Lo MK. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology. 2002;296:219–233. doi: 10.1006/viro.2002.1453. [DOI] [PubMed] [Google Scholar]
- Sirigulpanit W, Kinney RM, Leardkamolkarn V. Substitution or deletion mutations between nt 54 and 70 in the 5′ non-coding region of dengue type 2 virus produce variable effects on virus viability. J. Gen. Virol. 2007;88:1748–1752. doi: 10.1099/vir.0.82455-0. [DOI] [PubMed] [Google Scholar]
- Stocks CE, Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinantsin C, the signal peptide, and prM. J. Virol. 1998;72:2141–2194. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanudji M, Hevi S, Chuck SL. J Cell Sci. Vol. 115. The Company of Biologists Ltd; 2002. Improperly folded green fluorescent protein is secreted via a non-classical pathway; pp. 3849–3857. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway inEscherichia coli. Mol. Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Varnavski AN, Young PR, Khromykh AA. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 2000;74:4394–4403. doi: 10.1128/jvi.74.9.4394-4403.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Qin C, Chen S, Jiang T, Yu M, Yu X, Qin E. Attenuated dengue 2 viruses with deletions in capsid protein derived from an infectious full-length cDNA clone. Virus Res. 2007;126:226–232. doi: 10.1016/j.virusres.2007.03.004. [DOI] [PubMed] [Google Scholar]