The human Mediator complex controls RNA polymerase II (pol II)function in ways that remain incompletely understood. Activator–Mediator binding alters Mediator structure, and these activator-induced structural shifts appear to play key roles in regulating transcription. A recent cryo-EM analysis revealed that pol II adopted a stable orientation within a Mediator–pol II–TFIIF assembly in which Mediator was bound to the activation domain of VP16. Whereas TFIIF was shown to be important for orienting pol II within this assembly, the potential role of the activator was not assessed. To determine how activator binding might affect pol II orientation, we isolated human Mediator–pol II–TFIIF complexes in which Mediator was not bound to an activator. Cryo-EM analysis of this assembly, coupled with pol II crystal structure docking, revealed that pol II binds Mediator at the same general location; however, in contrast to VP16-bound Mediator, pol II does not appear to stably orient in the absence of an activator. Variability in pol II orientation might be important mechanistically, perhaps to enable sense and antisense transcription at human promoters. Because Mediator interacts extensively with pol II, these results suggest that Mediator structural shifts induced by activator binding help stably orient pol II prior to transcription initiation.
The large size of the transcription initiation machinery and the inherent flexibility of its components present formidable challenges for structural analysis. These challenges are compounded by the fact that large-scale structural shifts are triggered upon activator binding to human Mediator complexes.1–3 Electron microscopy is suited to study large, conformationally flexible assemblies because it enables individual complexes to be grouped into structurally distinct classes. This circumvents potential problems with structural heterogeneity, provided the majority of complexes are structurally uniform.
Unlike other general transcription factors (e.g. TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and pol II) that comprise the Pre-Initiation Complex (PIC), human Mediator was discovered in the late 1990s.4–13 As a consequence, Mediator has not been studied as extensively as other PIC factors, such as pol II or even TFIID. Many fundamental questions regarding Mediator function remain unknown, such as how it helps control pol II activity or how it regulates PIC function. The 26-subunit, 1.2 MDa human Mediator complex interacts directly with the pol II enzyme and is a general target for DNA-binding transcription factors (a.k.a. activators). Thus, Mediator appears to function as a “molecular bridge” to allow activators to communicate regulatory signals to the pol II enzyme.14,15 Interestingly, activator-Mediator binding appears to stabilize the PIC16 and triggers major structural shifts within the human Mediator complex, including shifts within regions that bind the pol II enzyme.1,2 These activator-induced structural shifts appear to regulate pol II activity,3,17 but the precise mechanisms involved remain unclear.
To address whether activator-Mediator binding might impact PIC structure, we initiated a cryo-negative stain EM analysis (hereafter referred to as cryo-EM) of the Mediator–pol II–TFIIF assembly, which represents approximately half the protein density of the entire human PIC. Because Mediator interacts extensively with the pol II enzyme and is generally required for pol II-dependent transcription, we hypothesized that activator-induced structural shifts might serve to alter PIC structure at the promoter. We recently completed cryo-EM analysis of Mediator–pol II–TFIIF, in which Mediator was bound to the activation domain of viral protein 16 (VP16).18 Structural comparison of this VP16-Mediator–pol II–TFIIF assembly with an assembly that simply lacks the activation domain would help determine whether activators might serve additional structural roles during PIC assembly and transcription initiation.
Isolation of the Mediator–pol II–TFIIF assembly
Mediator, pol II, and TFIIF were each purified independently, as shown in Figure 1. Note that Mediator was isolated without an activator bound, using a monoclonal MED26 antibody column, followed by elution of the complex with peptide. To isolate the Mediator–pol II–TFIIF assembly, we used a strategy that was successful in purifying Mediator–pol II–TFIIF assemblies bound to VP16.18 We first ensured that the pol II CTD was thoroughly de-phosphorylated by incubating purified pol II over a phosphatase resin. This de-phosphorylated pol II sample was then mixed with TFIIF, followed by the addition of Mediator (Fig. 2A). This sample was then separated over a glycerol gradient, which effectively resolved free TFIIF and free pol II from fully-assembled Mediator–pol II–TFIIF complexes, which concentrate in the final fraction of the gradient (Fig. 2B). Western blot analysis confirmed the presence of Mediator, pol II and TFIIF in this final fraction (Fig. 2C). These data suggested that a stable Mediator–pol II–TFIIF assembly could form in the absence of an activator. To probe the molecular architecture of this assembly, we next examined this Mediator–pol II–TFIIF fraction using electron microscopy (EM).
