Abstract
We report herein a set of calculations designed to examine the effects of epigenetic modifications on the structure of DNA. The incorporation of methyl, hydroxymethyl, formyl and carboxy substituents at the 5-position of cytosine is shown to hardly affect the geometry of CG base pairs, but to result in rather larger changes to hydrogen-bond and stacking binding energies, as predicted by dispersion-corrected density functional theory (DFT) methods. The same modifications within double-stranded GCG and ACA trimers exhibit rather larger structural effects, when including the sugar-phosphate backbone as well as sodium counterions and implicit aqueous solvation. In particular, changes are observed in the buckle and propeller angles within base pairs and the slide and roll values of base pair steps, but these leave the overall helical shape of DNA essentially intact. The structures so obtained are useful as a benchmark of faster methods, including molecular mechanics (MM) and hybrid quantum mechanics/molecular mechanics (QM/MM) methods. We show that previously developed MM parameters satisfactorily reproduce the trimer structures, as do QM/MM calculations which treat bases with dispersion-corrected DFT and the sugar-phosphate backbone with AMBER. The latter are improved by inclusion of all six bases in the QM region, since a truncated model including only the central CG base pair in the QM region is considerably further from the DFT structure. This QM/MM method is then applied to a set of double-stranded DNA heptamers derived from a recent X-ray crystallographic study, whose size puts a DFT study beyond our current computational resources. These data show that still larger structural changes are observed than in base pairs or trimers, leading us to conclude that it is important to model epigenetic modifications within realistic molecular contexts.
Keywords: Epigenetics, DNA modifications, DNA methylation, Density functional theory, hybrid QM/MM calculations, DNA model systems
Introduction
The standard four-letter alphabet used to encode genetic information in DNA is a central tenet of molecular biology. However, in vivo chemical modification of bases can expand this alphabet markedly, giving rise to a host of important biological phenomena 1. Epigenetic modifications, most importantly DNA methylation and histone variation, have the potential to affect gene expression, and are believed to play a major role in the complex pattern of development and differentiation of multi-cellular organisms. Fascinatingly, such modifications may be heritable despite not affecting DNA sequence, although the mechanism(s) by which this could be achieved are currently unknown.
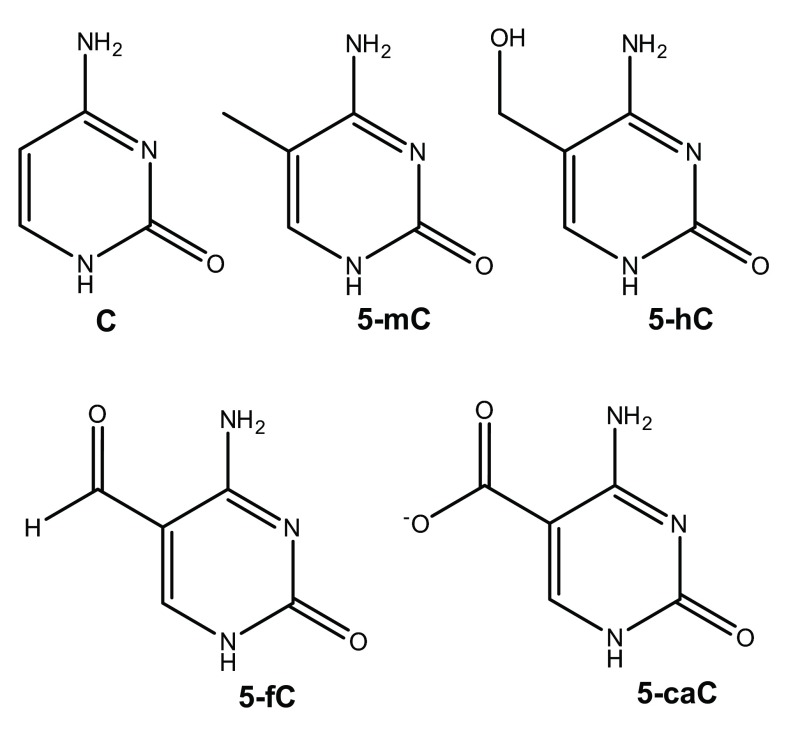
The most common and biologically important such modification involves methylation of the 5 position of cytosine (C) to form 5-methylcytosine (5-mC), illustrated in Figure 1. This does not strongly affect the ability of the base to pair with guanine (G), and in mammals is generally found in CpG sequences, though bacteria and plants display less sequence specificity 2. Oxidation of 5-mC can form 5-hydroxymethylcytosine (5-hmC), which is believed to be involved in regeneration of C via ten-eleven translocation (TET) proteins. Moreover, recent work has shown that 5-formylcytosine (5-fC), and 5-carboxycytosine (5-caC) are present in stem cells and organs of mice 3.
Figure 1. Structures of cytosine and its epigenetic modifications.
The structural consequences of cytosine methylation and related modifications were the focus of a recent study 4 that used X-ray crystallography to show that incorporation of 5-mC or 5-hmC at different points in the d(CGCGAATTCGCG) dodecamer has a negligible effect on both local (base pair) and global (helical) geometry, although specific preference for the orientation of the hydroxyl group in the latter was clearly evident. However, while elegant, the resolution of these studies (between 1.42 and 1.99 Å) may mean that subtle structural changes could go unnoticed. Therefore, molecular modelling, whether based on quantum or classical mechanics, has the potential to contribute significantly in this field. Quantum mechanical models, typically using density functional theory (DFT), have been used to examine the base pairing and stacking of both unmodified (wild-type) and 5-mC DNA. Many groups, including those of Fonseca-Guerra 5– 7, Šponer 8– 13, Leszczynski 14– 16 and others have used DFT to great effect in understanding the structure and properties of unmodified DNA. Regarding epigenetic modifications in particular, Acosta-Silva et al. 17 showed in this manner that methylation enhances stacking interactions, and can produce local distortions in base-pair step parameters, most notably slide. Yusufaly et al. used similar calculations to show that methylation can induce over-twisting as well as softer modes for distortion from the global energy minimum 18. We recently employed classical mechanics to examine not only the structure but also the flexibility of different DNA sequences with methyl and hydroxymethyl substituents 19. Through use of extended molecular dynamics (MD) simulations, we showed that structural effects are subtle, but that epigenetic modifications can give rise to changes in twist, roll and tilt angles that are markedly sequence-dependent. Moreover, introduction of 5-mC within a sequence that already contains hydrophobic groups in the major groove strongly affects hydration patterns, whereas an isolated 5-mC has a lesser effect on solvation and structure.
