Abstract
In this publication we report on the development of a quantitative enzymatic method for the detection of four botulinum neurotoxin (BoNT) serotypes responsible for human botulism by MALDI-TOF mass spectrometry. Factors that might affect the linearity and dynamic range for detection of BoNT cleavage products were initially examined, including the amount of peptide substrate and internal standard, the timing of cleavage reaction, and the components in the reaction solution. It was found that a long incubation time produced sensitive results, but was not capable of determining higher toxin concentrations, whereas a short incubation time was less sensitive so that lower toxin concentrations were not detected. In order to overcome these limitations, a two-stage analysis strategy was applied. The first stage analysis involved a short incubation period (e.g. 30 min). If no toxin was detected at this stage, the cleavage reaction was allowed to continue and the samples were analyzed at a second time point (4 hr), so that toxin levels lower than 1 mouse LD50 or 55 attomole/mL could be quantified. By combining the results from two-stage quantification, 4 or 5 orders of magnitude in dynamic range were achieved for the detection of the serotypes of BoNT/A, /B, /E, or /F. The effect of multiplexing the assay by mixing substrates for different BoNT serotypes into a single reaction was also investigated in order to reduce the numbers of the cleavage reactions and to save valuable clinical samples.
INTRODUCTION
Botulinum neurotoxins (BoNTs) are bacterial proteins that cause the life-threatening disease botulism 1. These toxins inhibit the release of neurotransmitters at the neuromuscular junction by cleaving soluble NSF attachment protein receptor (SNARE) complex proteins in nerve cells, which prevents the fusion of neurotransmitter containing vesicles to cell membranes 2. BoNTs have very high toxicity and are relatively simple to produce; therefore, they have been categorized as potential agents for bioterrorism 3. The only approved treatment for botulism is antitoxin. These antitoxins are often limited in serotype range, and they must be administered rapidly for optimal protection. In addition, the effectiveness of antitoxin treatment may vary depending on circulating toxin levels, so that rapid identification and quantification of BoNT proteins in clinical specimens and food is desirable for optimization of treatment.
BoNTs consist of seven confirmed serotypes (A through G), classified according to their antigenic properties. Human botulism is associated with exposure to BoNT serotypes A, B, E, and F and usually occurs through three clinical forms: intestinal colonization, foodborne, and wound4. BoNTs are dichain proteins consisting of a light chain of 50 kDa and a heavy chain of 100 kDa linked by a disulfide bond 5. The light chain is a zinc-metalloprotease which is responsible for the cleavage of three SNARE complex proteins including SNAP-25 (Synaptosome-associated protein of 25 kDa), synaptobrevin-2 (also termed VAMP-2) and syntaxin. Each serotype of BoNT hydrolyzes their protein substrates at different peptide bonds. BoNT/A, /E, and /C cleave SNAP-25 at different positions near its C-terminal region 6-9. BoNT/B, /F, /D, and /G have targets on VAMP-2 at distinct sites 10-13 and BoNT/C breaks both SNAP-25 and syntaxin molecules at specific locations 8,14. Cleavage sites are serotype-specific, with the exception of BoNT/F5, which cleaves between amino acids 54L and 55E, four amino acids different from the standard BoNT/F cleavage site of 58Q and 59K 13.
In addition to the standard mouse lethality bioassay, many in vivo and in vitro analytical methods have been recently developed for the determination of a BoNT's identity and concentration for the purposes of diagnosis and confirmation of botulism 15-17. Enzymatic cleavage assays represent an attractive approach where the functional active toxins are detected by monitoring the cleavage of their target substrates, or peptide mimics in most cases, by means of different techniques, such as immunological detection 18,19, capillary electrophoresis 20, fluorescence analysis 21, Forster resonance energy transfer (ALISSA) 22, and high performance liquid chromatography 23. Modern mass spectrometric technology (MS) has become a powerful analytical technique for the analysis of proteins and peptides enhanced by the invention of matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) mass spectrometers. Our laboratory has developed mass spectrometry-based in vitro Endopep-MS activity assays for BoNT identification and differentiation 24-26. In this method, the BoNT proteins are initially enriched by an antibody extraction, and the concentration or enzymatic activity is then determined by incubating affinity purified toxins with their specific peptide substrates, followed by analyzing the cleavage products using mass spectrometry techniques. This method evaluates both immunologic identity (antibody binding) and activity (enzymatic cleavage) of the toxin. Advantages of the Endopep-MS assay include rapid assay time, very sensitive detection, and serotype identification in a single experiment.
