Abstract
Autoantibodies targeting the H+/K+-ATPase proton pump of the gastric parietal cell (parietal cell antibodies [PCA]) are diagnostic of atrophic body gastritis (ABG) leading to pernicious anemia (PA). PCA, ABG, and PA occur in increased frequency in patients with type 1 diabetes and their relatives and are considered “minor” components of forms of autoimmune polyglandular syndrome (APS). A customized radioimmunoprecipitation assay was applied to 6,749 samples from the Type 1 Diabetes Genetics Consortium to measure ATP4A autoreactivity. Autoantibody prevalence was correlated with variants in HLA class II, PTPN22, and CTLA4 genes. With an ATP4A radioimmunoprecipitation assay, PCA were detected in sera from 20.9% of affected individuals. PCA prevalence increased with age and was greater in females (25.3%) than males (16.5%) and among Hispanics (36.3%) and blacks (26.2%) compared with non-Hispanic whites (20.8%) and Asians (16.7%). PCA and other organ-specific autoantibodies GAD65, IA-2, thyroid peroxidase (TPO), 21-hydroxylase (21-OH), and transglutaminase (TG) clustered within families with heritability estimates from 71 to 95%. PCA clustered with TPO, 21-OH, and persistent GAD65 autoantibodies but not with celiac (TG) or IA-2 autoantibodies. PCA-positive subjects showed an increased frequency of DRB1*0404, DPB1*0201, and PTPN22 R620W (rs2476601-T) and a decreased frequency of DRB1*0101, DPB1*0301, and CTLA4 CT60 (rs3087243-T). Genetic variants accounted for 4–5% of the heritable risk for PCA. The same alleles were associated with other autoantibody phenotypes in a consistent pattern. Whereas most of the heritable risk for PCA and other antibodies reflects genetic effects that are tissue specific, parietal cell autoimmunity is a major pathogenetic contributor in APS2.
Introduction
Autoantibodies targeting the H+/K+-ATPase proton pump on the plasma membrane of the gastric parietal cell (parietal cell antibodies [PCA]) are a specific and sensitive diagnostic of atrophic body gastritis (ABG). Chronic ABG may lead to pernicious anemia (PA) as a result of impaired production of intrinsic factor and presence of intrinsic factor autoantibodies, which hinder cobalamin uptake in the duodenum leading to cobalt deficiency. PCA and PA are common autoimmune conditions, affecting 2% and 0.15–1% of the general population, respectively. PCA prevalence increases with age, up to 12%, yet the true incidences of ABG and PA are uncertain, as ABG is generally asymptomatic and cobalt deficiency and its clinical manifestations are slow to develop. ABG is also associated with gastric malignant lesions in ∼10% of individuals (1). The identification of demographic, genetic, and immunological risk factors associated with ABG is vital to its diagnosis and early intervention to prevent the serious clinical sequela of PA.
Familial aggregation of PCA and PA is considered well established, although based on limited data in families of patients with PA (2,3). Parietal cell autoimmunity, in common with many other autoimmune disorders, may be determined by an interplay of genetic and environmental risk factors with involvement of HLA class I and/or HLA class II genes. Most of the evidence supporting a genetic basis for PCA comes from studies documenting the frequency of PCA and PA in patients and their relatives with other autoimmune diseases, including type 1 diabetes, autoimmune thyroid disease, and adrenal autoimmunity (1–5).
The gastric proton pump H+/K+-ATPase is a heterodimer composed of an eight transmembrane–spanning catalytic A subunit (ATP4A) and a single transmembrane highly glycosylated B subunit (ATP4B), both of which are antigenic targets for T and B cells (6,7). A mouse model of autoimmune gastritis, induced via thymectomy, mimics the human disease including submucosal infiltration, parietal cell destruction, and autoantibodies targeting both A and B H+/K+-ATPase subunits. Transgenic expression of the B subunit protects mice from developing autoimmune gastritis, implying tolerance induction within the thymus. Moreover, the initial immune response may target the B subunit, as the other autoantigen reactivates are avoided in the transgenic mouse (8). Detection of PCA has relied on immunocytochemistry (9) and ELISA, using biochemically purified pig stomach enzyme as the target antigen (10) with diagnostic confirmation by biopsy. Most autoantibodies target epitopes in the carbohydrate moiety of the B subunit, whereas in our study we use the ATPase A subunit protein as the antigen probe. We recently developed a radioimmunoprecipitation assay (RIA) to defined epitopes in ATP4A using in vitro translated human ATP4A cDNA to generate a “biochemical” assay of high sensitivity (95%) and specificity (100%) to measure antibodies in individuals with histologically confirmed ABG (11). In the 2012 International Autoantibody Study Program, this novel assay detected autoantibodies in 30% of type 1 diabetes samples and 4% of controls.
In the current study, the new radioimmunoprecipitation assay was used to characterize ATP4A autoreactivity in 6,749 subjects as part of the Type 1 Diabetes Genetics Consortium (T1DGC) Autoantibody Workshop. The primary objective was to characterize phenotypic and genetic differences between ATP4A sero-positive and sero-negative individuals with type 1 diabetes. We demonstrate that observed differences are consistent with a genetic basis. This result likely reflects predisposing and protective alleles primarily in gene(s) with relatively specific effects on gastric autoimmunity. Furthermore, there are likely strong contributions by HLA class II and non-HLA variants with pleiotropic effects on gastric, thyroid, and adrenal autoimmunity.
Research Design and Methods
Radioimmunoprecipitation Assay
A full-length gastric ATPase4A cDNA clone was generated by reverse transcription of human stomach RNA (iScript; Bio-Rad, Hercules, CA), and sequences encoding antigenic epitopes were subcloned into the pCMVTNT vector for coupled in vitro transcription/translation (cat. no. L1170; Promega TNT kit) to produce [35S]-methionine (PerkinElmer)–labeled antigen probes. Unincorporated [35S]-labeled amino acids were removed by G-25 columns (cat. no. 17-0853-02; NAP-5, GE HealthCare), and the labeled probe was quantified by trichloroacetic acid precipitation.
Serum samples (2.5 μL) were incubated overnight at 4°C with the antigen (20,000 counts per minute [CPM]) in 60 μL of 20 mmol/L Tris-HCl, pH 7.4, containing 150 mmol/L NaCl, 0.1% BSA, 0.15% Tween-20, and 0.1% sodium azide in 96-well plates. Immune complexes were captured by addition of 25 μL of a 50% Protein A Sepharose slurry (cat. no. 17-5280; 4 Fast Flow, GE HealthCare), transferred to 96-well filtration plates (cat. no. MSDVN6B50; Millipore, Billerica, MA) and washed sequentially with 150 μL assay buffer at 4°C using an automated, vacuum-operated 96-well plate washer (Tecan, Mannedorf, Switzerland). After air-drying, 25 μL of scintillation fluid (MicroScint-20; PerkinElmer, Waltham, MA) was added to each well and radioactivity monitored on a Wallac 1450 MicroBeta TriLux β-counter (PerkinElmer).
