Abstract
Botulinum neurotoxin (BoNT) is one of the most toxic substances known. BoNT is classified into seven distinct serotypes labeled A through G. Among individual serotypes, researchers have identified subtypes based on amino acid variability within a serotype and toxin variants with minor amino acid sequence differences within a subtype. BoNT subtype identification is valuable for tracing and tracking bacterial pathogens. A proteomics approach is useful for BoNT subtyping since botulism is caused by botulinum neurotoxin and does not require the presence of the bacteria or its DNA. Enzymatic digestion and peptide identification using tandem mass spectrometry determines toxin protein sequences. But with the conventional one-step digestion method, producing sufficient numbers of detectable peptides to cover the entire protein sequence is difficult and incomplete sequence coverage results in uncertainty in distinguishing BoNT subtypes and toxin variants because of high sequence similarity. We report here a method of multiple enzymes and sequential in-gel digestion (MESID) to characterize the BoNT protein sequence. Complementary peptide detection from toxin digestions has yielded near-complete sequence coverage for all seven BoNT serotypes. Application of the method to a BoNT-contaminated carrot juice sample resulted in the identification of 98.4% protein sequence which led to a confident determination of the toxin subtype.
Introduction
The botulinum neurotoxins (BoNT) produced by Clostridium botulinum, C. baratii, C. butyricum, and C. argentinese are among the most toxic substances known and exposure results in the deadly flaccid muscular paralysis known as botulism.1 Human botulism consists of three main types: foodborne, infant and adult intestinal colonization, and wound. Due to BoNTs’ high toxicity and ease of preparation, they are considered potential bioterrorism agents and BoNTs are classified as category A agents.2
Full-length BoNTs are synthesized as a single protein of 150 kDa. The active BoNT form consists of two polypeptide chains linked through a disulfide bond. The light chain (50 kDa) is responsible for BoNT’s zinc-endopeptidase activity, which cleaves one or more cellular targets essential for the release of a neurotransmitter (acetylcholine) in the nervous system. The heavy chain (100 kDa) is composed of an N-terminal translocation domain and a C-terminal receptor binding domain.3 These domains play an essential role in the toxin’s transport into synaptic cells.
BoNTs are divided into seven distinct serotypes designated as A to G, according to their antigenic properties. Human botulism is generally associated with exposures to BoNTs of the serotypes A, B, E, and F. In addition to distinct serotypes, many BoNT subtypes have been recently identified based on their sequence variations and antigenic differences.4–6 For example, gene sequence analysis has identified five BoNT/A subtypes (A1 through A5).7–9 Whereas the sequence similarity among different serotypes is relatively low, the subtypes within a BoNT serotype generally exhibit high sequence similarity. The BoNTs are futher divided into toxin variants that are based on minor sequence differences within a toxin subtype derived from different bacterial strains.
For public health emergencies and counter-bioterrorism purposes, rapid and accurate identification of the BoNT serotype, subtype, and variant can be critical for epidemiological purposes because these differences can rapidly identify similarities and differences between concurrent botulism outbreaks. Nucleotide sequencing of BoNT genes using polymerase chain reaction (PCR) or real-time PCR techniques can subtype BoNTs.10–15 More recently, researchers have reported a multiplex PCR assay for the detection and identification of BoNT subtypes.16–17 These PCR techniques require the presence of DNA, however. The disease botulism is caused by toxins and the toxin producing clostridial organisms and their DNA are not always present in outbreak associated samples.
Mass spectrometry (MS) has proven a powerful tool in the identification of amino acid sequences of proteins and peptides.18 In a bottom-up approach, proteolytic enzymes are used to digest proteins into peptides either in-solution or in-gel. Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) in combination with database searching is used to determine the primary peptide sequence. Due to this technique’s high efficiency, high-throughput, and capability with complex samples, it has become one of the most powerful tools for protein identification, profiling, and quantification. We previously reported the differentiation of BoNT/A1 and BoNT/A2 in milk with MS based proteomics. 19 In that study, the peptides unique to a specific BoNT subtype were identified through LC-MS/MS analysis, allowing the determination of the BoNT/A subtype from the toxin itself. A limitation of many proteomic studies, however, is that the use of a single enzyme (e.g. trypsin) can result in a limited number of tryptic peptides resulting in low sequence coverage. The subtypes of BoNT serotypes often show a high level of sequence similarity with as little as 2.6% sequence difference and the toxin variants can vary by as little as a single amino acid. 9 Thus, identification of the toxin subtype/variant, which can be a critical factor in responding to and preventing public health emergencies, requires high sequence coverage to expose subtype/variant-specific amino acid variable sites and provide confident discrimination.