Quantification of botulinum neurotoxins aids in treatment decisions, and it can be important when characterizing botulism outbreaks, and evaluating the effectiveness of medical countermeasures. Quantitative mass spectrometry provides a sensitive tool for identification and quantification of target analytes from complex biological samples. Liquid chromatography coupled ESI tandem mass spectrometry (LC-ESI-MS/MS) combined with a multiple reaction monitoring (MRM) technique and stable isotope labeled internal standards is commonly used as a basic quantification platform and is considered the “gold standard” technique for target quantification due to its high selectivity, reproducibility, sensitivity and accuracy. A MALDI-TOF method has its own advantage for quantitative analysis of peptides and proteins due to its high-throughput capacity and its tolerance for more complex sample matrices. The methods of MALDI-TOF MS and LC-ESI-MS/MS have been directly compared in our laboratory for an endopeptidase activity-based quantification of anthrax lethal factor 27. Similar quantitative results obtained from both MS platforms have demonstrated that isotope dilution MALDI-TOF-MS can be used as a robust and precise quantitative MS platform. Here we report the investigation of rapid and accurate quantification of the four BoNT serotypes that cause human botulism by MALDI-TOF mass spectrometry.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were obtained from Sigma–Aldrich (St. Louis, MO) except where otherwise indicated. Monoclonal antibodies were provided by Dr. James Marks at the University of California, San Francisco. Streptavidin-coupled Dynabeads were purchased from Invitrogen (Lake Success, NY). Botulinum neurotoxin complexes were obtained from Metabiologics (Madison, WI). Crude culture supernatants representing bivalent neurotoxin-secreting strains were produced by incubating subcultures of each strain for 5 days at 30 or 37°C. After centrifugation, supernatants were removed and filtered through 0.22 μm filters. The filtered supernatants were tested for upper limits of toxicity using the mouse bioassay, which indicated that the toxins were all present at concentrations of ≤ 10 μg/mL. Botulinum neurotoxins are highly toxic and appropriate safety measures are required. All BoNT neurotoxins were handled in a class 2 biosafety cabinet equipped with HEPA filters. Fmoc-amino acid derivatives and peptide synthesis reagents were purchased from EMD Chemicals, Inc. (Gibbstown, NJ) or Protein Technologies (Tucson, AZ). Peptides were purchased from Midwest Biotech (Fishers, IN) or prepared in house by a solid phase peptide synthesis method using Fmoc chemistry. The peptide substrate for BoNT/E was recently developed in our laboratory and its sequence will be published at a later date.The sequences of the peptide substrates for BoNT/A, /B, or /F, corresponding cleavage products and their internal standards are listed in the supplementary Table S1.
Activity assay
In-solution or on-bead endopeptidase activity assays were carried out as previously described28. In brief, the reaction was conducted in a 20 μL reaction volume containing toxin, peptide substrate, 10 μM ZnCl2, 1 mg/mL BSA, 10 mM dithiothreitol, and 200 mM HEPES buffer (pH 7.4) at 37°C for various lengths of time as indicated in the text. Individual peptides or a mixture of the peptide substrates were added to the reaction at the indicated concentration(s). For the in-solution assays the antibody-coated bead incubations were skipped and toxin (BoNT/A, /B, /E, or /F) was added directly into the reaction mixture. For the on-bead assay, the toxin was spiked or was present in buffer, cell culture, serum or stool extract and was incubated with antibodies immobilized on Streptavidin-coupled magnetic beads prior to the activity assessments. After washing, the beads were immersed into the 20 μL reaction solution for the assay as described above. Two microliters of the reaction were removed at various time points (0.5 to 4 hr) and were mixed with 2 μL of an internal standard peptide and 16 μL of α-cyano-4-hydroxy cinnamic acid (CHCA, 5 mg/mL in 50% acetonitrile/0.1% TFA/1 mM ammonium citrate). This mixture (0.7 μL) was then applied onto a MALDI plate and subjected to mass spectrometric analysis. The relative production of the cleavage fragment from any peptide substrate was measured as the ratio of the isotope cluster areas of the MS peak of the N- or C-terminal product versus its internal standard (Table S1). Triplicate samples were spotted on a MALDI plate and analyzed on a 4800 or 5800 MALDI-TOF/TOF instrument (Applied Biosystems, Framingham, MA). Mass spectra of each sample were obtained by scanning from 800 to 3000 m/z in MS-positive ion reflector mode. This instrument uses a Nd-YAG laser at 355 nm, and each spectrum is an average of 2400 laser shots. The data were usually an average of three experiments. CV values for the samples were typically below 20%.
Optimal assay condition
Samples were split into two equal volumes and subjected to the singleplex assay for BoNT/A and the multiplex assay for BoNT/B, /E, or F. The samples and calibrants of BoNT/A, /B, /E and /F having various toxin concentrations (activities) were captured on toxin-specific antibody-bound beads (20 μL bead suspension) followed by washing and immersion of toxin-bound beads into 20 μL reaction solutions which were incubated at 37°C for various time periods. The singleplex reaction solution contained 100 μM of the substrate of BoNT/A (SA) whereas the substrate mixture containing 100 μM SB, 50 μM SE and 100 μM SF was included in multiplex reaction solution. In order to avoid the appearance of premature cleavage reactions, beads and reaction solutions were quickly mixed on ice and the mixtures were then placed at 37° C. After a 30 min incubation period, the initial analysis was conducted by transferring 2 μL aliquots of each reaction solution into a new tube containing 16 μL of CHCA matrix and 2 μL of single or multiple internal standard. At this stage, the internal standards of high concentration were used including 5 μM IS-A (internal standard for BoNT/A cleavage product) for the singleplex assay and a mixture of 5 μM IS-B (for BoNT/B), 2 μM IS-E (for BoNT/E) and 5 μM IS-F (for BoNT/F) (Table S1) for the multiplex assay, respectively. If botulinum neurotoxin(s) were detected and quantitated at this stage, no further assessments were done. Otherwise, the remaining reaction solutions continued their incubation at 37°C until the 4 hour time point was reached and 2-μL aliquots were subjected to the second-stage analysis. The optimized concentration of the internal standards at this stage included 0.5 μM IS-A and the mixture of 1 μM IS-B, 0.4 μM IS-E, and 1 μM IS-F for singleplex and multiplex assay, respectively.