Duplicate positive standards (a pool of eight ATPase4A high-titer sera samples) and individual negative control sera were included in each assay set for calibration and results expressed as a binding index (sample CPM – negative control CPM)/(positive control CPM – negative control CPM). The upper limit of normal (index 0.020) was established as the 95th percentile from receiver operator characteristic curves generated from a panel of 220 type 1 diabetes autoantibody–negative first-degree relatives of subjects with type 1 diabetes attending the Barbara Davis Center (median age 6.32 years [range 1–52]; 75% Hispanic, 25% white Caucasian/non-Hispanic). These samples served as healthy controls for subsequent assays.
Sera
Sera from 6,749 individuals with type 1 diabetes were provided by the T1DGC Coordinating Center. These samples were from 3,384 males (50.14%) and 3,365 females (49.86%). A total of 5,223 subjects were non-Hispanic whites (NHWs) (77.39%), with the majority from 2,444 type 1 diabetes–affected sibling pair (ASP) families as well as a small subset of 299 singletons (case subjects with type 1 diabetes with no parental sample); the average age of the NHW subjects was 22.5 years. There were 450 Hispanic (Mexican) ancestry samples (6.67% of the total collection) from 62 ASP families as well as 324 singletons, with an average age of 14.0 years. There were 671 African ancestry participants (9.94% of the total collection) from 30 multiplex families with an additional 609 singletons, with an average age of 14.8 years. There were 365 Asian ancestry participants (5.41% of the total) from 32 ASP families with an additional 299 singletons, with an average age of 17.4 years. There were 21 mixed-ethnicity (0.31% of total) participants, all singletons, and 19 subjects of “other ethnicity” (0.28% of the total) from nine ASP families as well as 10 singletons, with an average age of 11 years.
Genotypes
The primary genetic risk for type 1 diabetes resides in the MHC, with specific risk and protective antigens defined by the HLA class I and HLA class II genes. HLA loci, HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, and -DPA1, were genotyped at 4-digit resolution in all T1DGC ASP and trio (two parents and one type 1 diabetes–affected child) families, as well as in collections of case subjects with type 1 diabetes and control subjects as previously described (12). Single nucleotide polymorphism (SNP) markers for the PTPN22 and CTLA4 genes were genotyped in a subset of T1DGC subjects as part of the T1DGC Rapid Response Project (13), including 2,520 of 6,749 subjects tested for ATP4A autoantibodies.
Statistical Analysis
The association of each autoantibody phenotype with age, age of onset of type 1 diabetes, duration of type 1 diabetes, sex, HLA genotypes, CTLA4 SNPs, and PTPN22 SNPs was tested using logistic regression methods with robust variance estimators, stratified by ethnicity, and clustered by pedigree to accommodate the correlation between affected siblings. Initial screening for genetic associations was restricted to 38 common (minor allele frequency [MAF] >3%) HLA alleles in 5,219 NHWs. Similar methods were used to test for association with 43 common PTPN22 and CTLA4 SNPs (MAF >10%) in 2,003 NHWs.
HLA class II haplotypes were reconstructed using MENDEL software to assign the parental origin of HLA-DRB1, -DQA1, and -DQB1 alleles. In most cases, parental origin of the alleles at all three HLA class II loci were assigned unambiguously from family data, including both parents and unaffected siblings. Haplotypes with unphased alleles were inferred from linkage disequilibrium patterns.
Estimates of the prevalence of ATP4A autoantibodies in subjects of Hispanic (Mexican), African, and Asian ancestry were standardized to the age and sex distribution of NHWs. Regression coefficients, SEs, significance levels (Wald test), and standardized prevalence estimates were calculated using STATA (version 11.2; StataCorp, College Station, TX). Statistical significance of genetic association tests was based on a P < 0.001 threshold, based on a Bonferroni correction allowing for 50 independent tests, a conservative criterion given the strong linkage disequilibrium between HLA alleles and SNP markers for both the CTLA4 and PTPN22 genes. Univariate and bivariate heritability analyses, allowing for phenotype-specific covariates, were conducted on 2,444 multiplex families (NHWs only), estimating heritability (h2) of each phenotype and genetic correlation (rg) between phenotypes, and significance levels using the liability threshold model for discrete traits (14) as implemented in SOLAR (15,16).
Results
Demographic Associations
Among 6,749 T1DGC subjects, autoantibodies to the ATP4A antigen were detected in 1,408 (20.9%), varying significantly by age, sex, and ethnicity (Table 1). The prevalence of ATP4A autoreactivity increased from 13.6% in subjects ≤10 years of age to 32.6% in those 40 ≥ years (P < 10−10). Prevalence of ATP4A autoantibodies differed by sex (16.5% in males vs. 25.3% in females; odds ratio [OR] 1.7; P < 10−10) after adjustment for age and ethnicity. ATP4A autoantibodies differed by ethnicity, with prevalence of 20.8% in NHWs compared with 36.3% of those of Hispanic/Mexican ancestry (P < 10−7), 26.2% of African ancestry (P = 0.05), and 16.7% of Asian ancestry (P = 0.10) after adjustment for age and sex. There was no evidence that the prevalence of ATP4A autoantibodies differed by disease duration after allowance for the effects of age (P = 0.212).
Table 1.
