Abstract
Background
Prevotella intermedia is a Gram-negative black-pigmenting oral anaerobe associated with periodontitis in humans, and has a haem requirement for growth, survival and virulence. It produces an iron porphyrin-containing pigment comprising monomeric iron (III) protoporphyrin IX (Fe(III)PPIX.OH; haematin). The bacterium expresses a 90-kDa cysteine protease termed interpain A (InpA) which both oxidizes and subsequently degrades haemoglobin, releasing haem. However, it is not known whether the enzyme may play a role in degrading other haem-carrying plasma proteins present in the gingival sulcus or periodontal pocket from which to derive haem. This study evaluated the ability of InpA to degrade apo- and haem-complexed albumin.
Results
Albumin breakdown was examined over a range of pH and in the presence of reducing agent; conditions which prevail in sub- and supra-gingival plaque. InpA digested haemalbumin more efficiently than apoalbumin, especially under reducing conditions at pH 7.5. Under these conditions InpA was able to substantially degrade the albumin component of whole human plasma.
Conclusions
The data point to InpA as an efficient “albuminase” with the ability to degrade the minor fraction of haem-bound albumin in plasma. InpA may thus contribute significantly to haem acquisition by P. intermedia under conditions of low redox potential and higher pH in the inflamed gingival crevice and diseased periodontal pocket where haem availability is tightly controlled by the host.
Keywords: Albumin, Protease, Periodontal disease, Haem, Interpain A, Prevotella intermedia
Background
Members of the genus Prevotella are associated with periodontal diseases in humans and other animals [1]. They have a growth requirement for iron protoporphyrin IX, and display a characteristic black-pigmenting phenotype due to cell-surface accumulation of haem (iron(III) protoporphyrin IX) in the monomeric form (Fe(III)PPIX.OH, haematin), derived via the breakdown of haemoglobin [2]. The haem-containing pigments of both Prevotella and Porphyromonas species are important defensively, since ferrihaems in both the monomeric and dimeric ([Fe(III)PPIX]2O) forms possess inherent catalase activity and thus can protect against hydrogen peroxide [3] which is produced by activated neutrophils.
In the primary habitats of the black-pigmenting anaerobes (the inflamed gingival sulcus and periodontal pocket), the most abundant haem source is haemoglobin. However, in healthy or minimally inflamed gingival crevices, in the absence of overt bleeding (and hence haemoglobin), their survival and growth may depend on haemalbumin and haem-haemopexin as primary haem sources. These proteins normally sequester any circulating free haem and transport it to the liver [4, 5], thus limiting its bioavailability to invading micro-organisms.
Albumin is the most predominant protein from both healthy and inflamed periodontal disease sites [6–8]. Although albumin has a 75-fold lower affinity for haem compared to haemopexin [9], its haem sequestering capability is compensated by its 40-80-fold greater concentration in serum [10], and therefore it binds the majority of any haem liberated into the circulation. In addition, in normal serum, approximately 1 in 5000 albumin molecules is haem liganded [11]. Importantly, these haemalbumin molecules may serve as a non-haemoglobin source of iron protoporphyrin IX under conditions of haem limitation for P. intermedia and other sub- and supra-gingival plaque bacteria.
We have investigated the mechanism of protease-mediated haem release from haemoglobin by P. intermedia [12]. This organism expresses a 90-kDa cysteine protease, termed interpain A (InpA) [13], an orthologue of periodontain (PrtP) of P. gingivalis and streptopain (SpeB) of Streptococcus pyogenes [14, 15]. InpA is likely to serve a major role in pigment production through oxidising oxyhaemoglobin and then fully degrading the methaemoglobin product to release haem [12]. In addition, it has been recently shown that InpA may serve to aid P. gingivalis haem acquisition by proteolytically mediating formation of methaemoglobin from which the P. gingivalis HmuY haemophore can extract haem [16]. Cell associated protease activity of P. intermedia can degrade albumin and haemopexin [17, 18], although the proteases mediating this have not been characterised. Incubation of the bacterium with albumin intensifies its proteolytic activity and growth [19]. Given that albumin represents an important source of haem under conditions of haem scarcity, we examined the ability of InpA to degrade albumin in both its apo- and haem-liganded forms under conditions which may prevail in the primary habitats of this bacterium, the inflamed gingival sulcus and diseased periodontal pocket.