RESULTS AND DISCUSSION
1. Two-stage quantification extends the dynamic range for detection of BoNT by MALDI-TOF mass spectrometry
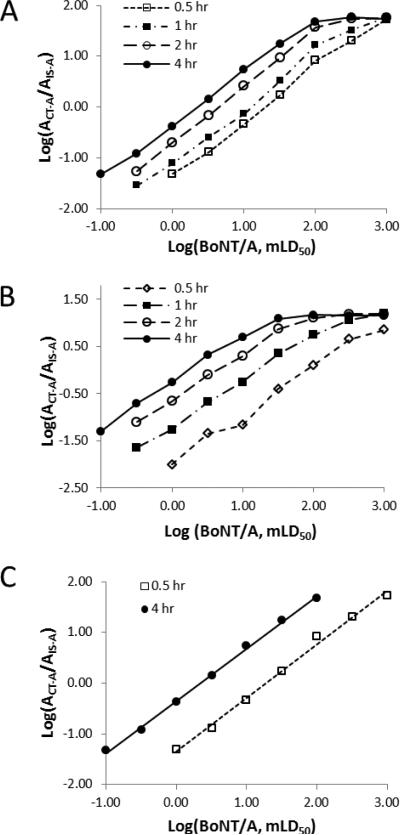
In a previous study for the quantitative detection of BoNT/A and /B activities using ESI mass spectrometry 29, it was observed that a nonlinear response occurs when the toxin concentration reached a certain point, e.g., 100 mouse LD50 (mouse lethal dose determined by standard lethality bioassay) for BoNT/A. This could be attributed to a reduced ratio of the substrate versus toxin, where a hydrolysis reaction no longer conforms to the Michaelis-Menten kinetics, and the formation of cleavage products no longer correlates linearly with BoNT concentration. A possible simple solution is to reduce the reaction time but leave the other parameters unchanged so that the appropriate relative amounts of enzyme and substrate can be maintained. To examine how the time of a BoNT hydrolysis reaction affects the linear dynamic range of BoNT quantification, we conducted toxin cleavage experiments at four different reaction times. Each of the assays was comprised of cleavage reactions with various BoNT/A concentrations ranging from 0.1 to 1000 mouse LD50 where the generation of the C-terminal product (CT-A) formed from the hydrolysis of the BoNT/A substrate (SA) was monitored by MALDI-TOF mass spectrometry. We chose MALDI-TOF rather than LC-ESI mass spectrometry in the present study due to its high throughput capability and significantly reduced sample analysis times compared to the latter. The detection of cleavage products from the reactions with various amounts of BoNT/A at different reaction times was displayed on a logarithmic scale (Figure 1A). Obvious differences were observed among the four data curves obtained from the assays terminated at 0.5, 1, 2, or 4 hours. The reactions, when stopped at 30 minutes, resulted in an apparent linear response in the range of 1 to 1000 -LD50 of BoNT/A toxin. When the reaction times were increased to one hour and higher, some data points at the high toxin concentrations did not fit a linear model, and the curve reached a plateau at the higher concentrations. The apparent bend point (about 100 mouse LD50) for a 4 hour incubation is consistent with the outcome described in a previous ESI-MS study 29. Longer reaction times are important for the analysis, however, because of the improved signal to noise at low toxin concentrations (< 1 - LD50) and the improved detection limits. Figure 1B depicts a similar pattern observed in the assay where the toxin was initially enriched and purified with antibody immobilized on magnetic beads followed by on-bead incubation, which is the Endopep-MS assay platform for the analysis of clinical samples. These results prompted a two-stage approach, in which the aliquots of the cleavage reaction can be analyzed at two different reaction times. For instance, two linear calibration curves could be produced by extracting data points in the above experiments from either the 30 min(1 – 1000 -LD50) or 4 hour reactions (0.1 – 100 LD50) (Figure 1C). By combining these two curves, the amount of BoNT toxin ranging from 0.1 to 1000 LD50 was quantitatively determined. Therefore, it is possible to extend the dynamic range of BoNT detection by preparing two or more calibration curves from measuring different time points in a single experiment. In comparison to a conventional method with serial dilutions of unknown samples to fit one standard curve, one of the potential advantages of a two-stage assay could be to maintain original concentration of an analyte and save precious samples, particularly those containing very low abundant analyte.