Fig. 1.
Purification of (A) pol II, (B) Mediator, and (C) TFIIF. In each case, a schematic of the purification protocol is shown at left, with gels showing the purified complex at the right. Pol II and Mediator gels were stained with silver whereas TFIIF was stained with coomassie. Identities of subunits are shown at the right of each gel.
Fig. 2.
Isolation of the Mediator–pol II–TFIIF assembly. (A) Schematic of the protocol. The gradient was set up such that the complete Mediator–pol II–TFIIF assembly would migrate and concentrate in the final fraction. Note that Mediator is not bound to an activator in these experiments. (B) Silver-stained gel of the glycerol gradient fractions. Free TFIIF, free Mediator, and free pol II migrate earlier in the gradient, as indicated. Subunit identities shown at the right. (C) Western blot analysis confirms the presence of Mediator, pol II, and TFIIF in the final gradient fraction, as expected. The pol II antibody was directed against RPB1, the Mediator antibody targeted MED23, and the TFIIF antibody RAP74.
Cryo-EM analysis of Mediator–pol II–TFIIF
Although we recently completed a cryo-EM analysis of Mediator–pol II–TFIIF in which Mediator was bound to the activation domain of VP1618 (hereafter referred to as VP16-Mediator–pol II–TFIIF), we wanted to independently evaluate the structure of activator-free Mediator–pol II–TFIIF. Therefore, we did not use the VP16-Mediator–pol II–TFIIF structure as a starting model. Instead, we used an unbiased, reference-free approach that was initiated by imaging complexes in negative stain (2% uranyl acetate) using random conical tilt methods. Examination of individual micrographs revealed particles of a size and shape consistent with Mediator–pol II–TFIIF (Supplemental Fig. 1A). Parallel data sets were obtained in untilted and tilted (30° – 45°) stage orientations, resulting in a data set of 26,718 single-particle images (13,359 untilted and 13,359 tilted, from 146 micrograph pairs). Alignment and 2D classification of the untilted images resulted in a majority of 2D projection class averages that resembled Mediator–pol II–TFIIF, based upon our previous work with VP16-Mediator–pol II–TFIIF. A fraction of the 2D classes correlated poorly with Mediator–pol II–TFIIF assemblies and appeared to represent Mediator itself (i.e. not bound to pol II–TFIIF). These classes were not included in the refinement of Mediator–pol II–TFIIF. Whereas a majority of 2D classes appeared to represent Mediator–pol II–TFIIF assemblies, many of these classes, when grouped together in an angular refinement, resulted in structures that were discontinuous within the body domain of Mediator (see Supplementary Methods). This suggested structural variability within this region, perhaps due to multiple distinct pol II orientations within the assembly. Indeed, similar discontinuous structures were observed in past studies of VP16-Mediator–pol II in the absence of TFIIF, a situation in which pol II does not adopt a stable orientation.18 A structurally homogeneous data set, representing 16% of the negative-stain data, was ultimately subjected to multiple angular refinement steps and resulted in a 3D reference volume suitable for further refinement with cryo-EM data (Supplemental Fig. 1B, C, D). Extra density was evident at the head domain of Mediator, indicative of pol II–TFIIF binding. Pol II–TFIIF also binds this region in the presence of VP16-Mediator.18 In addition, yeast pol II appears to bind the head domain of yeast Mediator, indicating some structural similarity between yeast and human Mediator–pol II complexes.19,20
For cryo-EM analysis, we used the same Mediator–pol II–TFIIF sample (Fig. 2B) that was used for analysis in negative stain. Micrographs were first screened for defocus and astigmatism; a representative micrograph and power spectrum is shown in Supplemental Figure 2A and 2B. Single-particle images (9,169) were then selected and windowed from 139 high-quality micrographs. The preliminary Mediator– pol II–TFIIF structure, generated from negatively-stained samples (Supplemental Fig. 1B), was low-pass filtered to 57 Å resolution and used as a starting reference for angular refinement with cryo-EM data. The refined 3D structure of the activator-free Mediator–pol II–TFIIF assembly is shown in Figure 3A (see also Supplemental Fig. 2C and D). Whereas the structure generally resembles that of the VP16-Mediator–pol II– TFIIF assembly, notable differences are apparent. These differences derive at least in part from varied pol II orientations within the activator-free assembly (see below).
Fig. 3.