We describe here a multiple enzyme and sequential in-gel digestion approach that we term MESID. It was designed for subtyping botulinum neurotoxins with high sequence coverage and may prove useful to differentiate other toxins or human pathogens to aid public health tracking and disease prevention. We show that a combination of 1) multiple proteolytic enzymes with different specificities (trypsin, chymotrypsin, Asp-N, and Lys-C) and 2) three sequential digestions on a single 1D-PAGE band of the toxin can significantly increase toxin-protein sequence coverage.
Experimental Procedures
Materials
All chemicals were obtained from Sigma–Aldrich (St. Louis, MO) except where otherwise indicated. Endoproteases including trypsin, chymotrypsin, Asp-N, Lys-C, Glu-C and Arg-C were purchased from Roche Applied Science (Indianapolis, IN). BoNT/A, /B, /C, /D, /E, /F, and /G complexes were obtained from Metabiologics (Madison, WI). BoNT contaminated carrot juice obtained from bottles of carrot juice that were implicated in a botulism outbreak in 2006,20 was provided by the U. S. Food and Drug Administration.
1D Gel electrophoresis
SDS-PAGE gel electrophoresis was performed on a NuPAGE Novex Bis-Tris gel following the standard procedure (Invitrogen). In brief, 1μL of the toxin complex (1 mg/mL) was mixed with 2.5 μL of 4X sample buffer and 6.5 μL of deionized water and heated at 90 °C for 10 min. The toxin in the carrot juice sample was extracted by an antibody immobilized on the Protein G coupled Dynabeads (Invitrogen) as previously described19 and was eluted by incubating the beads in 10 μL of the sample buffer and heated to 90°C for 10 min. The eluted proteins were loaded on a 4–12% gradient gel. The gels were stained with a Silver Stain kit (Protea Biosciences, Morgantown, WV) following manufacturer’s protocol.
In-gel digestion of BoNTs
The gel bands of BoNT were sliced and each band was further cut into four equal pieces followed by destaining. The protein was reduced with 10 mM dithiothreitol at 60°C for 30 min and alkylated with 55 mM iodoacetamide at room temperature in the dark for 30 min. For in-gel guanidination, the gels were incubated in a freshly prepared O-methylisourea solution (1.2M) at 65°C for 2 hours. The in-gel digestion was conducted in 20 μL of solutions containing 0.2 μg of the enzyme (trypsin, chymotrypsin, Asp-N, Lys-C, Arg-C or Glu-C) at various temperatures and reaction times as indicated. Sequential digestions were performed by redigesting each of the bands with a select number of enzymes after removing peptides extracted from previous digestion. In some cases, the particles of the same band were split for two subsequent in-gel digestions carried out separately using different enzymes and then recombined for the third digestion with a single protease. After each digestion, the supernatant was transferred to a new tube and the second extraction was conducted with 20μL of 50:50 water/acetonitrile solution. Extracted fractions were combined with the digestion supernatants. The peptides were vacuum-dried and resuspended in 15 μL of 0.1% formic acid.
LC-MS/MS analysis
NanoESI LC-MS/MS analysis was performed on an LTQ-FT mass spectrometer (Thermo Scientific) connected with a nanoAcquity UPLC (Waters, Milford, MA). The protein digest (7 μL) was injected on a C18 column (100 μm × 100 mm, 1.7 μm, BEH130Å) and the peptides were separated using a linear gradient of 5–35% of buffer B (acetonitrile, 0.1% formic acid) in buffer A (95:5 water/acetonitrile, 0.1% formic acid) over 80 min at a flow rate of 0.5 μL/min. The data acquisition was performed on an Xcalibur system, using data-dependent mode where the five highest-intensity precursors in an MS1 survey scan were isolated for collision-induced dissociation. The resulting MS/MS data were searched for protein candidates with a database searching against an in-house BoNT database using MASCOT software (Matrix Sciences, London, UK). The mass tolerance of precursor ions and fragment ions was 10 ppm and 0.8Da, respectively. Resulting peptides were filtered with a significance threshold of p<0.05 and an ion score cut-off of 40. The peptides with ion scores of less than 50 were validated by manual inspection.