Figure 1.
Analysis of the cleavage of the peptide substrate SA by BoNT/A toxin under various reaction times. (A) The reaction was conducted in buffer; and (B) on-beads where the toxin was enriched by immobilized antibody followed by addition of the peptide substrate and reaction buffer. (C) Selected data points extracted from the data set displayed in (A). The adjacent data points were linked by straight lines in (A) and (B) and the curves in (C) were obtained using a power least-square fitting.
Two factors should be considered in isotope dilution mass spectrometric (IDMS) quantification analyses such as the quantitative Endopep-MS assay: the amount of internal standard (IS) used in the analysis solution after toxin cleavage and the original concentration of the peptide substrate added to the hydrolysis reaction. In IDMS analysis, the relative or absolute quantity of a peptide is determined by measuring the peak ratio of the peptide and its isotopically labeled internal standard spiked into samples at a fixed concentration. It has been reported that a large difference between the amounts of unlabeled and isotopically labeled peptides affects the precision of a MALDI response peak ratio as well as the linearity of a standard curve 30. If a target peptide or its internal standard is much less abundant than the other, the less abundant one is likely to have a very weak signal or its MS peak could embed into the noise level in a MS spectrum, producing inaccurate or incorrect analyte/standard ratios. Use of less IS, for example, may improve assay sensitivity because the ratio of the cleavage product versus internal standard would not become too great to be accurately determined at very low BoNT quantity. On the other hand, less IS usage may affect accurate determination of high toxin concentration, because the very intense signal of a cleavage product peak may result in an undetectable IS peak. In contrast, more IS may improve the linearity on high toxin concentrations but could adversely reduce assay sensitivity. Therefore, it is necessary to find a balance of the proper concentration of a labeled internal standard in a BoNT quantitative assay. A similar principle can be applied to peptide substrates added in an activity assay as described below.
To determine an optimal amount of an internal standard, we prepared a series of mixtures of a reaction solution, IS and MALDI matrix, to mimic the analysis solution. Various amounts of a synthetic peptide (CT-A, Table S1) derived from the C-terminal cleavage product of BoNT/A were added in the mock reaction solution, whereas a fixed amount of the internal standard (IS-A, Table S1) corresponding to CT-A was added to the analysis solution. Figure 2A shows measured ratios of CT-A and IS-A peak areas as a function of varied CT-A concentration at three fixed IS-A concentrations (1, 10 or 100 μM). It was observed that the data points generated with 1 μM and 10 μM IS-A resulted in measured peak ratios linearly correlated with CT-A concentration while the peak ratios using 100 μM IS-A did not form a linear curve within the range of CT-A products tested. In the latter case, CT-A/IS-A ratio remained almost unchanged when the amount of CT-A become low, indicating the product/IS ratio could not be accurately determined at very low CT-A concentration (below 1 μM), due to very weak CT-A peaks embedded in the noise of the spectra as discussed previously. To achieve a desired level for detection sensitivity and dynamic range, we decided to use different IS amounts in two analysis stages, where less IS used for a short reaction time and more IS for the long reaction time. This did not add an extra step or interfere in the flow of an experiment because the IS was prepared at individual cleavage reaction time points. The optimal IS concentrations were determined for each of the two stages by the analysis of four human-botulism related serotypes of BoNTs (data not shown) and used in the experiments described. For instance, 5 μM of IS-A was applied for the initial, half-hour, stage analysis of BoNT/A and 0.5 μM of IS-A for the second, four-hour, stage.
Figure 2.
Effect of the internal standard (A) and the substrate (B) on the detection of the C-terminal product (CT-A) yielded from the BoNT/A cleavage reaction. The curves of some data sets were obtained using a power least-square fitting.
Similarly, the amount of peptide substrate added in a cleavage reaction can affect the quantitative detection of BoNT toxins by MALDI analysis. Unlike LC-ESI-MS where peptide substrate and cleavage products can be chromatographically separated prior to mass spectrometry, all components in a cleavage reaction, together with an internal standard, are present in a single spot and analyzed simultaneously in a MALDI method. Therefore, the peak intensity of an un-cleaved substrate may impact the detectability of cleavage product and internal standard. As discussed, relatively low concentrations of a peptide substrate in a BoNT assay could give rise to a nonlinear response for samples at high toxin levels. This limitation could be overcome by using more peptide substrate, but a higher amount of substrate could adversely affect the detection of very low amounts of cleavage products in the same way as internal standard versus cleavage product. In addition, the ions of larger amounts of the unreacted peptides in a sample may saturate the MS detector and negatively impact the product/standard ratio. Figure 2B displays the effect of three concentrations (50, 100, and 200 μM) of the peptide substrate (SA) in the activity assays for the detection of BoNT/A toxin. The response of the cleavage product appeared linear in all three sets of experimental data when the BoNT concentrations were lower than 2000 mLD50 (logarithmic value: 3.30). However, significant deviations in product formation were observed when a higher amount of the toxin was present and 50 μM or 100 μM SA was used. In contrast, linear responses could still be achieved at these high toxin levels through the use of 200 μM SA in the reactions. On the other hand, use of high substrate amounts produced a higher limit of detection than with relatively low added peptide substrate (data not shown). Unlike internal standards which can be added at a different concentration for the two individual analytical stages as previously described, it is practical to use a single concentration of substrate in both low- and high-stages for preparing a calibration curve and measuring unknown samples. Thus, the concentrations of individual peptide substrates were optimized for the analysis of each of the four BoNT serotypes tested based on a working quantitation range, and optimal substrate concentrations (100 μM SA, 100 μM SB, 50 μM SE, and 100 μM SF) were used in the experiments described below.