Demographic characteristics of the T1DGC data set and prevalence of ATP4A autoantibodies standardized to the age and sex distribution of NHWs
| Ethnicity | Subjects (n) | Multiplex sibships (n) | Median (range) age, years | % Female | Standardized prevalence (95% CI) |
|---|---|---|---|---|---|
| NHW | 5,223 | 2,444 | 19 (1–76) | 48.9 | 0.208 (0.198, 0.220) |
| Hispanic | 450 | 62 | 13 (2–60) | 49.3 | 0.363 (0.301, 0.434) |
| Black | 671 | 30 | 13 (1–64) | 56.2 | 0.262 (0.220, 0.309) |
| Asian | 365 | 32 | 16 (2–56) | 52.3 | 0.167 (0.126, 0.218) |
| Mixed | 21 | 0 | 13 (5–37) | 76.2 | 0.171 (0.032, 0.506) |
| Other | 19 | 9 | 10 (3–19) | 15.8 | 0.387 (0.101, 1.000) |
Risks to Siblings
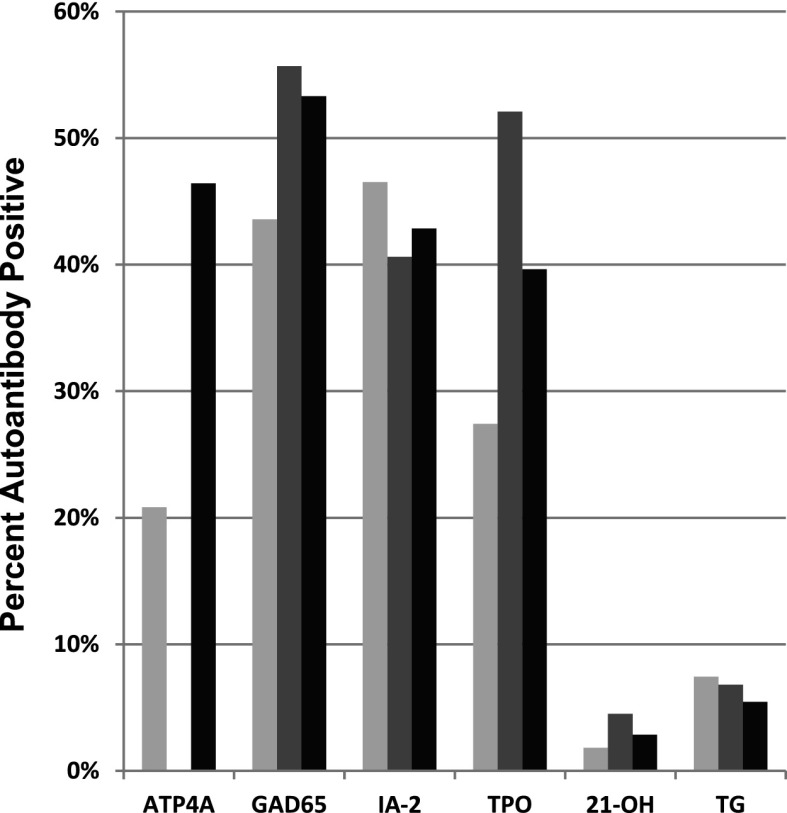
In analyses of 2,444 NHW type 1 diabetes ASP families, ATP4A autoantibodies were detected in 46.5% of siblings of ATP4A autoantibody–positive probands compared with 20.8% of all siblings (Fig. 1). Anti-ATP4A sero-positive probands and their siblings were more likely to have other autoantibodies. There was an increase in islet (anti-GAD65) autoantibodies (43.6% of all affected siblings, 55.7% of ATP4A sero-positive probands, and 53.3% of siblings of ATP4A sero-positive probands), thyroid (anti–thyroid peroxidase [TPO]) autoantibodies (27.4% of all affected siblings, 52.1% of ATP4A sero-positive probands, and 39.6% of siblings of ATP4A sero-positive probands), and adrenal (anti–21-hydroxylase [21-OH]) autoantibodies (1.8% of all affected siblings, 4.5% of ATP4A sero-positive probands, and 2.9% of siblings of ATP4A sero-positive probands). Those anti-ATP4A probands and their siblings were less likely to have other islet (anti–IA-2) autoantibodies (46.5% of all affected siblings, 40.6% of ATP4A sero-positive probands, and 42.9% of siblings of ATP4A sero-positive probands) or thyroid (anti-transglutaminase [TG]) autoantibodies (7.4% of all affected siblings, 6.8% of ATP4A sero-positive probands, and 5.4% of siblings of ATP4A sero-positive probands).
Figure 1.
Genetic associations between autoantibody phenotypes shown as an increased prevalence of ATP4A autoantibodies in siblings of ATP4A sero-positive probands (far left); an increased prevalence of GAD65, IA-2, and TPO autoantibodies; and decreased prevalence of 21-OH and TG autoantibodies in ATP4A sero-positive subjects (gray) and their siblings (black) compared with all T1DGC subjects (light gray). Analyses were restricted to NHW subjects.
Univariate (Heritability) and Bivariate (Genetic Correlation) Estimates
Family data for NHW subjects were used to estimate heritability assuming the liability threshold model (14). The liability threshold model assumes that an individual’s “liability” for developing ATP4A autoantibodies is the sum of their inherited risk and environmental exposures, with the expression of autoantibodies occurring in individuals with liabilities exceeding a threshold defined by the prevalence in the study population (20.8% for ATP4 autoantibodies in NHW subjects with type 1 diabetes). To the extent that genetic differences contribute to differences in liability, the mean liability of siblings of ATP4A autoantibody–positive probands will be higher than the mean liability in the study population, and thus a higher frequency of siblings will exceed the threshold for ATP4A autoantibody–positivity (43.6% vs. 20.8%). Taken together with the expected familial resemblance (genetic correlation) between siblings (siblings share one-half of their genome, on average, or 0.5), the shift in mean liability provides an estimate of heritability (h2).
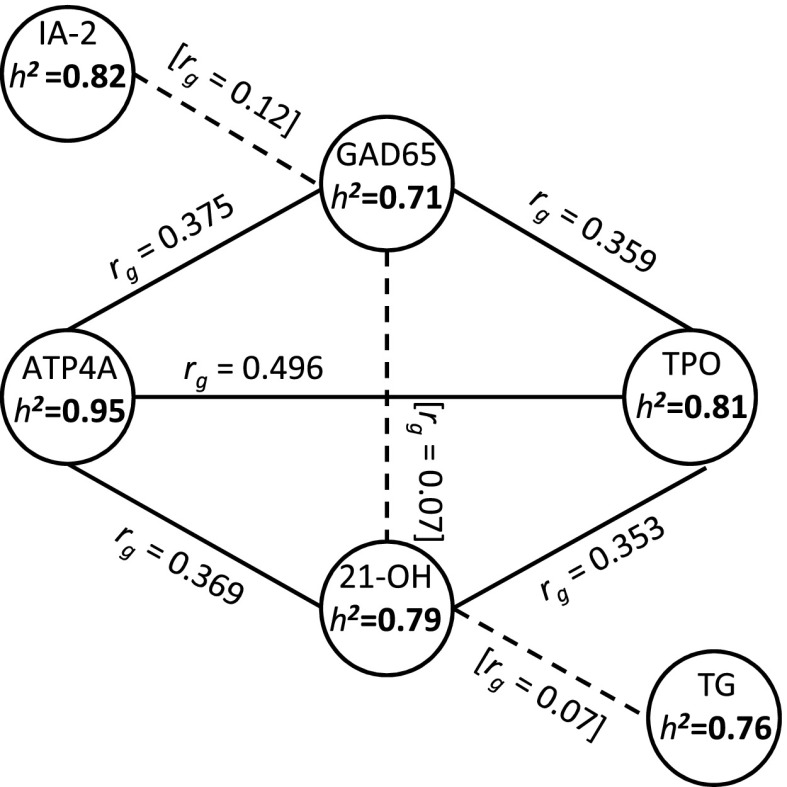
Heritability analyses were performed for all autoantibody phenotypes (ATP4A, IA-2, GAD65, TPO, 21-OH, and TG), with adjustment for phenotype-specific covariates. In this sample of participants with type 1 diabetes, ATP4A autoantibody positivity showed the highest heritability, with genetic variation accounting for 95% of differences in risk (P < 10−46) (Fig. 2). Similarly, the five other autoantibody phenotypes exhibited high heritability estimates, ranging from 71% for autoantibodies to GAD65 (P < 10−30) to 82% for autoantibodies to IA-2 (P < 10−43), with autoantibodies to TPO, 21-OH, and TG having heritability estimates of 81% (P < 10−33), 79% (P < 10−4), and 76% (P < 10−13), respectively.
Figure 2.
Heritabilities (h2) for ATP4A autoantibodies and five other autoantibody phenotypes (IA-2, GAD65, TPO, 21-OH, and TG) and genetic correlations (rg) between different autoantibody phenotypes in NHW subjects from T1DGC.