Results
In-gel digestion using multiple enzymes
To evaluate whether the multi-enzyme digestion might improve BoNT coverage, we initially used six common proteases including trypsin, Lys-C, Arg-C, chymotrypsin, Asp-N and Glu-C, for the in-gel digestion of BoNT/A1 protein. Digestion of the toxin embedded in six identical gel bands was carried out with individual enzymes followed by LC-MS/MS analysis. As expected, different sequence coverage (1–59%) for the toxin protein was observed, depending on individual enzyme specificity and digestion efficiency (Table 1). By combining the data of the six digestions, a 72% overlapped sequence coverage was obtained. This result demonstrates that the multi-enzyme strategy can, through LC-MS/MS analysis, improve significantly the sequence coverage of botulinum neurotoxins. Further analysis of the peptides derived from these digestions revealed that some but not all of the six proteases produced unique peptides that were absent in other digestions. This contributed to the merged sequence’s extension of coverage.
Table 1.
Sequence coverage and unique amino acid residues of BoNT/A1 detected by digestion with specific proteasesa.
| Enzyme | Condition | SC (%) | Unique residues | SC from in-silico digestion (%)b |
|---|---|---|---|---|
| Trypsin | 37°C/ON | 59 | 287 | 71 |
| Chymotrypsin | 25°C/ON | 27 | 75 | 50 |
| Asp-N | 37°C/ON | 24 | 55 | 44 |
| Lys-C | 37°C/ON | 15 | 11 | 53 |
| Arg-C | 37°C/ON | 7 | 0 | 25 |
| Glu-C | 37°C/ON | 1 | 0 | 30 |
| Overlapped | 72 |
Six gel bands containing the same amount (loaded 0.14 μg toxin complex) of the BoNT/A protein were digested with different proteases separately. SC: sequence coverage; ON: overnight.
Sequence covered by the peptides derived from theoretical digestion of the toxin protein, based on enzyme specificity and protein sequence. Peptides ranging from 800 Da to 3000 Da were selected for covered sequence.
In this study, we defined a “unique peptide” as one produced exclusively by a given enzyme and whose sequence contained amino acid residues not determined by other enzyme digestions. In-gel digestions by trypsin, chymotrypsin, Lys-C, or Asp-N added to the overall coverage 11 to 287 “unique peptide”-determined unique amino acid residues. In contrast and under the experimental conditions used, Arg-C and Glu-C digestion did not contribute any unique residues towards the overall sequence coverage. In-silico digestion of BoNT/A1 also indicates the sequence coverage derived from the theoretical digestion by either of those two proteases would be lower compared with the other four enzymes (Table 1). Therefore, Arg-C and Glu-C were excluded from further studies as described below.
Sequential in-gel digestion
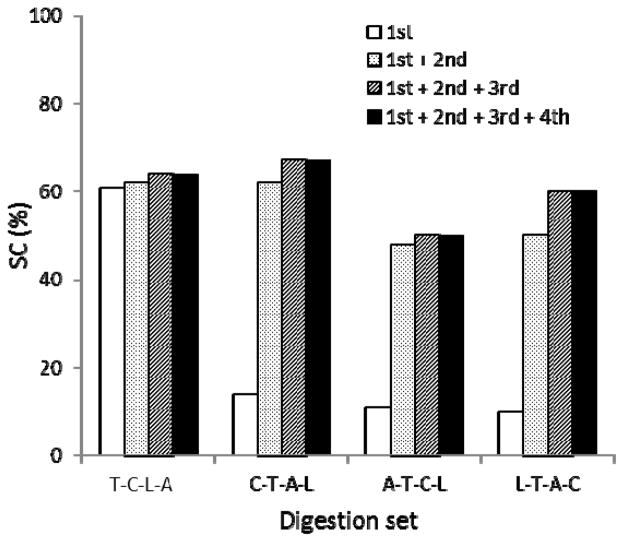
The efficiency of in-gel digestion is sometimes lower than in-solution digestion which can result in fewer peptides being identified by LC-MS/MS following in-gel digestion. This phenomenon may be caused by a variety of factors including low enzyme accessibility to the proteins embedded inside the gel, peptide loss during post-digestion extraction, or a combination of both. Consequently, undigested intact protein and large fragment pieces resulting from partial digestion could possibly remain within the gel matrix. We theorized that additional digestion applied to an already digested gel band might yield new MS detectable peptides from the cleavage of the intact or partial protein fragments remaining in the gel matrix. To investigate this hypothesis, we examined the sequential digestions on the same gel band of a BoNT/A1 protein using various digestion hierarchies with identical or different proteases. Each of the first in-gel digestions on four identical gel bands of BoNT/A1 was carried out with one of the selected individual proteases including trypsin, chymotrypsin, Asp-N, and Lys-C. After the completion of each digest reaction, the solution containing the extracted peptides from the gel was removed. The gel bands were then immersed into a new reaction solution for the second or further digestions using the next digestion enzyme. Figure 1 contains the results of the four sets of the sequential in-gel digestions, where the sequence coverage for each successive individual digestion is summed with the preceding digestion results. It was observed that the second and the third digestions resulted in elevated sequence coverage. This suggested that additional digestions indeed generated unique peptides that improve the sequence coverage. It was noted that under the tested conditions, the fourth digestion in all the sets yielded no additional sequence coverage improvement. To get in-depth information on the origin of the peptides