2. Multiplex detection using a mixture of substrates reduced the usage of clinical samples
In the current Endopep-MS assay platform, an unknown toxin sample is usually divided into four equal parts and then analyzed in separate activity assays with an single peptide substrate specific for each BoNT serotype A, B, E, or F, in each reaction solution. In other words, four experiments would be performed for any single sample. In order to save valuable samples and to reduce reagent usage and lab efforts needed for preparing calibration curves and sample analysis, we examined the feasibility of developing a multiplex analysis procedure, where a mixture of multiple substrates could be used in a single reaction. For this purpose, combinations of two, three or four peptide substrates for the four BoNT serotypes were mixed in a single reaction, and the cleavage of a serotype-specific substrate in any of the mixtures by an individual BoNT serotype was monitored. Figure 3 shows the effect of a substrate mixture on the detection of the cleavage products of a target substrate, in comparison with the results from the single-substrate reactions.
Figure 3.
Effect of substrate mixtures on the cleavage of a specific peptide substrate (SA, SB, SE, or SF) by its corresponding enzyme (A) BoNT/A; (B) BoNT/B; (C) BoNT/E; or (D) BoNT/F. (E) The hydrolysis of SA by BoNT/A toxin in the presence of SE at various concentrations.
For the hydrolysis of the BoNT/A substrate (SA), none of the combinations generated more cleavage product (CT-A) than that obtained in the reaction consisting of SA alone (Fig. 3A). While three substrate mixtures yielded 40 – 80% product relative to that of the single substrate, the other four combinations produced less than 10% product in comparison to the single substrate system. Several factors could have contributed to this lower intensity of the CT-A product in these experiments. First, the cleavage efficiency for SA might be decreased by the presence of other BoNT substrate(s) (SB, SE, and SF for BoNT/B, /E, and /F, respectively, Table S1); second, the mass spectrometry signal of CT-A and/or IS-A could be suppressed by the other peptide substrates included in the reactions and/or other foreign internal standards present in the analysis solutions. It was observed that all four substrate mixtures which produced less than 10% product detection included SE, a common foreign component in addition to SA, whereas three other combinations without SE yielded relatively high amount of BoNT/A cleavage product. This was consistent with results of a SE titration experiment in a BoNT/A detection assay where the increasing concentrations of SE caused a decrease in CT-A formation (Fig. 3E), indicating that SE inhibits the cleavage of SA. MS signal suppression of SE on the peak of CT-A and/or IS-A could be ruled out because the relative ratio of IS-A versus other internal standards in subsequent assays involving substrate mixtures remained unchanged (data not shown). This inhibition effect might be attributed to the fact that both peptide substrates, SA and SE, were developed using sequences from the same native protein substrate, SNAP-25, where the cleavage sites for BoNT/A and BoNT/E are only 17 amino acid residues apart. The entire sequence or a partial region of the SE peptide may interfere with the binding of SA to the catalytic domain of the BoNT protein and, hence, reduce its cleavage efficiency. In conclusion, these results revealed that the presence of SE in a substrate mixture negatively impacted the cleavage of the substrate SA; therefore, it is not appropriate to include BoNT/E substrate in the assay for the detection of type A botulinum neurotoxin. On the other hand, it seems not practical to use a single peptide substrate that contains both cleavage sites for BoNT/A and /E because such peptides generated lower sensitivity for both serotypes than the separate, serotype-specific, peptides (SA and SE) did (data not shown). For this reason, we decided to still use two different peptide substrates to achieve the best assay sensitivity for the detection of these two serotypes.
In contrast to BoNT/A, detection of the three other types of BoNTs was not reduced by the presence of other peptide substrates in the mixtures (Figure 3B, 3C, and 3D). In some combinations, the peak intensity of a specific cleavage product was even enhanced. It was interesting to observe that the presence of SB, SE and SF as a mixture in any BoNT type-specific assay either retained or improved the detection of the target products. This result demonstrated that it is viable to include the mixture of the substrates for BoNT/B, /E, /F, but not for BoNT/A, in a single cleavage reaction for the detection of an unknown BoNT sample. In other words, we are able to reduce the number of experiments from four individual assays for any one sample as established in the current Endopep-MS assay to two assays - one individual BoNT/A assay and a multiplexed BoNT/B, /E, and /F assay.