Bivariate analyses (estimating single trait heritability and cross-trait genetic correlation) were performed in 2,444 T1DGC NHW families (as above) to determine whether latent genetic factors shared by different autoantibody phenotypes account for a significant fraction of the observed heritability. The results supported extensive shared genetic factors, particularly for ATP4A, TPO, and 21-OH autoantibodies. The genetic correlation estimates were 0.496 for TPO and ATP4A (P < 10−57), 0.353 for TPO and 21-OH (P < 0.01), and 0.369 for ATP4A and 21-OH (P < 0.001) (Fig. 2). Significant genetic correlations were also observed for GAD65 autoantibodies with both TPO (0.359; P < 10−7) and ATP4A autoantibodies (0.375; P < 10−19). There was no significant genetic correlation estimated between GAD65 with 21-OH autoantibodies (0.07; P = 0.88). The genetic correlation between the islet autoantibodies GAD65 and IA-2 was relatively modest (0.12; P = 0.05). There was no evidence of a genetic association significantly different from zero for IA-2 or TG autoantibodies with any other autoantibody phenotype.
HLA Class II Association With Autoantibody Phenotypes
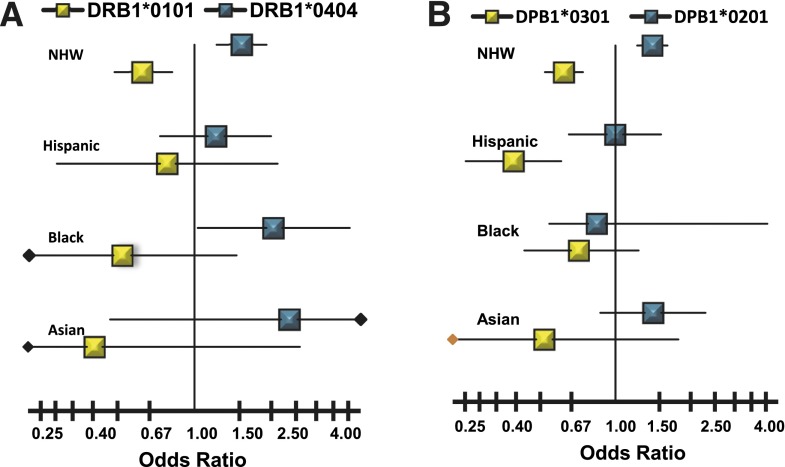
Of 38 common HLA class I (HLA-A, -B, and -C) and HLA class II (HLA-DR, -DQ, and -DP) alleles tested for association with ATP4A autoantibodies in NHWs, four HLA class II alleles exhibited increased risk for ATP4A autoantibodies, while four HLA class II alleles had decreased risk. There was no evidence of association of ATP4A autoantibodies with any HLA class I alleles. The HLA class II alleles associated with increased risk are DRB1*0404 (OR 1.53; P < 3 × 10−4), DQA1*0301 (OR 1.40; P < 3 × 10−7), DQB1*0302 (OR 1.38; P < 1.5 × 10−6), and DPB1*0201 (OR 1.40; P < 1.6 × 10−6). The class II alleles associated with decreased risk are DRB1*0101 (OR 0.62; P < 1.7 × 10−4), DQA1*0101 (OR 0.63; P < 2.4 × 10−5), DQB1*0501 (OR 0.63; P < 3.2 × 10−5), and DPB1*0301 (OR 0.62; P < 2.1 × 10−8). The ATP4A associations with DPB1*0201 and DPB1*0301 are independent of the ATP4A associations with HLA-DR and HLA-DQ alleles; however, the effects of the HLA-DR and HLA-DQ alleles cannot be distinguished owing to strong linkage disequilibrium among the high-risk alleles DRB1*0404, DQA1*0301, and DQB1*0302 and low-risk alleles DRB1*0101, DQA1*0101, and DQB1*0501. In addition to the results observed in NHWs, the decreased risk associated with DPB1*0301 is also significant in subjects of Hispanic/Mexican ancestry (OR 0.39; P < 3.1 × 10−5). Further, the increased risk associated with DRB1*0404 attained nominal significance in subjects of African ancestry (OR 2.04; P < 0.05). Overall, the effects of the HLA class II alleles appear similar across ethnicities, except for DPB1*0201, which is heterogeneous across ethnic groups (Fig. 3).
Figure 3.
Ethnicity-specific ORs and 95% CIs for HLA alleles identified as positively (blue) or negatively (yellow) associated with ATP4A autoantibodies in NHWs. A: HLA-DRB1 alleles associated with ATP4A autoantibodies in NHW subjects. B: HLA-DPB1 alleles associated with ATP4A autoantibodies in NHW subjects.
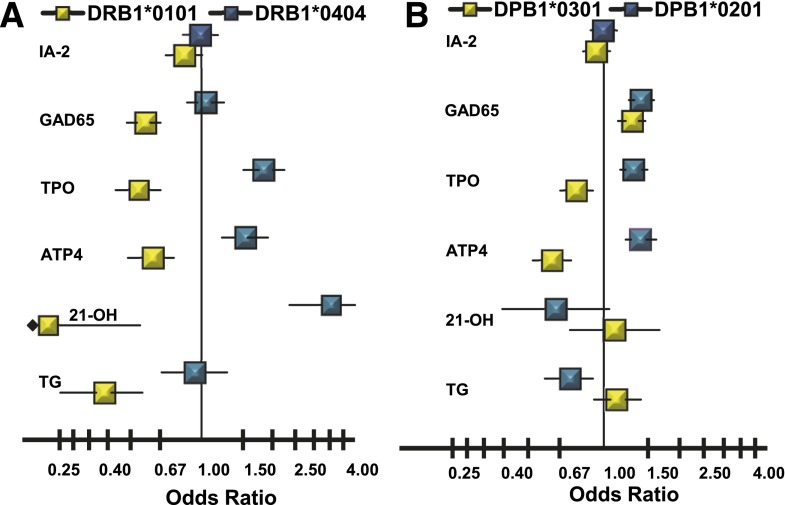
The HLA class II alleles associated with ATP4A autoreactivity are also associated with other autoantibody phenotypes (Fig. 4). The pattern of associations is consistent with the pattern of familial clustering (Fig. 1) and strength of the genetic correlation between phenotypes (Fig. 2), particularly for HLA-DRB1*0404 and HLA-DPB1*0201. DRB1*0404 is associated with ATP4A autoantibodies (OR 1.53; P < 3.0 × 10−4), TPO autoantibodies (OR 1.84; P < 1.2 × 10−9), and 21-OH autoantibodies (OR 3.55; P < 1.0 × 10−9). DRB1*0404 is not associated, however, with GAD65 autoantibodies (OR 1.04; P = 0.67), IA-2 autoantibodies (OR 0.99; P = 0.87), or TG autoantibodies (OR 0.93; P = 0.68) (Fig. 4).
Figure 4.
Autoantibody-specific ORs for HLA alleles identified as positively (blue) or negatively (yellow) associated with ATP4A autoantibodies in NHWs. A: HLA-DRB1 alleles associated with ATP4A autoantibodies in NHW subjects. B: HLA-DPB1 alleles associated with ATP4A autoantibodies in NHW subjects.