In this work, we use DFT and QM/MM methods to examine model systems containing modified cytosines. These range from individual base pairs, through double-stranded trimers, to heptamers. By including the sugar-phosphate backbone, sodium counterions and solvent we suggest that these are more realistic models than previous work using similar methods. However, a trimer of DNA brings us close to the size limit for application of DFT with the computing resources available to us. We therefore test and employ hybrid QM/MM methods for larger systems, in which the central bases are treated with dispersion-corrected DFT, while outer bases, sugar-phosphate backbone and solvent (where appropriate) with a molecular mechanics approach, thus allowing accurate and efficient description of systems consisting of hundreds of atoms.
Computational methodology
The initial structures of model systems were built in the canonical B-DNA geometry, using the w3DNA server 20. Hydrogen atoms were added to the system according to expected protonation states at physiological pH using the Molecular Operating Environment (MOE) software package, and Na + were added manually in the vicinity of each phosphate group to produce an overall neutral structure. Where relevant, the central cytosine was also manually modified, and the results of all simulations were analysed using the X3DNA software package 21, 22. Atomic coordinates of wild-type, methylated and hydroxymethylated DNA dodecamers were obtained from X-ray structures deposited in the Protein Data Bank (PDB IDs: 1BNA, 4GJU, 4GLG, 4GLH and 4GLC) 23, and truncated to 5´-ATTCGCG-3´ heptamers containing a single modification on the central C. All DNA termini were capped with methyl groups for simplicity.
All DFT calculations were performed with the Gaussian09 simulation package 20, using Grimme’s B97-D functional 24, that includes an explicit correction for the missing dispersion term in conventional DFT functionals, with either def2-TZVP or 6-31+G(d,p) basis set. This was previously recommended after thorough benchmarking for thermochemistry, kinetics, and non-covalent interactions 25. All such calculations took advantage of the density fitting approximation, and where appropriate included the effect of aqueous solvation via the use of the polarized continuum model (PCM) 26. Binding energies are corrected for the effects of basis set superposition error using the counterpoise method 27.
Hybrid QM/MM calculations were performed using the ONIOM approach with electrostatic embedding 28, as implemented in Gaussian09. The boundary between the quantum and classical regions was chosen as the N-C1’ glycosidic bond in the relevant nucleotide. The QM regions were saturated by the use of a “link” hydrogen atom placed along the N-C1’ vector at an idealized distance, and were modelled at the B97-D/6-31+G(d,p) level of theory, again within PCM water. The MM part of these calculations employed the AMBER force field parm96 29, as defined within Gaussian09. The subtractive nature of the ONIOM method means that undefined terms in the MM expression do not contribute to the overall energy if the relevant atoms are entirely within the QM region, making it ideally suited for the purposes of the current study. We note that this approach has been widely adopted for QM/MM studies of DNA and related structures 30– 32. Pure molecular mechanics (MM) geometry optimisation was also performed using the GROMACS simulation package 33 and the AMBERParmbsc0 force field 34, including RESP charges derived for modified bases in our previous work 19, in explicit aqueous phase, specifically TIP3P water 35 with Na + and Cl - counter ions to create a neutral system.
Results and discussion
Gas-phase base pairs
To examine the effect of modifications on base pairing we examined the structure and energy of gas-phase CG pairs in both hydrogen bonded and stacked orientations, with results reported in Table 1 and Table 2 respectively. These data show that methylation has little effect on the geometry or stability of the Watson-Crick base pair. The presence of a hydroxymethyl slightly weakens the N 4-H 4…O 6 H-bond, perhaps due to the proximity of CH 2OH and NH 2 groups, reported as X…H 4 in Table 1. Formyl has a larger effect overall, lengthening N 3…H 1-N 1 and O 2…H 2-N 2 H-bonds and hence reducing binding by over 3 kcal/mol. The pattern of changes induced by carboxylate is different from all other modifications, lengthening the peripheral H-bonds N 4-H 4…O 6 and O 2…H 2-N 2 markedly, but shortening N 3…H 1-N 1. Despite this weakening, the carboxylate-substituted cytosine binds most strongly to guanine, presumably due to ion-dipole interactions within the anionic system. Both formyl and carboxylate contain close O…H 4 contacts, but overall the proximity of these groups does not appear to be related to strength or geometry of binding.
Table 1. Hydrogen bond lengths and binding energies of CG Watson-Crick base pairs from B97-D/def2-TZVP (Å and kcal/mol).
| N 4-H 4…O 6 | N 3…H 1-N 1 | O 2…H 2-N 2 | X…H 4 a | Binding
Energy |
|
|---|---|---|---|---|---|
| C | 1.663 | 1.819 | 1.835 | 2.455 | -31.19 |
| 5-mC | 1.660 | 1.817 | 1.822 | 2.375 | -31.70 |
| 5-hC | 1.689 | 1.822 | 1.835 | 2.145 | -28.63 |
| 5-fC | 1.670 | 1.834 | 1.884 | 1.990 | -28.07 |
| 5-caC | 1.698 | 1.778 | 1.874 | 1.674 | -34.62 |
a X refers to the atom of the substituent on position 5 closest to H 4.
Table 2. Geometry and binding energies of stacked CG base pairs from B97-D/def2-TZVP (Å, ° and kcal/mol).
| Cent…
Cent a |
Dihedral a | Binding
Energy |
|
|---|---|---|---|
| C | 3.381 | 9.0 | -16.07 |
| 5-mC | 3.310 | 12.5 | -17.52 |
| 5-hC | 3.361 | 9.4 | -22.12 |
| 5-fC | 3.451 | 2.9 | -14.65 |
| 5-caC | 3.823 | 32.6 | -15.56 |
a Cent…Cent refers to the distance between centroids of 6-membered rings; Dihedral refers to the angle between mean planes of rings.