Results and discussion
Since the redox potential of diseased periodontal pockets can be as low as −300 mV [20] and that P. intermedia may experience wide fluctuations of pH within dental plaque, it was considered appropriate to examine the effect of these two variables on the activity of InpA. Our previous study [13] showed that InpA activity was stimulated by dithiothreitol (DTT) and so we examined the effect of various concentrations of DTT on InpA activity versus the fluorescent synthetic substrate Z-Arg-Arg-AMC (250 μM). This revealed that enzyme activity began to plateau at 2 mM (Fig. 1a). We then determined the influence of the thiol reducing agents cysteine, reduced glutathione and β-mercaptoethanol (all at 2 mM) on InpA activity using the above substrate, and this showed that DTT gave the greatest stimulatory effect (Fig. 1b). However, in subsequent experiments to examine the influence of reducing conditions on albumin and haemalbumin degradation, we employed 3 mM DTT to ensure full enzyme activation.
Fig. 1.
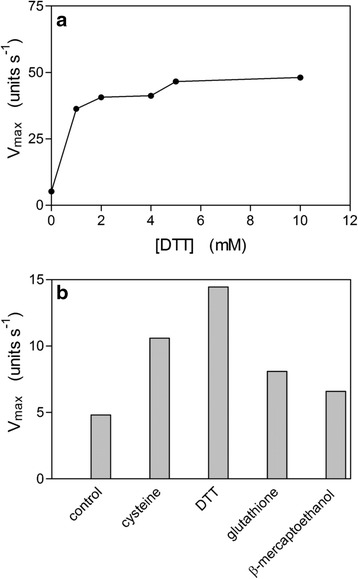
Effect of dithiothreitol (a) and other thiol reducing agents (b) on the enzyme activity of InpA. The enzyme activity was assayed in triplicate using Z-Arg-Arg-AMC at 37 °C in 0.1M Tris–HCl, pH 7.5 and the concentrations of the reducing agents in Fig. 1b were 3 mM. Due to the high reproducibility of the replicates, no SD is apparent. Control, activity in the absence of reducing agent. InpA concentrations in (a) and (b) were 12.5 μg ml−1. See text for details
InpA displayed a broad activity against Z-Arg-Arg-AMC over the pH range from 6 to 10 (Fig. 2a). However, for azo-albumin, the maximal activity was centred at pH 5.5, with little or no activity beyond pH 4 or above 8 (Fig. 2b). This may be important in terms of protein breakdown since P. intermedia can generate acid during growth in both liquid and solid media [2, 21, 22], and may thus depress the pH locally in vivo. We also investigated the temperature activity profile for InpA and thermal stability of the enzyme using azocoll and the synthetic substrate Z-Arg-Arg-AMC, respectively. The enzyme showed maximal activity in the temperature range from 30 to 40 °C versus azocoll (data not shown). However, the enzymatic activity was found to be drastically decreased after the protein was pre-incubated for 30 min at temperatures above 50 °C, followed by assay with Z-Arg-Arg-AMC (data not shown). This indicates that InpA should be highly active at elevated temperatures which may be encountered in vivo in the inflamed gingival sulcus or periodontal pocket during periods of inflammation. .
Fig. 2.
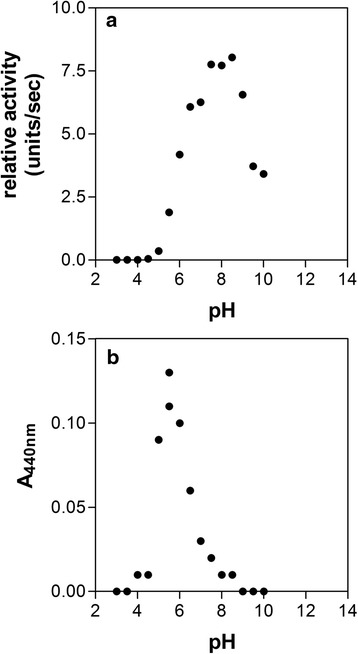
Effect of pH on InpA activity towards Z-Arg-Arg-AMC (a) and azoalbumin (b). Protease activity was assayed at 37 °C in PBS, pH 7.4, containing 3 mM DTT. InpA concentration was 50 μg ml−1. Azoalbumin concentration was 3 % (w/v). Enzyme activity is depicted as absorbance of azo dye (A440) released from the protein. See text for details
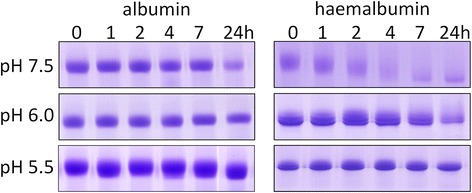
In the absence of DTT, there was little or no degradation of albumin (Fig. 3), and densitometry showed that total apoalbumin degradation over 24 h was less than 5 % at all three pHs studied. In contrast however, for haemalbumin, three lower molecular weight peptide fragments of Rf 0.68, 0.83 and 0.94, were generated at pH 7.5 and 6.0 (arrowed). The two fastest migrating of these bands, which stained with TMB-H2O2 and hence retained bound haem, were further degraded with time and also showed progressive loss of haem staining, as did the main 66-kDa haemalbumin band, especially at pH 7.5 after 4 h. Little protein degradation was observed at pH 5.5. However, after 24 h, TMB-H2O2 staining of haemalbumin had decreased by circa 45 % at all pHs examined, showing that extensive proteolysis was not necessary to effect haem release from the molecule. In contrast however, the percentage of haemalbumin protein degraded after 24 h was greater at pH 7.5 (35 % ± SD 13), compared to either pH 6.0 (20 % ± SD 6) or pH 5.5 (4 % ± SD 6) (n = 4 in all cases). It should be noted that both apo- and haemalbumin were stable over 24 h at all pH conditions tested in the absence of InpA (data not shown).