HLA-DPB1*0201 is associated with ATP4A autoantibodies (OR 1.40; P < 1.6 × 10−6), TPO autoantibodies (OR 1.32; P < 3.0 × 10−5), and GAD65 autoantibodies (OR 1.41; P < 9.0 × 10−9) but not with IA-2 autoantibodies (OR 1.00; P = 0.97). In addition, DPB1*0201 shows negative association (protection) with TG autoantibodies (OR 0.73; P < 0.009) but not with 21-OH autoantibodies (OR 0.65; P < 0.08). In contrast, the low-risk HLA-DRB1*0101 allele is negatively associated with many autoantibody phenotypes: ATP4 autoantibodies (OR 0.62; P < 1.7 × 10−4), GAD65 autoantibodies (OR 0.58; P < 8.0 × 10−9), TPO autoantibodies (OR 0.54; P < 2.0 × 10−7), 21-OH autoantibodies (OR 0.07; P < 0.01), and TG autoantibodies (OR 0.38; P < 2.0 × 10−5) but not IA-2 autoantibodies (OR 0.85; P < 0.07). The effects of the low-risk HLA-DPB1*0301 allele appear specific to ATP4A autoantibodies (OR 0.62; P < 2.1 × 10−8) and TPO autoantibodies (OR 0.77; P < 4.0 × 10−4), with an opposite effect for GAD65 autoantibodies (OR 1.30; P < 5.0 × 10−5). The DPB1*0301 allele has no effect on IA-2 autoantibodies (OR 0.94; P = 0.30), 21-OH autoantibodies (OR 1.10; P = 0.63), or TG autoantibodies (OR 1.13; P = 0.27).
CTLA4 and PTPN22 Associations
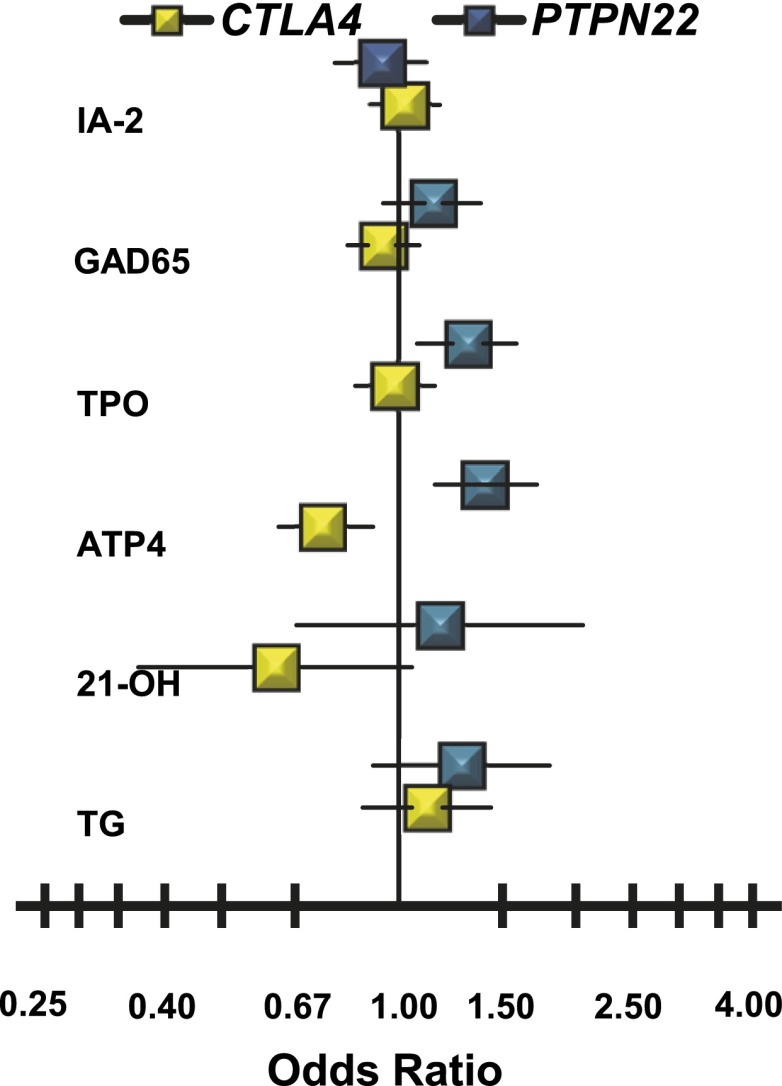
Genotypes for SNPs in the PTPN22 region on human chromosome 1p13.2 (24 SNPs with MAF >0.05) and in the CTLA4 region on chromosome 2q33.3 (19 common SNPs) were available in a subset of 2,003 NHWs. Of the PTPN22 region SNPs, only the rs2476601 variant in the coding region of PTPN22 (R620W), known to be associated with type 1 diabetes risk, is significantly associated with ATP4A autoantibodies (OR 1.40; P < 0.001) (Fig. 5). The association of rs2476601 with TPO autoantibodies is relatively weak (OR 1.31; P < 0.01). There is no evidence of association of PTPN22 rs2476601 with GAD65 autoantibodies (OR 1.13; P = 0.18), 21-OH autoantibodies (OR 1.17; P = 0.57), TG autoantibodies (OR 1.27; P = 0.16), or IA-2 autoantibodies (OR 0.93; P = 0.45).
Figure 5.
Autoantibody-specific ORs for the CTLA4 CT60 variant (rs3087243-T) identified as negatively (yellow) associated with ATP4A autoantibodies and the PTPN22 R620W variant (rs2476601-T) identified as positively (blue) associated with ATP4A autoantibodies in NHW subjects.
In contrast, four of the CTLA4 region SNPs, rs3087243, rs15231811, rs11571293, and rs11571316, are negatively associated with ATP4A autoantibodies. CTLA4 SNP rs3087243 (CT60) has the strongest association with ATP4A autoantibodies (OR 0.74; P < 3.0 × 10−4). Results of conditional analyses suggest that the ATP4A associations with rs15231811, rs11571293, and rs11571316 are most likely due to linkage disequilibrium with rs3087243. Although none of the other five autoantibody phenotypes show significant association with rs3087243, there are suggestive associations for 21-OH autoantibodies (OR 0.61; P < 0.08), but given the low effect size (OR 0.62), wide confidence limits, and relatively small sample size (38 21-OH autoantibody–positive NHWs with SNP genotypes), caution must be taken in this suggestive interpretation of effect. The effect size of CTLA4 SNP rs3087243 is not far from neutrality for IA-2 autoantibodies (OR 1.02; P = 0.71), GAD65 autoantibodies (OR 0.94; P = 0.39), TPO autoantibodies (OR 0.99; P = 0.84), and TG autoantibodies (OR 1.11; P = 0.39).