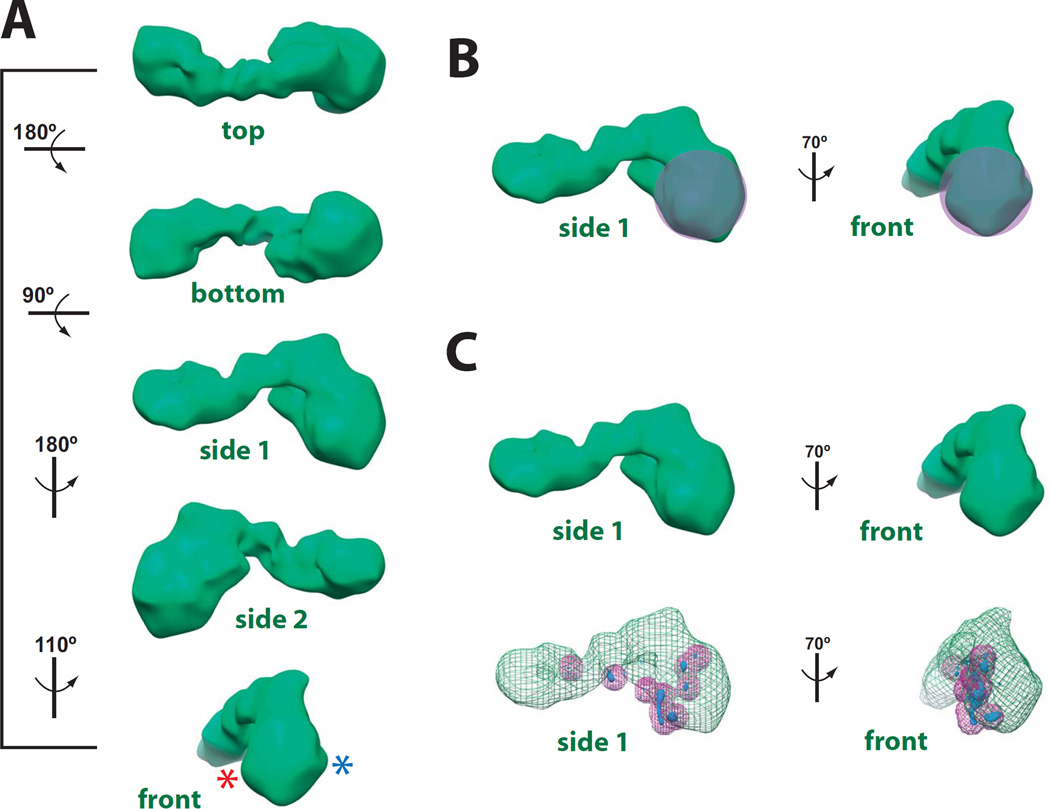
Three-dimensional structure and pol II docking results for the activator-free Mediator–pol II–TFIIF assembly. (A) Different views of the cryo-EM reconstruction, rendered to 1.8 MDa. Rotation of the structure shown at left. The resolution of this structure is 32 Å, based upon both the Fourier Shell Correlation (0.5 cutoff) and the half-bit threshold.54,55 Asterisks for the “front” view represent sites where the pol II stalk protrudes from the structure, based upon 1) extra protein density observed at higher mass thresholds (not shown), and 2) the top pol II docking results based upon cross-correlation coefficients with 8 different pol II crystal structures (see Supplemental Table 1). The absence of clear pol II stalk density at 1.8MDa rendering is consistent with multiple pol II orientations within the sample (see text). The cryo-EM map has been deposited in the EM Databank (http://www.ebi.ac.uk/pdbe/emdb/), entry number 5344.(B) General location of pol II, based upon the docking experiments. (C) The 3D variance map for Mediator–pol II–TFIIF. The variance map is displayed in blue and superimposed upon the Mediator–pol II–TFIIF cryo-EM map, which is shown in green mesh. To enhance visualization, peaks in the 3D variance map are also marked with purple spheres with a radius of 5 pixels (21 Å).
A separate cryo-EM angular refinement was completed using a multi-reference approach.21,22 Because the random conical tilt, negative stain EM data (described above) revealed free Mediator was present together with Mediator–pol II–TFIIF in the sample, we utilized a multi-reference refinement strategy that separated particles corresponding to Mediator alone from Mediator–pol II–TFIIF complexes. This analysis partitioned approximately 19% of complexes to free Mediator and 60% of single-particle images to the Mediator–pol II–TFIIF assembly, which was consistent with the single-reference angular refinement described above. Notably, the structure of the Mediator–pol II–TFIIF assembly was essentially identical relative to the single-reference refinement; furthermore, similar pol II docking results were obtained from this structure (see below).