On the basis of the results described above, a desired workflow for a two-stage multiplex MALDI quantification of multiple BoNT toxins was developed as shown in Figure 4. In this method, a clinical sample can be split for the detection and quantification of BoNT/A in one set and BoNT/B, /E, and /F in another set. Accurate calibration curves can be generated in separate reaction samples including SA alone and a mixture of SB, SE, and SF. The use of two rather than four samples for each evaluation reduces the use of reagents and can enable higher sensitivity by testing larger sample volumes. After the initial 30 min reaction, aliquots of sample and calibrants are withdrawn, mixed with internal standards and MALDI matrix, and subjected to MS analysis. If toxin is detected and quantified at this stage, the assay is stopped, avoiding the need for additional incubation time and reagents. This further improves assay efficiency and significantly decreases the time required for reporting of results. This is particularly important when responding to an outbreak, where every minute may be important for treatment. If toxin is not detectable after 30 minutes, it will be tested after a 4 hour reaction time. This prolonged assay time further improves toxin detection sensitivity while providing results within an eight hour timeframe. Standard curves were prepared in two stages with appropriate concentrations of toxin calibrants such that overlap of calibrant concentrations are included in both calibration curves. This provides consistency of results between the two reaction stages.
Figure 4.
Flow chart of two-stage quantitative detection of botulinum neurotoxins type A, B, E, and F by isotope dilution MALDI-TOF mass spectrometry. IgG represents specific antibodies aganinst according serotypes of BoNT protein.
3. Quantification of BoNTs in biological matrices
Botulinum neurotoxins usually exist in complex biological matrices. Serum and stool are two of the most common clinical specimens collected from botulism patients for disease diagnosis. To evaluate the two-stage method for toxins present in biological matrices, we examined the dynamic range for detection of BoNTs spiked in serum and stool. Figure 5 shows two typical calibration curves for the quantitative detection of a range of BoNT/A concentrations spiked in serum, obtained from either the initial (0.5 hr) or second stage (4 hr) analyses. A linear curve from the 4 hour reactions was achieved with a dynamic range from 0.05 to 50 mouse LD50 in 0.5 mL matrix, while analysis of the 30 minute reactions created a linear curve ranging from 10 to 3000 mouse LD50, demonstrating that 0.05 - 3000 -LD50 BoNT/A in serum matrix can be effectively and quantitatively measured using this two-stage strategy. The amounts of toxin in four QC samples in the two stages were accurately measured with a moderate error range (Table 1). Table 2 summarizes the dynamic ranges of the quantitative detection of botulinum neurotoxins A, B, E and F in either serum or stool matrices. Although the values of the maximum and minimum linear detection limitation differ among the four BoNTs and sometimes between the two matrices, four orders of magnitude in dynamic range were achieved for the quantitation of all four types of botulinum neurotoxins in both biological matrices. By combining the results of the two calibration curves from the initial and second steps, the linear ranges for BoNT/A and BoNT/B were determined to be 0.1 to 6000 and 0.1 to 3000 mouseLD50/mL, respectively. This represents a significant improvement in comparison with the results determined by a previous one-stage (4 hr) method, which yielded the linear ranges of 0.3 to 180 for BoNT/A and 0.3 to 64 mLD50/mL for BoNT/B 29.
Figure 5.
Calibration curve of the hydrolysis of SA by BoNT/A measured at high-end stage (top) and low-end stage (bottom). Insets represent zoomed areas in each plot at lower range of BoNT concentrations.
Table 1.
Blind QC samples (BoNT/A) spiked in 0.5 mL serum and analyzed by the two-stage quantitative Endopep-MS method.
| Sample | Analysis Stage | Calculated amt (mLD50) | Measured amt (mLD50)* | Diff (%) | |
|---|---|---|---|---|---|
| Average | CV (%) | ||||
| Blind QC1 | High | 2200 | 2122 | 18.6 | 3.6 |
| Blind QC2 | High | 160 | 159 | 19.9 | 0.9 |
| Blind QC3 | Low | 7.50 | 6.67 | 19.0 | 2.3 |
| Blind QC4 | Low | 0.35 | 0.41 | 19.4 | 11.1 |
The measured numbers are the average of three independent experiments.
Table 2.