Conclusions
Gastric autoimmunity, defined as presence of autoantibodies targeting the H+/K+-ATPase proton pump (PCA) of the gastric parietal cell, is understudied, lacking data or consensus about relationships with age, sex, ethnicity, and key genetic markers. The vast majority of previous studies of gastric autoimmunity used indirect immunofluorescence for detecting PCA in a relatively small number of samples from patients with PA or other autoimmune diseases (17). Our results demonstrate that the prevalence of ATP4A autoantibodies increases with age, is higher in females than in males, and is higher in subjects of Hispanic/Mexican and African ancestry and lower in those of Asian ancestry than in NHWs (Table 1). ATP4A autoantibodies are also more common among those with HLA-DRB1*0404-DQA1*0301-DQB1*0302, HLA-DPB1*0201, or the PTPN22 coding variant, R620W (rs2476601-T). ATP4A autoantibodies are less common among those with HLA-DRB1*0101-DQA1*0101-DQB1*0501, DPB1*0301, or the CTLA4 variant rs3087243-T (CT60) (Figs. 3–5). Except for DPB1*0201, the HLA effects are consistent across ethnicity (Fig. 3). Although we did not confirm previous reports of an association of PCA with DQA1*0501-DQB1*0301 (18) in those of NHW (OR 1.02; P = 0.91) or Asian (OR 1.07; P = 0.91) ancestry, there is suggestive evidence of association with DQA1*0501-DQB1*0301 in those of Hispanic/Mexican (OR 2.13; P = 0.038) and African (OR 1.91; P = 0.023) ancestry (data not shown).
There are few, if any, reports of ethnic differences in the frequency of PCA in subjects with type 1 diabetes of appreciable size. An early study observed similar frequencies in blacks and whites (19), whereas we observed an increased frequency in blacks (26.2%) and Hispanics (36.3%) compared with NHWs (20.8%). The decreased frequency of ATP4A autoantibody positivity seen in Asians (16.7%) is consistent with studies showing that ABG and PA are extremely rare among Asians in general (20), but the increased frequencies of ATP4A autoantibody positivity seen in blacks and Hispanics conflict with studies showing that ABG and PA are also less common in these groups. However, previous studies of PA focused on elderly subjects. Differences seen in the relatively young T1DGC cohort could relate to atypical clinical and immunological expression of PA in Hispanics and blacks, including accelerated onset of PA (21).
The T1DGC Autoantibody Workshop, providing data collected on NHW families with two or more type 1 diabetes–affected siblings, is the first systematic investigation of the heritability of ATP4A autoreactivity and other autoantibodies as well as bivariate heritability (genetic correlation) of ATP4A with other autoantibodies (Figs. 1 and 2). In addition, these data permitted evaluation of the influence of genetic variation of the HLA class I, HLA class II, CTLA4, and PTPN22 genes on the autoantibodies. The results demonstrate that the difference between ATP4A sero-positive (20.8%) and sero-negative (79.2%) subjects is primarily genetic (∼95% after allowance for age and sex), as is also the case for each of five other organ-specific autoantibodies targeting the GAD65 and IA-2 islet autoantigens, thyroid TPO autoantigen, adrenal 21-OH autoantigen, and celiac-associated TG autoantigen (h2 > 70%). Although a significant fraction of the heritability of ATP4A autoreactivity is shared (the genetic correlation) with TPO (24.6%), 21-OH (13.6%), and persistent GAD65 (14.1%) autoantibodies, most of the heritability is explained by genetic influences that do not relate to other autoantibody phenotypes and may be more specific to parietal cell autoimmunity (>75%).
The combined effects of class II HLA (DRB1*0404, DRB1*0101, DPB1*0201, DPB1*0301), PTPN22 (rs2476601-T), and CTLA4 (rs3087243-T) variants account for ∼4–5% of PCA heritability, much of which is shared with one or more autoantibody phenotypes in a pattern consistent with observed genetic correlations (Figs. 2, 4, and 5). A recent genome-wide association study of PCA in participants with type 1 diabetes who were unselected for family history identified a strong negative association (OR 0.34) with a marker tagging ABO blood type O (22,23). These results could relate to early reports of an increased frequency of blood type A in patients with PA (24). Since no other autoantibody phenotype or autoimmune disease has been found to be associated with ABO, the observed association with PCA may contribute to the parietal cell–specific component of PCA heritability. That same study did not detect an association with either rs2476601 (PTPN22) or rs3087243 (CTLA4), even using a less stringent level (P < 0.01) of statistical significance. This could be due to a lower frequency of PCA (∼10% vs. ∼20% in the T1DGC Autoantibody Workshop samples), younger-aged subjects with type 1 diabetes (13 years vs. 22 years in T1DGC), or genetic differences between T1DGC ASP families versus unselected cases.
The genes and variants identified by screening for association with ATP4A autoantibodies may be involved in lowering the threshold for developing autoimmunity or in determining the specific organs and tissues targeted by the autoimmune response. Most autoimmune diseases have been found to be associated with one or more HLA genes, particularly the HLA class II genes HLA-DRB1 and HLA-DQB1, and to a lesser extent HLA-DPB1 (25–27). The HLA class II genes encode the HLA-DR, -DQ, and -DP proteins residing on the surface of antigen-presenting cells. Antigen bound to the groove of an HLA class II molecule enables recognition by the T-cell receptor/CD3 complex, with additional stimuli required for T-cell activation and proliferation provided by genes regulating the T-cell response, such as CTLA4 and PTPN22. Different HLA class II alleles encode different peptide-binding structures, thus impacting the binding affinity for different tissue-specific autoantigens. Often viewed as primary determinants of tissue specificity in autoimmunity (25,26,28), the association of the same HLA alleles with multiple organ-specific autoimmune phenotypes could be due to linkage disequilibrium between different HLA class II genes. Experimental data suggest that the HLA-DQβ chain may be most critical for binding immunogenic peptides derived from pancreas in autoimmune diabetes (29–31), whereas the HLA-DRβ chain may be most critical for binding immunogenic peptides derived from thyroid in autoimmune thyroid disease (32,33). Thus, tight linkage disequilibrium between the genes encoding HLA-DR and -DQ could explain the increased risk for both thyroid and pancreatic autoimmunity associated with specific DR-DQ haplotypes such as DRB1*0404-DQ8. Nevertheless, commonality in HLA associations for a spectrum of autoimmune diseases suggests a more general effect on susceptibility and resistance to autoimmunity, particularly for the high-risk diabetes haplotypes, DR3-DQ2 and DR4-DQ8 (34). Specific residues in both DRβ (35,36) and DPβ (37,38) binding domains have been implicated in multicellular autoimmunity, possibly by influencing T-cell receptor docking, autoantigen recognition by CD4+ T-helper cells, or both (39).
The non-HLA genes, CTLA4 and PTPN22, influence the T-cell response to antigen presentation, and thus their role in multicellular autoimmunity is more obvious than for the multiple genes and variants in the HLA complex. The CTLA4 gene is expressed on activated CD4+ and CD8+ T cells, where the gene product downregulates T-cell activation, proliferation, and differentiation by interacting with costimulatory molecules on the surface of antigen-presenting cells. Screening 19 common polymorphisms in CTLA4 identified an association of ATP4A autoantibodies with the CTLA4 rs3087243 SNP. Subsequent analyses showed little or no evidence for association with other autoantibody phenotypes. The same CTLA4 variant rs3087243 (CT60) is associated with several autoimmune diseases, including autoimmune thyroid disease, type 1 diabetes, Addison disease, and celiac disease (40). Initial structural and functional studies suggest that the rs3087243 variant may modify the expression of alternative mRNA splice variants of CTLA4 in unstimulated CD4+ T cells (41), though a subsequent study failed to confirm these results (42). Similar to CTLA4, the association of ATP4A with the PTPN22 rs2476601 (R620W) SNP was identified by screening 24 common polymorphisms. In contrast, a trend in association was seen consistently for multiple autoantibody phenotypes. This variant is also associated with multiple autoimmune diseases, and functional studies suggest that the autoimmunity-predisposing variant decreases T-cell receptor–mediated signaling and subsequent T-cell activation (43).