As well as the effect on H-bonding, epigenetic modifications can alter the stacking behaviour of DNA bases. Table 2 reports geometrical details, as well as binding energies, of the five modified cytosines considered here stacked with guanine. All such calculations started from the idealised B-DNA orientation (Cent…Cent = 4.390 Å, Dihedral = 4.9°), and overall this is retained in our gas-phase DFT optimisation. Table 2 shows that methylation leads to closer contact and greater stabilisation between bases, as might be expected due to the increased polarizability of this modified base. Hydroxymethylation leads to the most stable pair considered here, largely due to a strong H-bond between the H—O of hydroxymethyl and O6 of guanine (H…O = 1.770 Å), whereas formylation leads to longer, weaker interaction between bases. Carboxylate-substituted cytosine is the only case considered here that loses the approximately parallel orientation of bases. This appears to be driven as much by repulsion between the carboxylate group and C=O 6 of guanine as by H-bonding.
Double-stranded DNA trimers
While these gas-phase dimers give useful information on the intrinsic effect of modifications on cytosine’s ability to interact with guanine, environmental effects including the DNA sequence, sugar-phosphate backbone and solvent will play a major role in determining their effect in real systems. In order to better simulate the behaviour of modified cytosines in real systems, structures of double-stranded d(GCG) and d(ACA), as well as epigenetic modifications to the central cytosine were optimized using DFT in continuum solvent (PCM), and the resulting geometries of the local base pairs were analysed in the coordinate frame recommended by Olson et al. 36. Unlike the free dimers considered above, modifications have only subtle effects on this larger structure, which retains the overall canonical B-DNA shape of the unmodified WT structure.
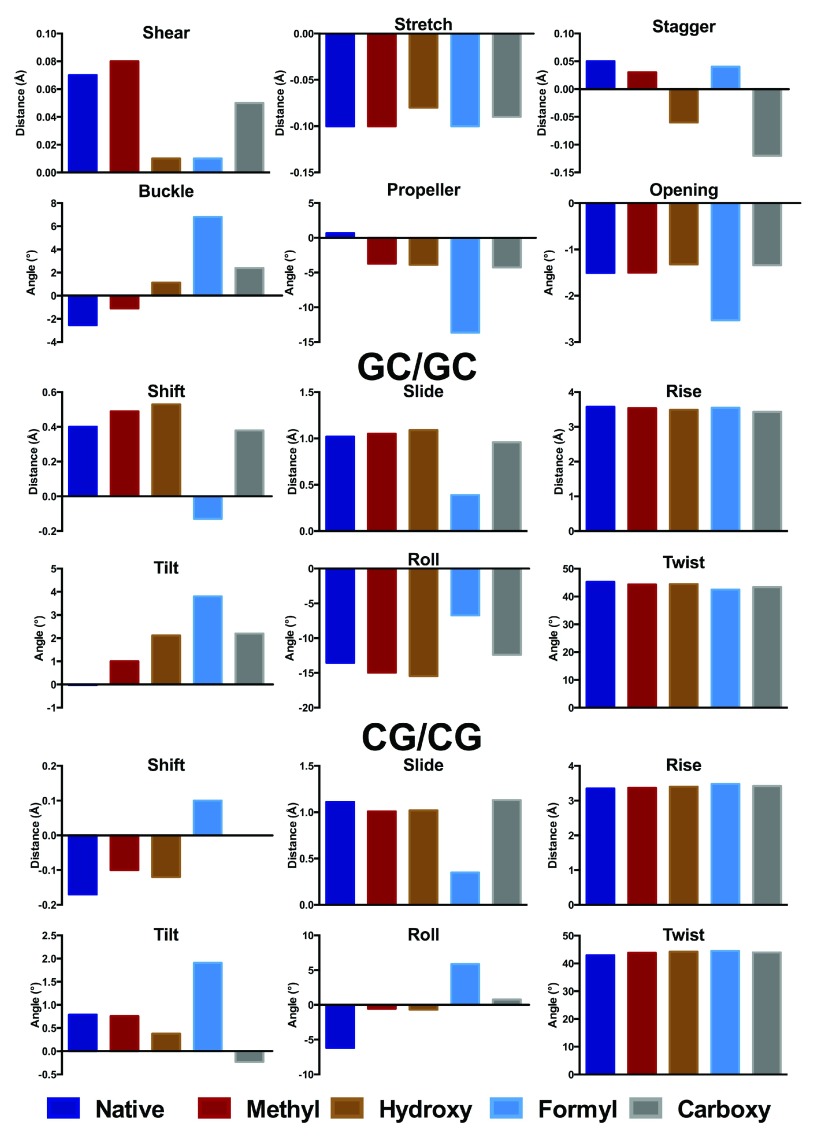
Following Zubatiuk et al. 37, we summarise key aspects of trimer structure, which are displayed graphically in Figure 2 and Figure 3. The corresponding values are tabulated in Table S1 of the Supporting Information, with the base step and local helical parameters tabulated in Table S2. As with Zubatiuk et al. 14, base pair step parameters are averaged over 3´ and 5´ directions. In the GCG oligomer, methylation has only a small effect on base pair distances, but does alter the propeller angle by over 4°. Hydroxymethylation has a larger effect on the GCG oligomer, especially on the stagger, buckle and propeller, whereas the stretch and opening parameters are much less affected. Formyl does not strongly affect base pair distances but does change angles substantially, especially buckle and propeller, which change by as much as 10°. In contrast, carboxylate induces a large change in stagger but only small changes in angular geometry. Base pair step parameters for d(GCG) in general are less affected than those for the base pair noted above, with the exception of formyl which exhibits smaller slide and less negative roll values than unmodified DNA.
Figure 2. Base pair (A) and step (B, C) parameters for central GC in d(GCG) and modifications (Å and °).
The corresponding data are provided in Table S1.