extracted from the second and third digestion, we examined the specificity of the peptides yielded from one sequential digestion set (Chymotrypsin-Trypsin-AspN-LysC) by checking their sequences and terminal residues at both ends. All the second-digestion peptides were identified as unique tryptic peptides that were not detected in the first digestion with chymotrypsin, whereas 31 of the 49 peptides identified in the third Asp-N digestion were new peptides (Table 2). These data reveal that most of the detected peptides after the second or third digestion were created by the cleavage of the intact protein or fragments retained in the gels. In addition, the newly generated peptides in the second or third digestion displayed different specificities through their terminal residues. For example, 22 of the 78 peptides yielded in the second digestion contained one trypsin-specific terminus. The other terminus was from a specific chymotrypsin cleavage, implying that those should be the peptides digested from the large fragments yielded by the first chymotrypsin digestion. A similar phenomenon was observed in the third digestion, where 20 of the 31 new peptides had an Asp-N specific terminus and a trypsin or chymotrypsin-specific residue at the other terminus. These results confirm that the low efficiency or sensitivity of conventional in-gel digestion might arise from incomplete cleavages of the target proteins and extraction of large digests. Thus, sequential digestion performed on a previously digested gel piece is a useful technique to increase further cleavages of undigested or partially digested proteins and newly generated peptides will result in more sequence information.
Figure 1.
In-gel digestion of BoNT/A (~0.1μg per set) with 4 sets of protease combinations. Each set of the digestions was performed sequentially on the same gel band. All digestion reactions were carried out at 52°C for 10 min. T: Trypsin, C: Chymotrypsin, L: Lys-C, A: Asp-N.
Table 2.
Number of the peptides identified in one digestion set and the distribution of their cleavage specificity (see Figure 2 for experimental condition).
| Digestion Sequence | Enzyme | Identified Peptides | Peptides with specific terminusb | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Total | Newa | Two | One | Zero | ||
| First | Chymotrypsin | 15 | 15 | 14 | 1 | 0 |
| Second | Trypsin | 78 | 78 | 55 | 22 | 1 |
| Third | Asp-N | 49 | 31 | 7 | 20 | 4 |
Number of peptides not identified in previous digestion(s)
Number of new peptides with different types of terminus.
Two: specific cleavage at both termini by present enzyme; One: specific cleavage at one end by present enzyme and another end by previous digestion; Zero: both termini were formed from the specific cleavage of previous enzyme(s) or non-specific cleavage.
Combination of multi-enzyme and sequential in-gel digestion
A systematic study was conducted to assess the effect of different combinations of enzymes, sequential orders, and reaction conditions on the characterization of the BoNT/A1 protein. In an attempt to obtain complementary results from a single sample, we used two different proteases, each to a split sample for the second digestion. To reduce the time required for sample preparation and instrument operation, split samples were combined for the third digestion with trypsin. Although one of the samples (S1, Table S1) underwent trypsin cleavage in the first digestion, this protease was used again in the third digestion with the expectation that the enzyme could access and cleave the newly formed protein fragments produced in the second digestion. For the same enzyme in each experimental sequence, different reaction times and temperatures were used to assure that the overall experiment including gel electrophoresis and in-gel digestions were optimized and could be finished in 2 days. As expected, in almost all the samples the second and third digestions of each individual combination set led to sequence coverage expansion (Table S1). These data further confirm the enhancement of multi-enzyme and sequential digestion in protein sequence coverage as described above. An interesting point was to see that in some cases, the second split sample digestion improved the sequence coverage for BoNT. For instance, sample S4’s identified sequence region increased from 79% of the first Asp-N digestion to 88% after the second chymotrypsin digestion on one half of the gel band, and to 85% of the Lys-C cleavage on the other half of the gel band, respectively. Overlapping regions from the two combinations further extended the sequence coverage to 90%, indicating the usefulness of splitting one gel band for different digestion sequences.