The T1DGC Autoantibody Workshop and the current analyses demonstrate that the pathogenesis of parietal cell, thyroid, and adrenal autoimmunity in those with type 1 diabetes involves common genetic pathways, perhaps also shared with persistent GAD65 autoimmunity, but not with persistent IA-2 or celiac autoimmunity. The results are consistent with many studies showing an increased frequency of PCA and other organ-specific autoantibodies in families with type 1 diabetes, including reported associations between PCA and persistent GAD65 antibody positivity, but not persistent IA-2 antibody positivity (1,17), possibly reflecting a broader tissue distribution for GAD65 than for IA-2, and in line with studies showing that GAD65 autoantibodies are less specific markers of pancreatic autoimmunity than are IA-2 autoantibodies (44,45). The comorbidity of type 1 diabetes and other chronic inflammatory diseases, particularly Addison disease, chronic lymphocytic thyroiditis, and gastric autoimmunity, was one of the first clues pointing to an autoimmune basis for type 1 diabetes, ultimately distinguishing type 1 and type 2 diabetes, as well as autoimmune and nonautoimmune forms of Addison disease and ABG (2,46). Family studies showing overt or latent autoimmunity also occurred in patients’ relatives and led to the recognition of autoimmune polyglandular syndromes and to the notion that multiple autoimmune diseases can arise from the same inherited immunological defect (18).
Two distinct genetic syndromes, APS1 and APS2, were described based on autosomal recessive inheritance of candidiasis-associated hypoparathyroidism and polyendocrine failure in some families (APS1) and autosomal dominant or complex HLA-associated inheritance of polyendocrine failure without candidiasis or hypothyroidism in others (APS2) (47,48). Whereas adrenal insufficiency with polyendocrine failure was initially considered the cardinal manifestation of both APS1 and APS2, more recent data show a much wider range of familial autoimmune disease associations, encompassing both organ-specific and systemic disorders such as systemic lupus erythematosus and rheumatoid arthritis (49). In contrast to adrenal autoimmunity, gastric autoimmunity and PCA have been considered a “minor” component of virtually all validated (APS1 and APS2) and proposed (APS3 and APS4) (50) APS subtypes. However, results of the T1DGC ASP family study indicate that PCA is a major component of a genetically distinct autoimmune cluster that includes both thyroid and adrenal autoimmunity and may well represent a pathogenetic subtype of type 1 diabetes. Our results point to the important role of family studies in dissecting the complex genetics of autoimmunity targeting multiple cells and tissues.
Article Information
Funding. This research uses resources provided by the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases, the National Human Genome Research Institute, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and JDRF and supported by U01 DK062418. The study was also supported in part by grants from the BDC-JDRF Autoimmunity Prevention Center (4-2007-1056); the National Institutes of Health (NIH) Diabetes and Endocrine Research Center (NIDDK P30 DK57516); and the NIH (DK32083 and AI50864).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.M.W. researched data and wrote the manuscript. P.R.F. wrote the manuscript and was responsible for the statistical analyses. T.J.G., L.M.F., and B.A. researched data. J.C.H. conceived the study, researched data, contributed to discussion, and wrote, reviewed, and edited the manuscript. J.C.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This publication is based on the presentations from the Type 1 Diabetes Genetics Consortium (T1DGC) Autoantibody Workshop, which was held on 7 June 2011 in Bethesda, MD. The publication of this supplement was made possible by resources provided by the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, the National Human Genome Research Institute, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and JDRF and supported by grant U01 DK062418.
References
- 1.De Block CEM, De Leeuw IH, Van Gaal LF. Autoimmune gastritis in type 1 diabetes: a clinically oriented review. J Clin Endocrinol Metab 2008;93:363–371 [DOI] [PubMed] [Google Scholar]
- 2.Whittingham S, Mackay IR. Autoimmune gastritis: historical antecedents, outstanding discoveries, and unresolved problems. Int Rev Immunol 2005;24:1–29 [DOI] [PubMed] [Google Scholar]
- 3.Banka S, Ryan K, Thomson W, Newman WG. Pernicious anemia - genetic insights. Autoimmun Rev 2011;10:455–459 [DOI] [PubMed] [Google Scholar]
- 4.Baxter AG, Jordan MA, Silveira PA, Wilson WE, Van Driel IR. Genetic control of susceptibility to autoimmune gastritis. Int Rev Immunol 2005;24:55–62 [DOI] [PubMed] [Google Scholar]
- 5.Johnston C, Millward BA, Leslie RD, Pyke DA, Bottazzo GF. Are thyrogastric autoantibodies associated with an increased susceptibility to developing type 1 (insulin-dependent) diabetes? A study in identical twins. Autoimmunity 1990;6:195–201 [DOI] [PubMed] [Google Scholar]
- 6.Karlsson F, Burman P, Lööf L, Mårdh S. Major parietal cell antigen in autoimmune gastritis with pernicious anemia is the acidproducing H+,K+adenosine triphosphatase of the stomach. J Clin Invest 1988;81:475479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toh BH, Sentry JW, Alderuccio F. The causative H+/K+ ATPase antigen in the pathogenesis of autoimmune gastritis (Abstract). Immunol Today 2000;21:348354 [DOI] [PubMed] [Google Scholar]
- 8.Alderuccio F, Toh BH, Tan SS, Gleeson PA, van Driel IR. An autoimmune disease with multiple molecular targets abrogated by the transgenic expression of a single autoantigen in the thymus. J Exp Med 1993;178:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher JM, Taylor KB. A comparison of autoimmune phenomena in pernicious anemia and chronic atrophic gastritis. N Engl J Med 1965;272:499–503 [DOI] [PubMed] [Google Scholar]
- 10.Chuang JS, Callaghan JM, Gleeson PA, Toh BH. Diagnostic ELISA for parietal cell autoantibody using tomato lectin-purified gastric H+/K(+)-ATPase (proton pump). Autoimmunity 1992;12:1–7 [DOI] [PubMed] [Google Scholar]
- 11.Wenzlau JM, Gardner TJ, Frisch LM, Davidson HW, Hutton JC. Development of a novel autoantibody assay for autoimmune gastritis in type 1 diabetic individuals. Diabetes Metab Res Rev 2001;8:887–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mychaleckyj JC, Noble JA, Moonsamy PV, et al.; T1DGC . HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin Trials 2010;7(Suppl.):S75–S87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown WM, Pierce JJ, Hilner JE, Perdue LH, Lohman K, Lu L, de Bakker PIW, Irenze K, Ziaugra L, Mirel DB. Overview of the Rapid Response data. Genes Immun 2009;10:S5–S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falconer D. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet 1965;29:51–76 [Google Scholar]