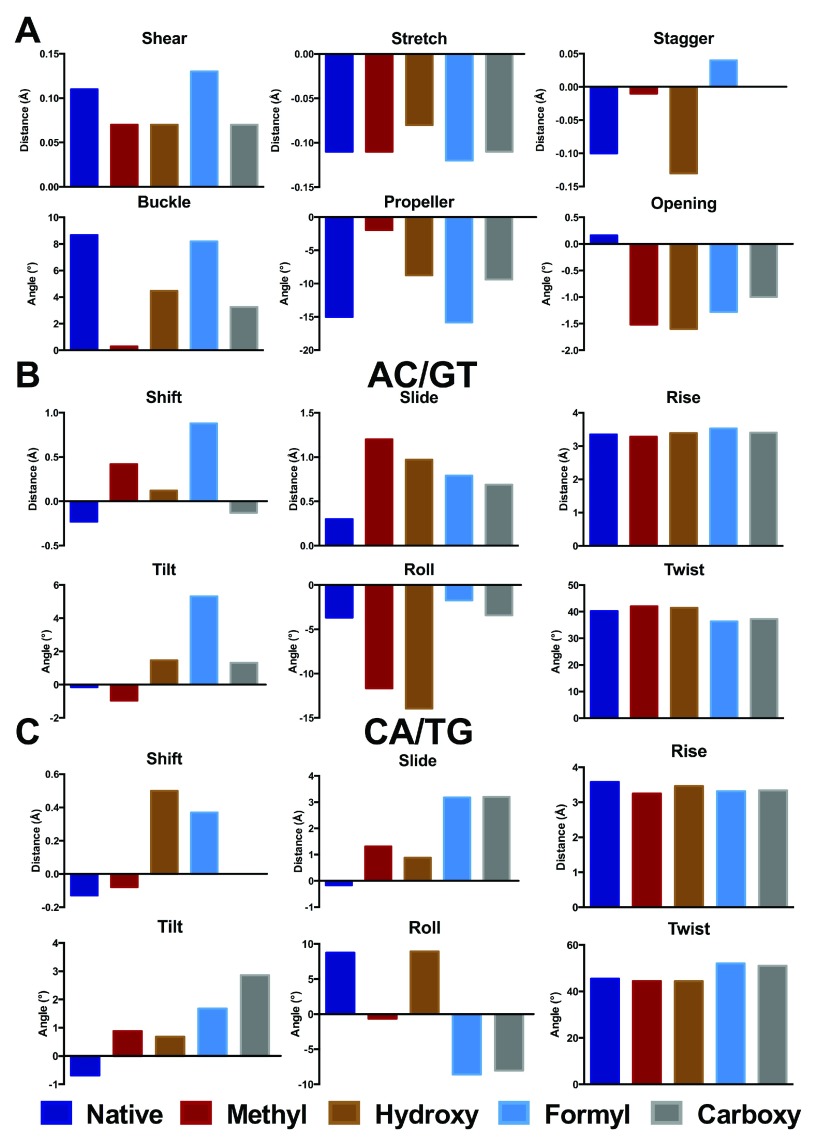
Figure 3. Base pair (A) and step (B, C) parameters for central GC in d(ACA) and modifications (Å and °).
The corresponding data are provided in Table S1.
Rather larger changes are evident on modification of d(ACA), as shown in Figure 3. In this case, even methylation induces significant changes in distances, especially stagger which increases by 0.1 Å, and angles (buckle and propeller change by 8 and 13°, respectively). At the base pair step level, methylation gives rise to substantial increase (0.9 and 1.5 Å) in slide and more negative roll in both 3´ and 5´ directions. Less apparent in Figure 3, but still notable, are changes in rise that are 0.1 and 0.3 Å smaller in the methylated structure, reflecting the greater stacking that results from addition of a methyl group. Other modifications induce different patterns of structural change: for the central base pair these changes are typically smaller than for methylation, but for base pair steps much larger changes are found in some parameters. Most notable of these are slide, which changes by over 3 Å and roll (up to 17°) in the 3´ direction, in a similar way to that reported previously for smaller systems 17, 18. Other parameters such as the width of the DNA strand, measured as the distance between C1´ nuclei, and virtual angles λ Y and λ R, which describe the pivoting of complementary bases in the base-pair plane, vary only slightly from the idealised values for B-DNA.
QM/MM studies of double-stranded oligomers
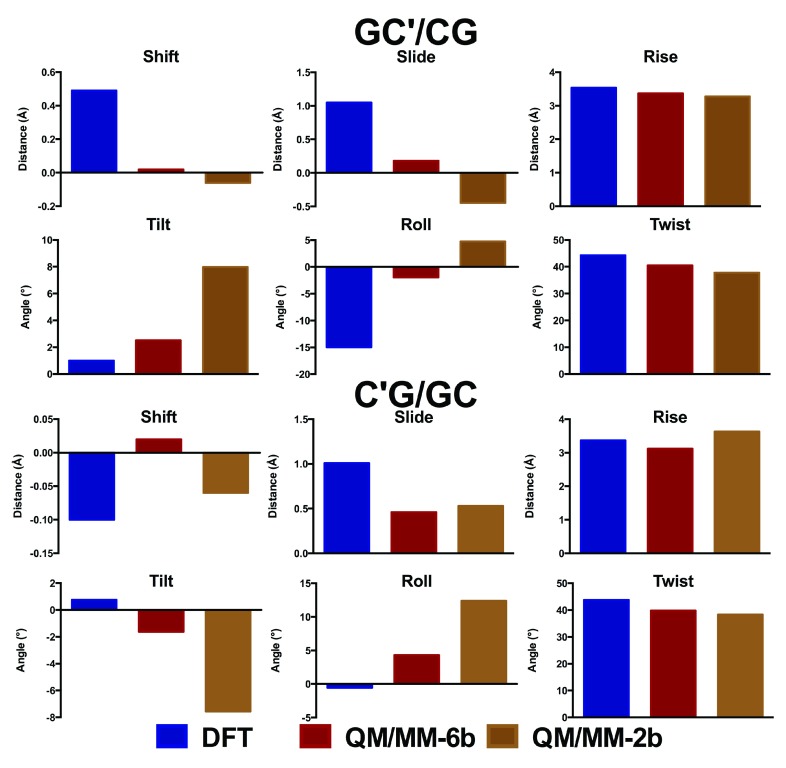
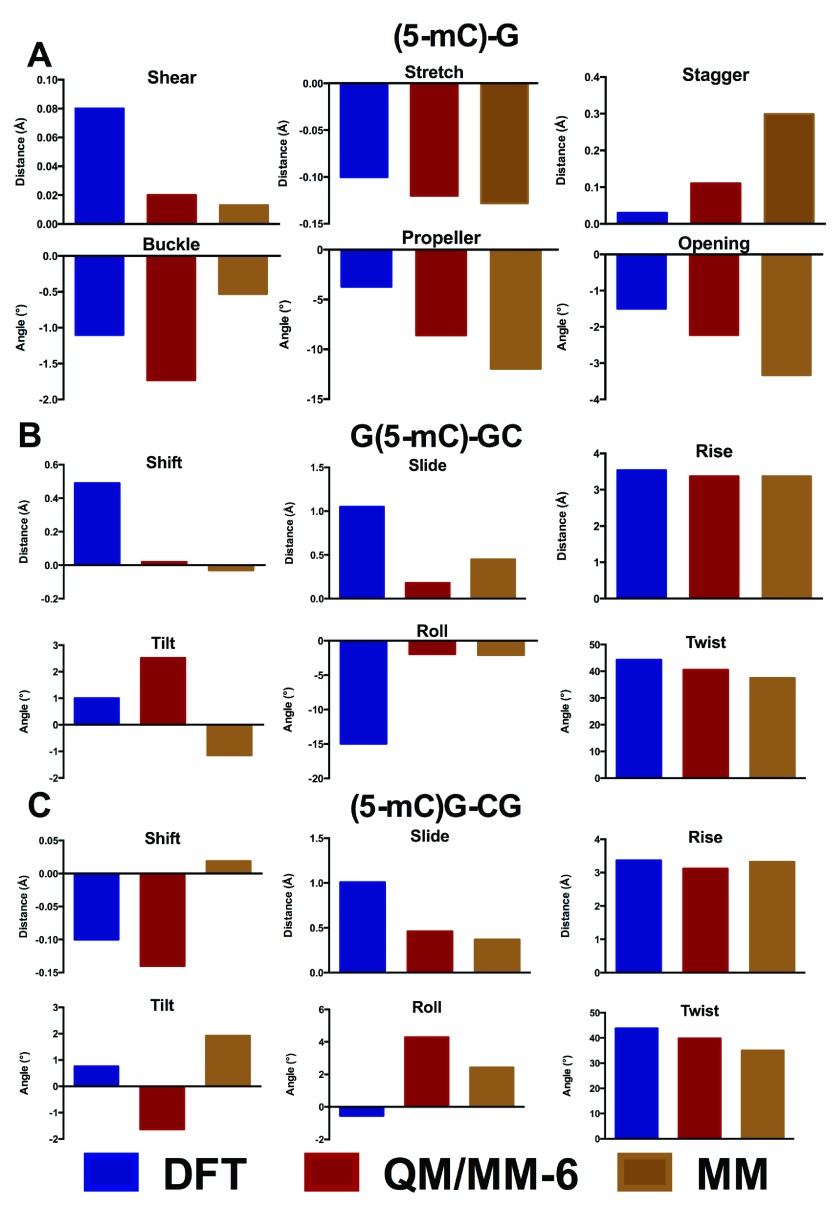
The oligomers considered so far are close to the limit of our computational capabilities of current DFT methods (the largest structure, carboxylated d(ACA), has 962 electrons in 2743 basis functions), such that longer sequences cannot currently be routinely studied in this manner. However, they are too small to correctly represent how DNA behaves in a real system, where the conformations adopted by