A combined effort on the use of multiple enzymes and sequential digestion of a single gel band resulted in a higher sequence coverage (up to 93%) than any single enzyme used for a single digestion event. The combination of two or more sequential digestion sets further elevated the sequence coverage (Table 3). For example, overlapped coverage of the toxin in the gel band S2 (initially digested by chymotrypsin) and in the band S4 (first digested by Asp-N) after the first (81%), second (96%) and third digestion (97%) were higher than corresponding values of either S2 or S4 alone. Moreover, different combinations could obtain similar coverage. The combination of the first digestions of S1, S2, and S4 generated a sequence coverage (97%) identical to that obtained from merging the data of S1 and S4, or S3 and S4 after the first two digestions. This indicates potential flexibility in experiment design based on the availability of sample quantity or on other factors that might need consideration. As a result of combing all data from the four experimental sets, 1285 of 1295 total amino acid residues of the 150 kDa BoNT/A1 protein were covered by MS-identified peptides. In contrast to 76% coverage of the protein from a single step trypsin digestion, this near-complete (99%) sequence identification should enable highly confident determinations of known or unknown BoNT subtypes and toxin variants.
Table 3.
Sequence coverage of different in-gel digestion combinations. Data were obtained from the same experiments described in Table S1.
| Combination | Digestion
|
||
|---|---|---|---|
| 1st | 2nd | 3rd | |
| S1+S2 | 88 | 89 | 89 |
| S1+S3 | 83 | 92 | 92 |
| S1+S4 | 93 | 97 | 97 |
| S2+S3 | 86 | 92 | 94 |
| S2+S4 | 81 | 96 | 97 |
| S3+S4 | 89 | 97 | 97 |
| S1+S2+S3 | 92 | 94 | 95 |
| S1+S2+S4 | 97 | 98 | 98 |
| S1+S3+S4 | 94 | 97 | 97 |
| S2+S3+S4 | 95 | 98 | 98 |
| S1+S2+S3+S4 | 97 | 98 | 99 |
Chemical derivatization by guanidination
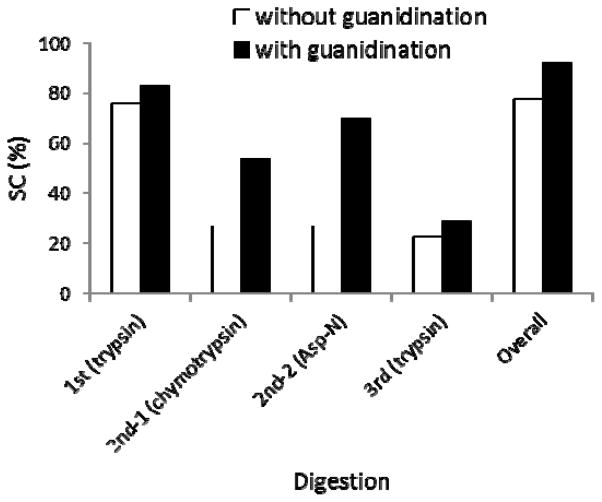
Previous studies reported that guanidination of lysine residues in tryptic peptides enhance their ionization efficiency and hence facilitate the detection of such peptides by mass spectrometry.21 To study the effect of this derivatization for improving sequence coverage in the present method, we applied guanidination to a gel band of the toxin before digestion. We observed that the derivatization resulted in more identified peptides and led to improved sequence coverage in a sequential digestion initiated with trypsin digestion (Figure 2). In comparison with the result from the sample without modification, digestion of guanidinated protein yielded higher sequence coverage at each individual digestion stage and produced higher overall sequence coverage as well. Note, however, that guanidination on the samples initially digested by other enzymes did not result in changes as significant as on the first trypsin sample (data not shown). Yet these data do demonstrate that to enhance protein sequencing further, guanidination of lysine residues of a protein could be a useful part of the MESID method.
Figure 2.
Sequence coverage yielded from three sequential in-gel digestion on the two identical BoNT/A gel bands, with and without guanidination treatment. Digestion conditions were identical for two samples (see table 3).
Application on other serotypes of BoNTs
We examined next the suitability of the MESID method to other botulinum neurotoxins. Seven serotypes of commercially available BoNT complex were loaded on a one-dimensional SDS gel, and the toxins were well separated from associated protein (Figure S1). Referring to the results of previous studies described above, we selected three sets of different enzyme combinations for each of the BoNT/A-G bands (Figure S2). As shown in Table 4, all of seven serotypes of BoNT proteins were identified with high sequence coverage (90–99%). Very high MASCOT scores (> 10,000) confirmed the confident identification. These results suggest that the new method can become a useful tool for subtyping all botulinum neurotoxins.