Fig. 3.
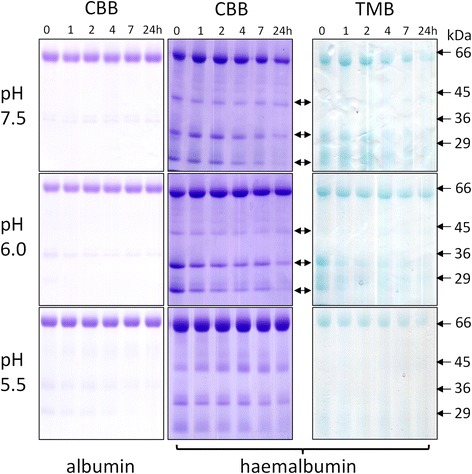
Albumin and haemalbumin breakdown by InpA at different pH under non-reducing conditions. Albumin and haemalbumin (1:1 haem:protein ratio) (16 μM) were incubated with InpA (100 μg ml−1) at 37 °C and aliquots of the incubations were sampled periodically at 1, 2, 4, 7 and 24 h and InpA activity inhibited by E-64 (0.5 mM) and subjected to SDS-PAGE on 10 % non-reducing gels. Samples were solubilised at 37 °C for 1 h in non-reducing application buffer. The haemalbumin gels were stained with TMB to reveal haem-associated peroxidise activity and counterstained with CBB for protein, whilst albumin gels were stained only with CBB. Protein loading was ≈ 20 μg per lane. Arrows denote digestion fragments carrying bound haem
Despite InpA displaying optimum activity towards azo-albumin at pH 5.5 (Fig. 2) and being more active against aquomethaemoglobin at this pH [12], it is interesting that haemalbumin degradation by InpA was negligible at pH 5.5 but greatest at the slightly alkaline pH of 7.5. This apparent increase may not reside in InpA activity per se, but may result from structural changes in the albumin molecule occurring in solution at high pH [23, 24]. Albumin displays a greater thermal stability at pH 6.4 than at pH 7.4 [25], undergoing a conformational change around pH 7.5 referred to as the N → B transition [26], pointing to the B form being more susceptible to InpA, as has been demonstrated for trypsin [27]. This may be physiologically relevant as diseased periodontal pockets and the inflamed gingival sulcus are slightly alkaline [28, 29, 30]. However, SDS-PAGE revealed (Fig. 3) that haem loss also occurred at acid pH despite the low level of proteolysis, suggesting that only limited proteolysis by InpA is required to loosen the haem-albumin association.
Compared to apoalbumin, haemalbumin is more susceptible to InpA. Haem complexing to bovine serum albumin also leads to greater proteolysis by trypsin and chymotrypsin [30] although proteolysis of haemalbumin by whole cells of P. gingivalis is decreased, particularly as the ratio of bound haem to protein increases [31]. Nevertheless, conformational changes occurring upon haem ligation [11] may account for InpA attack, enabling InpA to differentiate between apoalbumin and the scarcer haem-liganded form, which represents approximately 1/5000 of the total albumin molecules in serum under normal physiological conditions [12, 33].
In stark contrast to non-reducing conditions, albumin degradation was greatly increased under reducing conditions (3 mM DTT) at all pHs examined, but especially at pH 7.5, where complete breakdown was observed after 4 h (Fig. 4). The presence of DTT also increased albumin susceptibility at pH 6.0 (86 % after 24 h). However, at pH 5.5 less than 10 % was broken down even when DTT was present. Albumin was found to be stable under reducing conditions in the absence of InpA as judged by minimal depreciation in CBB staining observed only after 24 h incubation at pH 7.5 (Fig. 5). In contrast to reducing conditions, apoalbumin was less susceptible to InpA (Fig. 3), and densitometric scans (data not shown) indicated that approximately 20 % had been degraded after 7 h at each pH.