each base pair step depend on the neighbouring step. Moreover, simulations of nucleic acids are known to suffer problems due to greater elasticity of the terminal part of the structure (the so called “end-effect” 38). For these reasons, these small oligomers are inadequate models to probe the effects of epigenetic modifications on the structure of DNA. We therefore turn to hybrid QM/MM methods, in which a subset of the atoms in the system is treated with DFT, and the remainder of the system with much faster molecular mechanics methods. In order to test the validity of this approach, methylated GCG was optimized using either only two or six bases in the QM region ( Figure 4). These tests show that including only two bases in the QM region leads to significant differences in geometry to that obtained from DFT, particularly in the stagger and buckle coordinates. In contrast, including six bases in the QM region reproduces the DFT structure reasonably well. Similar observations were made from analogous treatment of methylated ACA (data not shown).
Figure 4. Comparison of QM/MM with DFT geometry (Å and °).
The corresponding values are shown in Tables S3 and S4.
As a further test, we also compared DFT and QM/MM derived structures with those optimised using the force field parameters developed in our previous work. Figure 5 shows the base-pair parameter values of the methylated structure d(GC´G) for the different methods. The MM structures provide very close values to those obtained by both QM/MM and DFT approaches, showing slight difference only in the stagger and propeller angle. We can therefore conclude that for small DNA oligomers, DFT, QM/MM and MM methods can all produce almost equally adequate DNA structures, but that QM/MM and MM approaches are more similar to one another than those obtained from DFT alone.
Figure 5. Comparison of DFT, QM/MM and molecular mechanics energy minimised (EM) geometries of d(GC´G) (Å and °).
The corresponding values are shown in Tables S3 to S6.