Dynamic range of the quantitative detection of botulinum neurotoxins spiked in 0.5 mL serum or stool extract.
| Toxin | Analysis Stage | Serum | Stool | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Linear Range (mouseLD50/mL) | R2 | Linear Range (mouseLD50/mL) | R2 | ||||||
| BoNT/A | Initial (0.5 hr) | 20 | - | 6000 | 0.990 | 20 | - | 6000 | 0.989 |
| Second (4 hr) | 0.1 | - | 100 | 0.999 | 0.1 | - | 20 | 0.999 | |
| Combined | 0.1 | - | 6000 | 0.1 | - | 6000 | |||
| BoNT/B | Initial (0.5 hr) | 10 | - | 3000 | 0.995 | 2 | - | 3000 | 0.994 |
| Second (4 hr) | 0.1 | - | 20 | 0.998 | 0.1 | - | 20 | 0.999 | |
| Combined | 0.1 | - | 3000 | 0.1 | - | 3000 | |||
| BoNT/E | Initial (0.5 hr) | 10 | - | 4000 | 0.999 | 10 | - | 6000 | 0.996 |
| Second (4 hr) | 0.1 | - | 20 | 0.999 | 0.1 | - | 20 | 0.998 | |
| Combined | 0.1 | - | 4000 | 0.1 | - | 6000 | |||
| BoNT/F | Initial (0.5 hr) | 1 | - | 1000 | 0.994 | 0.2 | - | 500 | 0.995 |
| Second (4 hr) | 0.02 | - | 2 | 0.998 | 0.01 | - | 1 | 0.983 | |
| Combined | 0.02 | - | 1000 | 0.01 | - | 500 | |||
4. Determination of the changes of toxin activities in culture media under different growing temperatures
This novel two-stage method was applied to a study of temperature-induced change in the BoNT toxin activity of a bivalent bacterial strain. It is known that the production of the toxin proteins in some bivalent strains that express two types of botulinum neurotoxins may be affected by growth temperature during inoculation. The level of the activity of the two toxins produced in a bivalent strain secreting both BoNT/A and BoNT/B (Ba) was evaluated under different temperature conditions. The bacteria were cultured at two temperatures (30°C and 37°C), and BoNT/A and BoNT/B activities were analyzed using this two-stage quantitative Endopep-MS method. As shown in Table 3, the activity of the two different BoNT toxins was successfully determined. The amount of BoNT/A in the samples was too low to be detected after the initial reaction, but was detected and quantified after the 4 hour cleavage reaction. In contrast, the activity levels of BoNT/B were very high and could only be quantitatively determined from the initial stage analysis. This study shows that the production of both BoNT/A and BoNT/B toxins are affected by temperature, with an enhancement of toxin production occurring at 37° C. However, the relative amounts (ratio) of BoNT/A versus BoNT/B remained constant at ~2500:1 B:a. This analysis further demonstrates the feasibility and usefulness of this two-stage quantitative method.
Table 3.
Measurement of the toxin activities of two BoNT toxins expressed in the cell culture of the bivalent BoNT strains (Ba) under different growing temperatures by the two-stage method.*
| Culture Temp. (°C) | BoNT/A | BoNT/B | ||||
|---|---|---|---|---|---|---|
| Rx Time (hr) | Measured Amt (mLD50) | Amt30°C/Amt37°C | Rx Time (hr) | Measured Amt (mLD50) | Amt30°C/Amt37°C | |
| 30 | 4.0 | 0.09 | 0.8 | 0.5 | 243.0 | 0.9 |
| 37 | 4.0 | 0.11 | 0.5 | 257.8 | ||
The cell culture samples were diluted by 100 fold and 5μl of the resulting solutions were subjected to toxin enrichment and two-stage quantitative Endopep-MS assay.
CONCLUSIONS
A two-stage multiplex quantitative Endopep-MS method was developed for the detection and quantification, by MALDI mass spectrometry, of human botulism-causing serotypes of botulinum neurotoxins (BoNT/A, /B, /E, /F). By measuring the cleavage product at two time points in a BoNT activity assay (Endopep-MS), two standard curves covering separate detection ranges of toxin concentrations were generated, so that the dynamic range was extended compared to a conventional single stage method. In order to reduce the number of experiments and save valuable clinical samples, the effect of multiplexing the assay by mixing substrates for different BoNT serotypes into a single reaction was investigated. The results revealed that the substrates specific for BoNT/B, /E, and /F could be multiplexed in a single experiment with cleavage efficiencies that are similar to individual assays containing only one substrate. Our data demonstrated that this novel two-stage and multiplex strategy significantly improved the quantification of BoNT/A, /B, /E, and /F using MALDI-TOF analysis by extending the dynamic range of detection. Four orders of magnitude in dynamic range were achieved for the quantitation of four serotypes of BoNTs in biological matrices (e.g. from 1 pg/mL to 6.0×104 pg/mL for BoNT/A).