Table 4.
Proteins identified from commercially available samples of the seven BoNT serotypes (in complex forms).
| Protein name | M.W. (kDa) | GenBank Accession No | MASCOT score | SC (%) |
|---|---|---|---|---|
| Botulinum neurotoxin type A1 | 149 | YP_001386738 | 42468 | 99 |
| Botulinum neurotoxin B1 | 151 | YP_001693307 | 20159 | 95 |
| Botulinum neurotoxin type C1 | 149 | YP_398516 | 13242 | 90 |
| Neurotoxin D | 147 | BAA90661 | 34227 | 96 |
| Botulinum neurotoxin type E | 143 | BAB86845 | 53001 | 92 |
| Neurotoxin type F | 147 | AAA23210 | 18551 | 97 |
| Botulinum neurotoxin type G | 149 | CAA52275 | 10471 | 91 |
Subtyping of a BoNT-contaminated carrot juice sample
We used the MESID method to characterize the sequence of the BoNT present in a toxin-contaminated carrot juice sample associated with botulism outbreaks in the US and Canada. The sample was previously determined to contain BoNT A by the Endopep-MS assay22 and the mouse assay and was subjected to MESID to obtain further information on the toxin subtype and variant on protein level. The toxin was extracted with antibody-coated beads and three identical samples were loaded into a 1D SDS gel (Figure S3). Following our optimized procedure (Figure S2), a pool of MS-detected peptides generated from the three sets of multi-enzyme, sequential in-gel digestions of the toxin containing gel bands confirmed that the target toxin was serotype A of BoNT (Table S2). The detected 98.6% sequence coverage determined by the 1082 unique peptides enabled identification of this toxin as the subtype BoNT/A1 (GeneBank accession no: ABY56330), consistent with the result of DNA sequencing23. Figure 3 shows the sequence alignment of the identified toxin and two homologues of the BoNT/A1 toxin variants (Accession no: ABZ80564 and YP_001383190). The identified sequence shared 99.9% and 99.6% identity with the two variants, respectively. Among the 1295 amino acid residues, only one variation is present at position 540 between the toxins ABY56330 and ABZ80564, while only five different residues are present between the toxin variants ABY56330 and YP_001383190. The identification of the unique peptides that covered those variable regions enabled the differentiation of such highly similar proteins and thus allowed not only the determination of the BoNT subtype but also toxin variant (ABY56330) as well.
Figure 3.
Sequence alignment of identified BoNT/A1 of ABY56330 (top row), and its two homologue toxin variant ABZ80564 (middle row) and YP_001383190 (bottom row). Bold letters represent identified sequence region. Red bold indicates the variable amino acid residues.
Discussion
Unequally distributed proteolytic cleavage sites over the BoNT molecules, however, make it almost impossible to accomplish complete sequence coverage using only a single protease in an MS-based subtyping method. For example, trypsin cleaves proteins specifically at the C-terminal sites of lysine and arginine residues. This almost always generates some tryptic fragments that are either too small, too large, or peptides with low ionization efficiency for detection by LC-MS/MS. A multiple enzyme digestion strategy is an effective way to overcome the limitations of single enzyme cleavage and a similar strategy has been successfully used in identification of post-translational modifications and in complete sequencing of some purified proteins.24–26 In this method, a sample can be divided into two or more parts and each part is digested with a protease that has different specificity. The digestion by multiple proteases usually generates overlapping peptides spanning a broader region of the target protein through cleavages at different residue-specific sites.
We examined here the effects of the multi-enzyme digestion on the gel bands of the BoNT/A1 protein. The combination of the individual digestions with four different enzymes resulted in the identification of more detectable peptides than by a single enzyme digestion. This in turn led to a more than 10% increase in sequence coverage under the experimental conditions used. Such an increase is particularly important for differentiating the subtypes and toxin variants of BoNT, where high-coverage of the BoNT sequence is often required for the confident determination of a small number of amino acid variations between different subtypes and variants.
In-gel digestion is one of the most commonly used methods to produce peptides for proteomic analysis, and gel electrophoresis followed by in-gel digestion using trypsin or other proteases can concentrate and separate proteins in a mixture or in a complex. Advantages of in-gel digestion include the separation of proteins of interest from others to reduce sample complexity and the removal of interfering contaminants that may affect LC-MS/MS analysis.