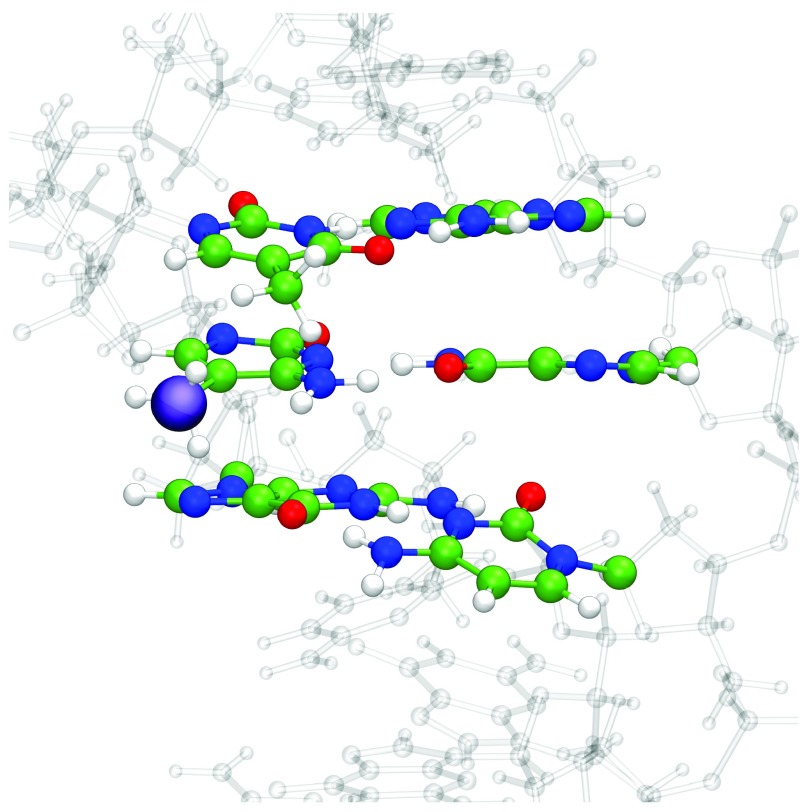
QM/MM geometry optimization with six bases in the QM region was then applied to a set of larger DNA sequences. The experimental structure of Renčiuk et al. 4 obtained using X-ray diffraction (PDB Entry 4GLG) was truncated to a sequence of 7 base pairs, i.e. 5´-ATT CGCG-3´, and the central 6 bases (TCG//CGA) assigned as QM atoms. The remaining atoms, including crystallographic water molecules and counterions, were assigned to the MM layer, and the entire system was geometry optimized. The resulting optimized structure of the system with methylated C in the central position is shown in Figure6. Base pair and base pair step geometries of wild type, methylated, hydroxymethylated structures optimised with QM/MM, along with experimental values for methylated C, are shown in Figure 7.
Figure 6. QM/MM optimised structure of 5´-ATTCGCG-3´ with 5-mC in central position, and the bases defined as QM atoms shown in CPK.
A purple sphere highlights the methylation position, and water molecules and counterions have been omitted for clarity.
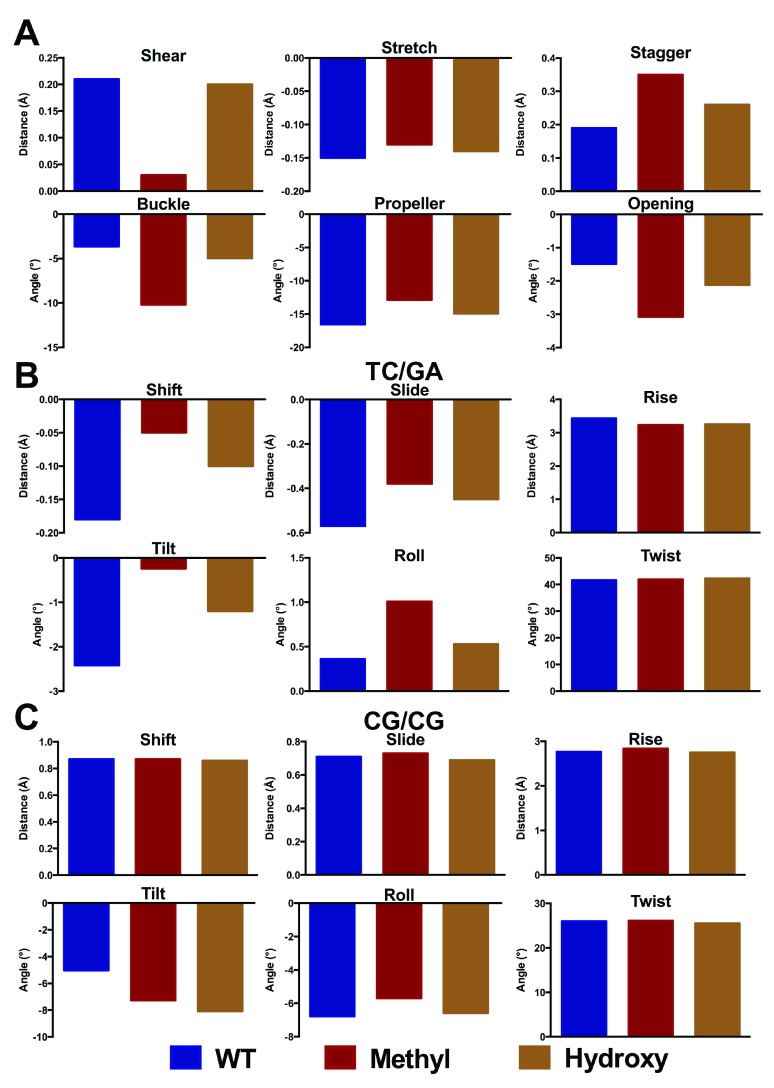
Figure 7. Base pair step parameters for central GC in 5´-ATT CGCG-3´ and modifications (Å and °).
The corresponding values are shown in Table S2.
We find that the structural effect of methylation is larger in this longer sequence than in the trimers considered above. Particularly, the optimized values of shear, stagger and buckle of the central base pair differ markedly between the methylated and WT forms of DNA. In contrast, the base pair step parameters exhibit rather smaller changes. For the hydroxymethylated structures, we observe similar profiles to the methylated structures. Furthermore, our simulations also allow us to probe the preferred orientation of the hydroxymethyl group: our DFT calculations predict a slight preference for the OH group to point in 3´ over 5´ and an optimized of C6-C5-C5A-O5 torsion angle of 118.4°, while previous MD simulations show this torsion to vary between 85 and 120° over 100 ns of simulation 19. This is in good agreement with the experimental and theoretical results of Renčiuk et al. 39, who reported values between 72 and 133° using X-ray diffraction methods.
Data file 1 Local base pair parameters for the central GC in d(GCG) and d(ACA) modifications. The shear, stretch and stagger parameters are measured in Å, and the buckle, propeller and opening parameters are measured in ˚.
Data file 2 Base pair step and local helical parameters for the central GC in d(GCG) and d(ACA) modifications. The slide and X-displacement parameters are measured in Å, and the roll, twist and inclination parameters are measured in ˚.