Supplementary Material
ABBREVATIONS
- BoNT
botulinum neurotoxin serotype
- SNARE
soluble NSF attachment receptor
- SNAP-25
soluble NSF attachment protein 25
- VAMP-2
vesicle associated membrane protein 2
- SA
peptide substrate of BoNT/A
- SB
peptide substrate of BoNT/B
- SE
peptide substrate of BoNT/E
- SF
peptide substrate of BoNT/F
REFERENCES
- 1.Hatheway C. Botulism. In: A B, WHJ H, M O, A T, editors. Laboratory Diagnosis of Infectious Diseases: Principle and Practice. Vol. 1. Springer; New York: 1988. [Google Scholar]
- 2.Simpson L. Annual Review of Pharmacology and Toxicology. 2004;44:167. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- 3.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K, Biodefense, f. t. W. G. o. C. JAMA: The Journal of the American Medical Association. 2001;285:1059. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 4.Hatheway CL. Laboratory diagnosis of infectious diseases: principles and practice. In: A B, WH H, M O, MA T, editors. Botulism. Springer-Verlag; New York: 1988. p. 111. [Google Scholar]
- 5.DasGupta BR, Dekleva ML. Biochimie. 1990;72:661. doi: 10.1016/0300-9084(90)90048-l. [DOI] [PubMed] [Google Scholar]
- 6.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Südhof TC, Jahn R, Niemann H. Journal of biological chemistry. 1994;269:1617. [PubMed] [Google Scholar]
- 7.Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Südhof TC, Niemann H, Jahn R. Nature. 1993;365:160. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 8.Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R. EMBO journal. 1993;12:4821. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiavo G, Rossetto O, Catsicas S, Polverino de Laureto P, DasGupta BR, Benfenati F, Montecucco C. Journal of biological chemistry. 1993;268:23784. [PubMed] [Google Scholar]
- 10.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Nature. 1992;359:832. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 11.Schiavo G, Malizio C, Trimble WS, Polverino de Laureto P, Milan G, Sugiyama H, Johnson EA, Montecucco C. Journal of biological chemistry. 1994;269:20213. [PubMed] [Google Scholar]
- 12.Yamasaki S, Baumeister A, Binz T, Blasi J, Link E, Cornille F, Roques B, Fykse EM, Südhof TC, Jahn R. Journal of biological chemistry. 1994;269:12764. [PubMed] [Google Scholar]
- 13.Kalb S, Baudys J, Webb R, Wright P, Smith T, Smith L, Fernández R, Raphael B, Maslanka S, Pirkle J, Barr J. FEBS letters. 2012;586:109. doi: 10.1016/j.febslet.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foran P, Lawrence GW, Shone CC, Foster KA, Dolly JO. Biochemistry. 1996;35:2630. doi: 10.1021/bi9519009. [DOI] [PubMed] [Google Scholar]
- 15.Singh A, Stanker L, Sharma S. Critical reviews in microbiology. 2013;39:43. doi: 10.3109/1040841X.2012.691457. [DOI] [PubMed] [Google Scholar]
- 16.Cai S, Singh B, Sharma S. Critical reviews in microbiology. 2007;33:109. doi: 10.1080/10408410701364562. [DOI] [PubMed] [Google Scholar]
- 17.Capek P, Dickerson T. Toxins. 2010;2:24. doi: 10.3390/toxins2010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones RGA, Ochiai M, Liu Y, Ekong T, Sesardic D. Journal of immunological methods. 2008;329:92. doi: 10.1016/j.jim.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Hallis B, James BA, Shone CC. Journal of clinical microbiology. 1996;34:1934. doi: 10.1128/jcm.34.8.1934-1938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler M, Shafer H, Manley H, Hackley B, Nicholson J, Keller J, Goodnough M. Journal of Protein Chemistry. 2003;22:441. doi: 10.1023/b:jopc.0000005459.00492.60. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt J, Stafford R. Applied and environmental microbiology. 2003;69:297. doi: 10.1128/AEM.69.1.297-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagramyan K, Barash J, Arnon S, Kalkum M. PLoS ONE. 2008;3:e2041. doi: 10.1371/journal.pone.0002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowe B, Schmidt J, Smith L, Ahmed SA. Analytical biochemistry. 2010;396:188. doi: 10.1016/j.ab.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Barr J, Moura H, Boyer A, Woolfitt A, Kalb S, Pavlopoulos A, McWilliams L, Schmidt J, Martinez R, Ashley D. Emerging infectious diseases. 2005;11:1578. doi: 10.3201/eid1110.041279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer A, Moura H, Woolfitt A, Kalb S, McWilliams L, Pavlopoulos A, Schmidt J, Ashley D, Barr J. Analytical chemistry. 2005;77:3916. doi: 10.1021/ac050485f. [DOI] [PubMed] [Google Scholar]
- 26.Kalb S, Moura H, Boyer A, McWilliams L, Pirkle J, Barr J. Analytical biochemistry. 2006;351:84. doi: 10.1016/j.ab.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 27.Kuklenyik Z, Boyer A, Lins R, Quinn C, Gallegos Candela M, Woolfitt A, Pirkle J, Barr J. Analytical chemistry. 2011;83:1760. doi: 10.1021/ac1030144. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Baudys J, Kalb S, Barr J. Analytical biochemistry. 2011;412:67. doi: 10.1016/j.ab.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Parks B, Shearer J, Baudys J, Kalb S, Sanford D, Pirkle J, Barr J. Analytical chemistry. 2011;83:9047. doi: 10.1021/ac201910q. [DOI] [PubMed] [Google Scholar]
- 30.Anderson NL, Razavi M, Pearson T, Kruppa G, Paape R, Suckau D. Journal of proteome research. 2012;11:1868. doi: 10.1021/pr201092v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.