- 15.Duggirala R, Williams JT, Williams-Blangero S, Blangero J. A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol 1997;14:987–992 [DOI] [PubMed] [Google Scholar]
- 16.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 1997;14:953–958 [DOI] [PubMed] [Google Scholar]
- 17.de Graaff L, Smit J, Radder J. Prevalence and clinical significance of organspecific autoantibodies in type 1 diabetes mellitus. Neth J Med 2007;65:235–247 [PubMed] [Google Scholar]
- 18.De Block CEM, De Leeuw IH, Vertommen JJF, et al.; Belgian Diabetes Registry . Beta-cell, thyroid, gastric, adrenal and coeliac autoimmunity and HLA-DQ types in type 1 diabetes. Clin Exp Immunol 2001;126:236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maclaren NK, Riley WJ. Thyroid, gastric, and adrenal autoimmunities associated with insulin-dependent diabetes mellitus. Diabetes Care 1985;8(Suppl. 1):34–38 [DOI] [PubMed] [Google Scholar]
- 20.Irvine WJ, McFadzean AJ, Todd D, Tso SC, Yeung RT. Pernicious anaemia in the Chinese: a clinical and immunological study. Clin Exp Immunol 1969;4:375–386 [PMC free article] [PubMed] [Google Scholar]
- 21.Carmel R. Reassessment of the relative prevalences of antibodies to gastric parietal cell and to intrinsic factor in patients with pernicious anaemia: influence of patient age and race. Clin Exp Immunol 1992;89:74–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plagnol V, Howson JMM, Smyth DJ, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 2011;7:e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aird I, Bentall HH, Bingham J, et al. An association between blood group A and pernicious anaemia: a collective series from a number of centres. BMJ 1956;2:723–724 [PMC free article] [PubMed] [Google Scholar]
- 24.Callender ST, Denborough MA, Sneath J. Blood groups and other inherited characters in pernicious anaemia. Br J Haematol 1957;3:107–114 [DOI] [PubMed] [Google Scholar]
- 25.Trowsdale J. The MHC, disease and selection. Immunol Lett 2011;137:1–8 [DOI] [PubMed] [Google Scholar]
- 26.Goris A, Liston A. The immunogenetic architecture of autoimmune disease. Cold Spring Harb Perspect Biol 2012;4:a007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varney MD, Valdes AM, Carlson JA, et al.; Type 1 Diabetes Genetics Consortium . HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: analysis of the Type 1 Diabetes Genetics Consortium families. Diabetes 2010;59:2055–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michels AW, Gottlieb PA. Autoimmune polyglandular syndromes. Nat Rev Endocrinol 2010;6:270–277 [DOI] [PubMed] [Google Scholar]
- 29.Kwok WW, Domeier ME, Johnson ML, Nepom GT, Koelle DM. HLA-DQB1 codon 57 is critical for peptide binding and recognition. J Exp Med 1996;183:1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu B, Gauthier L, Hausmann DHF, Wucherpfennig KW. Binding of conserved islet peptides by human and murine MHC class II molecules associated with susceptibility to type I diabetes. Eur J Immunol 2000;30:2497–2506 [DOI] [PubMed] [Google Scholar]
- 31.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol 2001;2:501–507 [DOI] [PubMed] [Google Scholar]
- 32.Sawai Y, DeGroot LJ. Binding of human thyrotropin receptor peptides to a Graves’ disease-predisposing human leukocyte antigen class II molecule. J Clin Endocrinol Metab 2000;85:1176–1179 [DOI] [PubMed] [Google Scholar]
- 33.Huber A, Menconi F, Corathers S, Jacobson EM, Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev 2008;29:697–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando MMA, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet 2008;4:e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer J, Gallazzi F, Bono E, et al. Peptide binding specificity of HLA-DR4 molecules: correlation with rheumatoid arthritis association. J Exp Med 1995;181:1847–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nepom BS, Nepom GT, Coleman M, Kwok WW. Critical contribution of beta chain residue 57 in peptide binding ability of both HLA-DR and -DQ molecules. Proc Natl Acad Sci USA 1996;93:7202–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson I, Varney M, Kanaan C, et al. Alloresponses to HLA-DP detected in the primary MLR: correlation with a single amino acid difference. Hum Immunol 1997;55:163–169 [DOI] [PubMed] [Google Scholar]
- 38.Díaz G, Catálfamo M, Coiras MT, et al. HLA-DPbeta residue 69 plays a crucial role in allorecognition. Tissue Antigens 1998;52:27–36 [DOI] [PubMed] [Google Scholar]
- 39.Gough SC, Simmonds MJ. The HLA region and autoimmune disease: associations and mechanisms of action. Curr Genomics 2007;8:453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gough SC, Walker LS, Sansom DM. CTLA4 gene polymorphism and autoimmunity. Immunol Rev 2005;204:102–115 [DOI] [PubMed] [Google Scholar]
- 41.Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511 [DOI] [PubMed] [Google Scholar]
- 42.Mayans S, Lackovic K, Nyholm C, et al. CT60 genotype does not affect CTLA-4 isoform expression despite association to T1D and AITD in northern Sweden. BMC Med Genet 2007;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanford SM, Mustelin TM, Bottini N. Lymphoid tyrosine phosphatase and autoimmunity: human genetics rediscovers tyrosine phosphatases. Semin Immunopathol 2010;32:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuomi T, Björses P, Falorni A, et al. Antibodies to glutamic acid decarboxylase and insulin-dependent diabetes in patients with autoimmune polyendocrine syndrome type I. J Clin Endocrinol Metab 1996;81:1488–1494 [DOI] [PubMed] [Google Scholar]
- 45.Söderbergh A, Myhre AG, Ekwall O, et al. Prevalence and clinical associations of 10 defined autoantibodies in autoimmune polyendocrine syndrome type I. J Clin Endocrinol Metab 2004;89:557–562 [DOI] [PubMed] [Google Scholar]
- 46.Betterle C, Lazzarotto F, Presotto F. Autoimmune polyglandular syndrome type 2: the tip of an iceberg? Clin Exp Immunol 2004;137:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirfält A. Genetic heterogeneity in autoimmune polyglandular failure. Acta Med Scand 1981;210:7–13 [DOI] [PubMed] [Google Scholar]
- 48.Neufeld M, Maclaren NK, Blizzard RM. Two types of autoimmune Addison’s disease associated with different polyglandular autoimmune (PGA) syndromes. Medicine (Baltimore) 1981;60:355–362 [DOI] [PubMed] [Google Scholar]
- 49.Weetman AP. Diseases associated with thyroid autoimmunity: explanations for the expanding spectrum. Clin Endocrinol (Oxf) 2011;74:411–418 [DOI] [PubMed] [Google Scholar]
- 50.Betterle C, Zanchetta R. Update on autoimmune polyendocrine syndromes (APS). Acta Biomed 2003;74:9–33 [PubMed] [Google Scholar]