Data file 3 Comparison between the obtained base pair parameters for the d(GCG) using different levels of theory (DFT and the QM/MM with 2bps or 6bps in the high level layer). The shear, stretch and stagger parameters are measured in Å, and the buckle, propeller and opening parameters are measured in ˚.
Data file 4 Comparison between the obtained base step parameters for the d(GCG) using different levels of theory (DFT and the QM/MM with 2bps or 6bps in the high level layer). The slide, shift and rise parameters are measured in Å, and the tilt, roll and twist parameters are measured in ˚.
Data file 5 Values of local base pair parameters of central GC pair of d(GCG) obtained after energy minimization. The shear, stretch and stagger parameters are measured in Å, and the buckle, propeller and opening parameters are measured in ˚.
Data file 6 Values of base pair step parameters of d(GCG), obtained after energy minimization. The shift, slide and rise parameters are measured in Å, and the tilt, roll and twist parameters are measured in °.
Data file 7 Cartesian coordinates of key species.
Copyright: © 2015 Carvalho ATP et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Conclusions
Through use of modern, dispersion-corrected DFT and hybrid QM/MM methods, we have examined the structural consequences of epigenetic modifications of DNA. Concentrating on methylation and related modifications of cytosine, we show that the overall Watson-Crick base-pairing is retained, with rather small changes to hydrogen bond and stacking geometries. Despite this, some modifications have a substantial effect on the strength of intermolecular interactions: hydroxymethyl and formyl groups reduce H-bonding strength, while carboxylate increases this markedly.
Situating these modifications within the double-stranded DNA trimers GCG and ACA allows us to examine the effects on the central CG base pair and base pair steps. Base pair geometries undergo rather larger changes within ACA than in GCG, with changes in buckle and propeller angles particularly apparent. Changes to base pair steps are smaller, although some changes in shift and slide values due to modifications are evident. Optimised geometries also act as a useful test of hybrid QM/MM methods. These can reproduce DFT structures if all six bases are included in the QM region, but if only the central base pair is treated with QM significant differences result. This approach is then applied to heptamers derived from a recent X-ray crystallography; here again, the central base pair is found to be significantly disrupted, whereas base pair step parameters are largely retained.
The studies reported here deal solely with static structures, but it is well-known that DNA is a flexible system that is in constant motion at biologically relevant temperatures. In previous work, we showed that long timescale molecular dynamics was able to highlight subtle differences in structure, flexibility and solvation resulting from incorporation of 5-mC and 5-hC in several different DNA sequences. The work reported here gives new insight into the intrinsic effects of epigenetic modification of cytosine, complementing our previous molecular dynamics study 19 as well as providing support for the molecular mechanics force field chosen for that work.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2015 Carvalho ATP et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Figshare: Data of theoretical modelling of epigenetically modified DNA sequences. Doi: 10.6084/m9.figshare.1310448 40
Acknowledgements
JAP is grateful to Advanced Research Computing @ Cardiff (ARCCA) for use of computing facilities.
Funding Statement
This work was supported by the Swedish Research Council (Vetenskapsrådet, 2010-5026) and funding from the Sven and Ebba Christina Hagbergs Foundation.
We confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 1 approved
References
- 1. Korlach J, Turner SW: Going beyond five bases in DNA sequencing. Curr Opin Struc Biol. 2012;22(3):251–61. 10.1016/j.sbi.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 2. Colot V, Rossignol JL: Eukaryotic DNA methylation as an evolutionary device. Bioessays. 1999;21(5):402–11. [DOI] [PubMed] [Google Scholar]
- 3. Ito S, Shen L, Dai Q, et al. : Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Renčiuk D, Blacque O, Vorlickova M, et al. : Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine. Nucleic Acids Res. 2013;41(21):9891–900. 10.1093/nar/gkt738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barone G, Guerra CF, Bickelhaupt FM: B-DNA structure and stability as function of nucleic acid composition: Dispersion-corrected DFT study of dinucleoside monophosphate single and double strands. ChemistryOpen. 2013;2(5–6):186–193. 10.1002/open.201300019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Wijst T, Guerra CF, Swart M, et al. : Performance of various density functionals for the hydrogen bonds in DNA base pairs. Chem Phys Lett. 2006;426(4–6):415–421. 10.1016/j.cplett.2006.06.057 [DOI] [Google Scholar]
- 7. Poater J, Swart M, Guerra CF, et al. : Solvent effects on hydrogen bonds in Watson–Crick, mismatched, and modified DNA base pairs. Comp Theoret Chem. 2012;998:57–63. 10.1016/j.comptc.2012.06.003 [DOI] [Google Scholar]
- 8. Mládek A, Šponer JE, Jurečka P, et al. : Conformational energies of DNA sugar-phosphate backbone: Reference QM calculations and a comparison with density functional theory and molecular mechanics. J Chem Theory Comput. 2010;6(12):3817–3835. 10.1021/ct1004593 [DOI] [Google Scholar]
- 9. Svozil D, Hobza P, Šponer J: Comparison of intrinsic stacking energies of ten unique dinucleotide steps in A-RNA and B-DNA duplexes. Can we determine correct order of stability by quantum-chemical calculations? J Phys Chem B. 2009;114(2):1191–1203. 10.1021/jp910788e [DOI] [PubMed] [Google Scholar]
- 10. Banáš P, Mládek A, Otyepka M, et al. : Can we accurately describe the structure of adenine tracts in B-DNA? Reference quantum-chemical computations reveal overstabilization of stacking by molecular mechanics. J Chem Theory Comput. 2012;8(7):2448–2460. 10.1021/ct3001238 [DOI] [PubMed] [Google Scholar]
- 11. Fonville JM, Swart M, Vokacova Z, et al. : Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment. Chemistry. 2012;18(39):12372–87. 10.1002/chem.201103593 [DOI] [PubMed] [Google Scholar]