In the absence of an activator, pol II does not stably orient within Mediator–pol II–TFIIF
To determine the orientation of pol II within the Mediator–pol II–TFIIF assembly, we performed independent docking calculations using several crystal structures of the yeast pol II enzyme (Supplemental Table 1). The structure corresponding to PDB 1Y1V was initially used for the docking calculations because this structure correlated best with a cryo-EM reconstruction of human pol II.23 Unlike pol II docking with the VP16-Mediator–pol II–TFIIF cryo-EM structure,18 a single, stable pol II orientation was not evident based upon pol II docking calculations. Whereas pol II consistently localized to the head/body interface of Mediator, its orientation was variable (Fig. 3B and Supplemental Table 1).
The major pol II structural features include the cleft (ca. 30×30×80Å long) and stalk (ca. 30×30×50Å). At the resolution of the 3D reconstruction (32 Å), these features drive the pol II docking result. In contrast to the VP16-Mediator–pol II–TFIIF cryo-EM structure,18 the pol II stalk was not resolved in the activator-free structure (Fig. 3A). The absence of a clearly defined pol II stalk domain suggests variability in the pol II orientation; indeed, because single-particle reconstruction requires alignment and averaging of thousands of individual images, alternate orientations of pol II within the population of images would yield a structure in which the pol II stalk domain would average out during angular refinement. The stalk domain was clearly resolved in the VP16-Mediator–pol II–TFIIF assembly structure due to a single, stable pol II orientation.18 The silver-stained gel of purified pol II (Fig. 1A) and mass spectrometry analysis18 indicates that RPB4 and RPB7 are present in this sample, as expected. The cryo-EM map indicates two putative regions of RPB4/7 stalk protein density (asterisks, Fig. 3A) at higher molecular mass thresholds (data not shown). These two pol II orientations are also suggested by pol II docking results, summarized in Supplemental Table 1. However, many plausible pol II docking results were observed, likely because pol II docking experiments were completed within a cryo-EM map containing an ensemble of pol II orientations.
To determine whether the cryo-EM data could be sub-classified into distinct populations, each representing a different pol II orientation, we completed a 3D variance analysis.21,24 Interestingly, the 3D variance map for Mediator–pol II–TFIIF indicated that structural variance concentrated mainly at the pol II binding site (Fig. 3C). Sub-classification of single-particle images within areas of peak variance, however, did not improve the spatial resolution and did not successfully resolve distinct pol II orientations within the population of images. This likely reflects the high degree of structural plasticity within Mediator and suggests that structural variability inherent within the entire assembly is more substantial in scope than a shift in pol II orientation. Such structural flexibility is well-documented for both human and yeast Mediator,14,25 and is also supported by peak variance regions outside the pol II-Mediator interface (Fig. 3C).With respect to cryo-EM analysis of activator-free Mediator–pol II–TFIIF, this structural variability precluded further sub-classification of the data into populations containing only one pol II orientation.
Structural comparison of VP16-bound vs. activator-free Mediator–pol II–TFIIF complexes
In contrast to the activator-free Mediator–pol II–TFIIF assembly described here, pol II adopted a stable orientation within the VP16-Mediator–pol II–TFIIF complex.18 Whereas different activators can induce different structural shifts upon binding human Mediator, the structural shift at the Mediator–pol II interface appears similar, regardless of the activator. In particular, a large pocket domain forms at the pol II binding site upon Mediator binding to the activation domains of several different transcription factors, including SREBP-1a, p53, VDR, TR, and VP16.1–3 This suggests a common means by which activators might help regulate pol II activity.14 Interestingly, the pol II orientation within VP16-Mediator–pol II–TFIIF appears compatible with transcription initiation (Supplemental Fig. 3A). For example, a complementary binding surface for the large, 1.1 MDa horseshoe-shaped TFIID complex is evident.26 TFIID assembly at this site is consistent with existing biochemical and structural studies, and would appropriately position TFIID, TFIIB and TFIIA upstream from the transcription start site.27,28 (TFIIB and TFIIA interact with the TBP subunit within TFIID.29–31) Moreover, upstream and downstream promoter sequences are accessible to DNA-binding transcription factors and/or TAF subunits within TFIID, which bind specific regulatory elements at the promoter.27,28,32 An open site for the multi-subunit, 480 kDa TFIIH complex is also apparent in the VP16-Mediator–pol II–TFIIF structure (Supplemental Fig. 3A). TFIIH contains an ATPase activity required to melt promoter DNA,33 and a helicase activity that facilitates pol II promoter clearance.34 The proposed location of TFIIH is consistent with these activities, and is also in agreement with crosslinking studies that positioned TFIIH at DNA sequences downstream from the transcription start site.35 Note that TFIIH is also positioned to physically interact with the Mediator head domain in Supplemental Figure 3A, in agreement with genetic and biochemical experiments.36 Furthermore, TFIIE physically interacts with TFIIH37,38 and helps regulate TFIIH function within the PIC.33,39–41 The model shown in Supplemental Figure 3A juxtaposes TFIIE and TFIIH, which is consistent with these observations. The model is also consistent with biophysical studies that localized the pol II binding sites for TFIIB, TFIIE, and TFIIF.42–47