The limitations of in-gel digestion include low digestion efficiency and peptide loss, presumably caused by limited accessibility of the enzymes to the proteins embedded in the gel matrix and inefficient peptide extraction, respectively.27–28 We speculated that a second digestion could further cleave both intact proteins and large fragments yielded from incomplete digestion and trapped within the gel. This hypothesis is supported by the detection of additional peptides after sequential in-gel digestion that contribute to improved sequence coverage of BoNT/A in a gel band (Figure 1, Table 2). As expected, substantial improvement in the protein sequence coverage resulted from a second tryptic digestion on the BoNT gel bands that had already undergone the first cleavage by chymotrypsin, Lys-C, and Asp-N, respectively. In addition, we observed that a total of three sequential digestions on a gel band was the optimal method to obtain the highest sequence coverage. In this way, several sequential digestions can efficiently overcome in-gel digestion’s main disadvantage while retaining the advantages.
Sequential digestions add a unique feature to the in-gel digestion method. It is not unusual to use a mixture of two proteases, such as trypsin and chymotrypsin, in an in-solution digestion for improving protein coverage. Typically two enzymes are added simultaneously in the same reaction solution, and the two sets of peptide digests cannot be separated. This may cause problems in that some of the MS detectable peptides resulting from the digestion with one protease may be cleaved to smaller, undetectable pieces by another. Therefore, the protein sequence covered by these peptides might become undetermined which is a particular concern in regions of the protein containing abundant cleavage sites. By contrast, sequential in-gel digestion allows multiple digestions to be carried out separately. This is particularly important for situations where determining post-translation modifications on a protein is desirable, or where only a limited sample quantity is available to differentiate protein isoforms. Because the MESID method is not restricted to a particular subject, it can be applied to the in-gel digestion of any protein sample and will likely find a broad application in proteomics.
We also investigated the effect of multi-enzyme digestion on the same sample at the second digestion step. On the assumption that the intact protein or large fragments from the first digestion are evenly embedded in the gel matrix, splitting of the gel particles of a given gel band for the second digestion with two different proteases might result in complementary results. In comparison with applying multiple enzymes on several identical samples, sequential digestion of one sample with different enzymes achieved similar effects but use less protein. In some sets of sequential digestions (Table 2), the combination of two separate second cleavages led to merged sequence coverage higher than those obtained from each of the digestions individually.
Conclusion
We have developed an efficient method for the subtyping of botulinum neurotoxins. In this study, multiple proteases used for in-gel digestion of the toxin produced complimentary results to increase sequence coverage of the toxin. Up to three sequential digestions on a single gel band with various enzymes significantly improved the BoNT sequence coverage. We applied the strategy of different combinations of multi-enzyme and sequential digestion to seven BoNT serotypes. More than 90% of the sequence coverage was accomplished for all of these proteins that have the size of approximately 150 kDa. The very high coverage of the toxin sequence should allow confident determination of BoNT subtypes or at least narrow down the range of potential toxin variant candidates to aid epidemiologic responses to botulism outbreaks. Application of the MESID method on a BoNT-contaminated carrot juice sample identified the 98.6% sequence of the toxin variant. The generation and identification of the peptides covering the amino acid residues unique to a specific subtype and toxin variant of BoNTs may provide useful candidates for the development of high-throughput, highly sensitive, and quantitative methods using advance techniques such as multiple-reactions-monitoring (MRM) for rapid BoNT subtyping.