- 12. Mládek A, Šponer JE, Kulhánek P, et al. : Understanding the sequence preference of recurrent RNA building blocks using quantum chemistry: The intrastrand RNA dinucleotide platform. J Chem Theory Comput. 2011;8(1):335–347. 10.1021/ct200712b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morgado CA, Svozil D, Turner DH, et al. : Understanding the role of base stacking in nucleic acids. MD and QM analysis of tandem GA base pairs in RNA duplexes. Phys Chem Chem Phys. 2012;14(36):12580–91. 10.1039/c2cp40556c [DOI] [PubMed] [Google Scholar]
- 14. Zubatiuk TA, Shishkin OV, Gorb L, et al. : B-DNA characteristics are preserved in double stranded d(A)3·d(T)3 and d(G)3·d(C)3 mini-helixes: conclusions from DFT/M06-2X study. Phys Chem Chem Phys. 2013;15(41):18155–66. 10.1039/c3cp51584b [DOI] [PubMed] [Google Scholar]
- 15. Gu J, Wang J, Xie Y, et al. : Structural and electronic property responses to the arsenic/phosphorus exchange in GC-related DNA of the B-form. J Comput Chem. 2012;33(8):817–21. 10.1002/jcc.22880 [DOI] [PubMed] [Google Scholar]
- 16. Gu J, Wang J, Leszczynski J: Stacking and H-bonding patterns of dGpdC and dGpdCpdG: Performance of the M05-2X and M06-2X Minnesota density functionals for the single strand DNA. Chem Phys Lett. 2011;512(1–3):108–112. 10.1016/j.cplett.2011.06.085 [DOI] [Google Scholar]
- 17. Acosta-Silva C, Branchadell V, Bertran J, et al. : Mutual relationship between stacking and hydrogen bonding in DNA. Theoretical study of guanine-cytosine, guanine-5-methylcytosine, and their dimers. J Phys Chem B. 2010;114(31):10217–27. 10.1021/jp103850h [DOI] [PubMed] [Google Scholar]
- 18. Yusufaly TI, Li Y, Olson WK: 5-Methylation of cytosine in CG:CG base-pair steps: a physicochemical mechanism for the epigenetic control of DNA nanomechanics. J Phys Chem B. 2013;117(51):16436–42. 10.1021/jp409887t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carvalho AT, Gouveia L, Kanna CR, et al. : Understanding the structural and dynamic consequences of DNA epigenetic modifications: Computational insights into cytosine methylation and hydroxymethylation. Epigenetics. 2014;9(12):1604–12. 10.4161/15592294.2014.988043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frisch MJ, Trucks GW, Schlegel HB, et al. : Gaussian Rev. C.01, Gaussian, Inc.: Wallingford, CT, USA,2009. [Google Scholar]
- 21. Lu XJ, Olson WK: 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31(17):5108–21. 10.1093/nar/gkg680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu XJ, Olson WK: 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3(7):1213–27. 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drew HR, Wing RM, Takano T, et al. : Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981;78(4):2179–2183. 10.1073/pnas.78.4.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grimme S: Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J Comput Chem. 2006;27(15):1787–99. 10.1002/jcc.20495 [DOI] [PubMed] [Google Scholar]
- 25. Goerigk L, Grimme S: A thorough benchmark of density functional methods for general main group thermochemistry, kinetics, and noncovalent interactions. Phys Chem Chem Phys. 2011;13(14):6670–88. 10.1039/c0cp02984j [DOI] [PubMed] [Google Scholar]
- 26. Miertuš S, Scrocco E, Tomasi J: Electrostatic interaction of a solute with a continuum. A direct utilization of AB initio molecular potentials for the prevision of solvent effects. Chemical Physics. 1981;55(1):117–129. 10.1016/0301-0104(81)85090-2 [DOI] [Google Scholar]
- 27. Boys SF, Bernardi F: The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys. 1970;19(4):553–566. 10.1080/00268977000101561 [DOI] [Google Scholar]
- 28. Bakowies D, Thiel W: Hybrid models for combined quantum mechanical and molecular mechanical approaches. J Phys Chem. 1996;100(25):10580–10594. 10.1021/jp9536514 [DOI] [Google Scholar]
- 29. Cornell WD, Cieplak P, Bayly CI, et al. : A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117(19):5179–5197. 10.1021/ja00124a002 [DOI] [Google Scholar]
- 30. Cerón-Carrasco JP, Requena A, Jacquemin D: Impact of DFT functionals on the predicted magnesium–DNA interaction: an ONIOM study. Theor Chem Acc. 2012;131:1188 10.1007/s00214-012-1188-9 [DOI] [Google Scholar]
- 31. Sundaresan N, Pillai CK, Suresh CH: Role of Mg 2+ and Ca 2+ in DNA bending: Evidence from an ONIOM-based QM-MM study of a DNA fragment. J Phys Chem A. 2006;110(28):8826–31. 10.1021/jp061774q [DOI] [PubMed] [Google Scholar]
- 32. Ahmadi F, Jahangard-Yekta S, Heidari-Moghadam A, et al. : Application of two-layer ONIOM for studying the interaction of N-substituted piperazinylfluoroquinolones with ds-DNA. Comp Theor Chem. 2013;1006:9–18. 10.1016/j.comptc.2012.11.006 [DOI] [Google Scholar]
- 33. Pronk S, Páll S, Schulz R, et al. : GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pérez A, Marchán I, Svozil D, et al. : Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys J. 2007;92(11):3817–3829. 10.1529/biophysj.106.097782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jorgensen WL, Chandrasekhar J, Madura JD, et al. : Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- 36. Olson WK, Bansal M, Burley SK, et al. : A standard reference frame for the description of nucleic acid base-pair geometry. J Mol Biol. 2001;313:229–237. 10.1006/jmbi.2001.4987 [DOI] [PubMed] [Google Scholar]
- 37. Olson WK, Gorin AA, Lu XJ, et al. : DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95(19):11163–8. 10.1073/pnas.95.19.11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dixit SB, Bevridge DL, Case DA, et al. : Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: Sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys J. 2005;89(6):3721–40. 10.1529/biophysj.105.067397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Renčiuk D, Kejnovská I, Školáková P, et al. : Arrangements of human telomere DNA quadruplex in physiologically relevant K + solutions. Nucleic Acids Res. 2014;37(19):6625–6634. 10.1093/nar/gku1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamerlin SCL, Platts J, Carvalho ATP: Data of theoretical modelling of epigenetically modified DNA sequences. Figshare. 2014. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]