Whereas the cryo-EM data indicate that pol II does not adopt a stable orientation in the absence of an activator, we did note in our docking experiments that two distinct pol II orientations were most consistently observed, based upon cross-correlation coefficients across multiple different pol II crystal structures (Supplemental Table 1). One of these pol II orientations is similar to that stabilized by VP16-Mediator binding (compare Supplemental Fig. 3A and B). This suggests that activator binding per se is not required for pol II to adopt this transcriptionally competent structural state, at least within this partial PIC assembly. This result might reflect the ability of Mediator to stimulate basal (activator free) transcription in vitro.48–50 A second putative pol II orientation does not appear compatible with transcription initiation (Supplemental Fig. 3C), but roughly resembles a Mediator–pol II orientation proposed with yeast factors in the absence of an activator.51
A means for variable pol II orientation may have important regulatory consequences. Previous structural studies with VP16-Mediator–pol II–TFIIF, combined with the data summarized here, indicate that TFIIF and activators are necessary to stabilize the pol II–Mediator interaction. The inability of pol II to stably orient in the absence of an activator or TFIIF may have implications for divergent (sense and anti-sense) transcription, which is common at human promoters.52,53 Variable pol II orientation may allow transcription in the sense or antisense direction in the absence of a key transcription factor and/or TFIIF.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 2.Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat StructMolBiol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD, Lin S, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat StructMolBiol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 6.Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 7.Boyer TG, Martin MED, Lees E, Riccardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1a protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 9.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 11.Naar AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R. Chromatin, TAFs, and a novel multiproteincoactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes & Development. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito M, Yuan C, Malik S, Gu W, Fondell JD, Yamamura S, Fu Z, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 13.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 14.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends BiochemSci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell DevBiol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Cantin GT, Stevens JL, Berk AJ. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. ProcNatlAcadSci U S A. 2003;100:12003–12008. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. ProcNatlAcadSci U S A. 2010;107:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 20.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 21.Penczek P, Frank J, Spahn CM. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Grob P, Cruse MJ, Inouye C, Peris M, Penczek P, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Penczek P, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J. Struct. Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Malleable machines in transcription regulation: the mediator complex. PLoSComputBiol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 27.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev BiochemMolBiol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 28.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat StructMolBiol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiger JH, Hahn S, Lee S, Sigler PB. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 30.Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–151. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 31.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 32.Juven-Gershon T, Hsu JY, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter--the gateway to transcription. CurrOpin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohkuma Y, Roeder RG. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 35.Kim T, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 36.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in Preinitiation Complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Maxon ME, Goodrich JA, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 38.Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Hayashi K, Tanaka A, Furumoto T, Hanaoka F, Ohkuma Y. The carboxy terminus of the small subunit of TFIIE regulates the transition from transcription initiation to elongation by RNA polymerase II. Mol Cell Biol. 2003;23:2914–2926. doi: 10.1128/MCB.23.8.2914-2926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serizawa H, Conaway JW, Conaway RC. An oligomeric form of the large subunit of transcription factor (TF) IIE activates phosphorylation of the RNA polymerase II carboxyl-terminal domain by TFIIH. J Biol. Chem. 1994;269:20750–20756. [PubMed] [Google Scholar]
- 41.Lu H, Zawel L, Fischer L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 42.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat StructMolBiol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostrewa D, Zeller ME, Armache K, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichner J, Chen H, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills J, Nilges M, Cramer P, Rappsilber J. Architechture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol. Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 51.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FA. Mediator head module structure and functional interactions. Nat StructMolBiol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 55.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.