References
- 1.Turton K, Chaddock JA, Acharya KR. Trends Biochem Sci. 2002;27:552–58. doi: 10.1016/s0968-0004(02)02177-1. [DOI] [PubMed] [Google Scholar]
- 2.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. JAMA. 2001;285:1059–1081. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 3.Schiavo G, Matteoli M, Montecucco C. Physiol Rev. 2000;80:717–66. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 4.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. J Bacteriol. 2006;189(3):818–32. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith TJ, Lou J, Geren IN, Forsyth CM, Tsai R, Laporte SL, Tepp WH, Bradshaw M, Johnson EA, Smith LA, Marks JD. Infect Immun. 2005;73:5450–457. doi: 10.1128/IAI.73.9.5450-5457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arndt JW, Jacobson MJ, Abola EE, Forsyth CM, Tepp WH, Marks JD, Johnson EA, Stevens RC. J Mol Biol. 2006;362:733–42. doi: 10.1016/j.jmb.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 7.Carter AT, Mason DR, Grant KA, Franciosa G, Aureli P, Peck MW. J Clin Microbiol. 2010;48(3):1012–3. doi: 10.1128/JCM.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dover N, Barash JR, Arnon SS. J Clin Microbiol. 2009;47:2349–2350. doi: 10.1128/JCM.00799-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson MJ, Lin G, Tepp W, Dupuy J, Stenmark P, Stevens RC, Johnson EA. Appl Environ Microbiol. 2011;77(12):4217–22. doi: 10.1128/AEM.00201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo EA, Pemberton JM, Desmarchelier PM. Appl Environ Microbiol. 1992;58(1):418–20. doi: 10.1128/aem.58.1.418-420.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo EA, Pemberton JM, Desmarchelier PM. Appl Environ Microbiol. 1993;59(9):3011–20. doi: 10.1128/aem.59.9.3011-3020.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fach P, Hauser D, Guillou JP, Popoff MR. J Appl Bacteriol. 1993;75(3):234–9. doi: 10.1111/j.1365-2672.1993.tb02771.x. [DOI] [PubMed] [Google Scholar]
- 13.Fach P, Gibert M, Griffais R, Guillou JP, Popoff MR. Appl Environ Microbiol. 1995;61(1):389–92. doi: 10.1128/aem.61.1.389-392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lövenklev M, Holst E, Borch E, Rådström P. Appl Environ Microbiol. 2004;70(5):2919–927. doi: 10.1128/AEM.70.5.2919-2927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macdonald TE, Helma CH, Ticknor LO, Jackson PJ, Okinaka RT, Smith LA, Smith TJ, Hill KK. Appl Environ Microbiol. 2008;74(3):875–82. doi: 10.1128/AEM.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umeda K, Seto Y, Kohda T, Mukamoto M, Kozaki S. J Clin Microbiol. 2009;47(9):2720–8. doi: 10.1128/JCM.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umeda K, Seto Y, Kohda T, Mukamoto M, Kozaki S. Microbiol Immunol. 2010;54(5):308–12. doi: 10.1111/j.1348-0421.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 18.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 19.Kalb SR, Goodnough MC, Malizio CJ, Pirkle JL, Barr JR. Anal Chem. 2005;77(19):6140–46. doi: 10.1021/ac0511748. [DOI] [PubMed] [Google Scholar]
- 20.Sheth AN, Wiersma P, Atrubin D, Dubey V, Zink D, Skinner G, Doerr F, Juliao P, Gonzalez G, Burnett C, Drenzek C, Shuler C, Austin J, Ellis A, Maslanka S, Sobel J. Clin Infect Dis. 2008;47(10):1245–51. doi: 10.1086/592574. [DOI] [PubMed] [Google Scholar]
- 21.Warwood S, Mohammed S, Cristea IM, Evans C, Whetton AD, Gaskell SJ. Rapid Commun Mass Spectrom. 2006;20(21):3245–56. doi: 10.1002/rcm.2691. [DOI] [PubMed] [Google Scholar]
- 22.Barr JR, Moura H, Boyer AE, Woolfitt AR, Kalb SR, Pavlopoulos A, McWilliams LG, Schmidt JG, Martinez RA, Ashley DL. Emerg Infect Dis. 2005;11(10):1578–83. doi: 10.3201/eid1110.041279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raphael BH, Luquez C, McCroskey LM, Joseph LA, Jacobson MJ, Johnson EA, Maslanka SE, Andreadis JD. Appl Environ Microbiol. 2008;74(14):4390–4397. doi: 10.1128/AEM.00260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chmelik J, Zidkova J, Rehulka P, Petry-Podgorska I, Bobalova J. Electrophoresis. 2009;30(3):560–7. doi: 10.1002/elps.200800530. [DOI] [PubMed] [Google Scholar]
- 25.Kang SU, Lubec G. Electrophoresis. 2009;30:2159–167. doi: 10.1002/elps.200900024. [DOI] [PubMed] [Google Scholar]
- 26.Ye Y, Mar EC, Tong S, Sammons S, Fang S, Anderson LJ, Wang D. J Virol Methods. 2010;163(1):87–95. doi: 10.1016/j.jviromet.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. Anal Biochem. 1992;203(1):173–9. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 28.Bornemann S, Rietschel B, Baltruschat S, Karas M, Meyer B. Electrophoresis. 2010;31(4):585–92. doi: 10.1002/elps.200900490. [DOI] [PubMed] [Google Scholar]