Fig. 4.
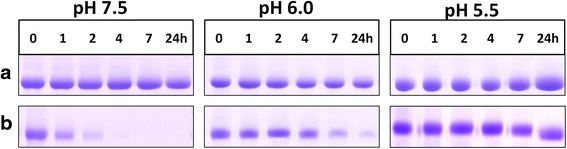
Chemical reduction increases the susceptibility of albumin to InpA. Albumin (16 μM) was incubated with InpA (100 μg ml−1) at different pH at 37 °C under non-reducing (a panels) and reducing conditions (3 mM DTT) (b panels). Gel lanes had a nominal loading of 20 μg albumin, and are stained with CBB. Samples were prepared and gels run as described for Fig. 3
Fig. 5.
SDS-PAGE showing the stability of albumin and haemalbumin under reducing conditions at different pH. Albumin and haemalbumin (16 μM) were reduced with 3 mM DTT and incubated at 37 °C. SDS-PAGE conditions were the same as those in Fig. 3. The gels were stained with CBB
In view the above, and the compositional similarities between gingival crevicular fluid and plasma [6], it was appropriate to examine the effect of InpA on whole blood plasma, under conditions commonly reflecting those in the inflamed sulcus and periodontal pocket i.e., alkaline pH and low redox potential. SDS-PAGE revealed that overnight incubation of whole human plasma with InpA at pH 7.5 and in the presence of 3 mM DTT, resulted in almost complete breakdown of the albumin band (Fig. 6a, arrowed), whilst leaving many other plasma proteins relatively undigested. In comparison, proteinase K, a broad spectrum serine endopeptidase without any peptide bond preference, degraded a large number of the proteins in whole plasma but was less efficient at degrading the albumin component when compared to InpA at the same enzyme concentration (Fig. 7a and b, arrowed). These data show that InpA is able to target albumin present in plasma.
Fig. 6.
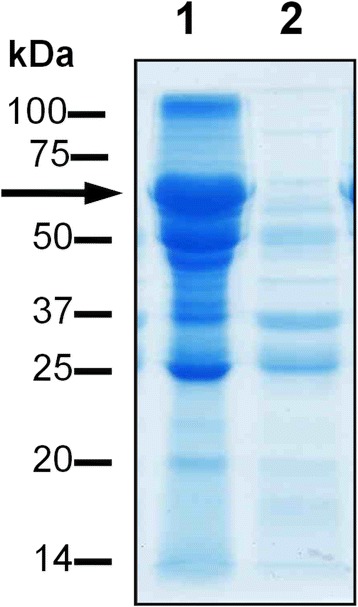
SDS-PAGE analysis of the breakdown of whole human plasma by InpA. Plasma (1 μl, ~60μg protein), was incubated overnight at 37 °C in PBS, pH 7.4, containing 3 mM DTT in absence (lane 1) or presence (lane 2) of active InpA (100 μg ml−1) in a final volume of 20 μl. ~ 50 μg total protein was loaded per lane. Arrow denotes albumin band. Samples were solubilised for 5 min at 100 °C in reducing application buffer, and gels were stained with CBB
Fig. 7.
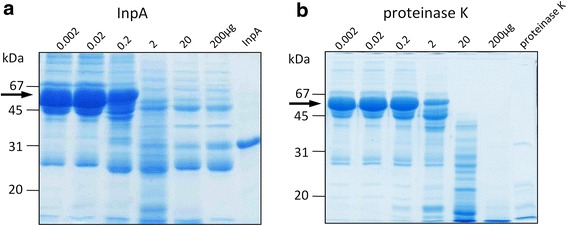
Comparative SDS-PAGE analysis of the digestion of whole human plasma by InpA and proteinase K. Whole human plasma (60 μg total protein) was incubated overnight with InpA and proteinase K (Sigma-Aldrich; product number P2308) at the indicated concentrations (μg ml−1) at 37 °C in PBS, pH 7.4 containing 3 mM DTT. Last lane of each gel was loaded with 10 μg of either InpA or proteinase K only. Samples were prepared and gels stained as in Fig. 6
The increased susceptibility of albumin to InpA in the presence of DTT may arise from structural changes brought about by chemical reduction of the disulphide bridges. Conformational changes of the local tryptophan environment in albumin are reflected in fluctuations of intrinsic fluorescence [32, 33]. When apoalbumin was treated with DTT there was a reduction of fluorescence intensity and moderate shift of λmax, indicating a change in the protein structure (Fig. 8). Fluorescent emission was not detected above 100 μM DTT, indicating that the concentrations used in the digestions (2–10 mM) were sufficient to induce structural alterations in albumin. Since InpA was pre-activated with an excess of DTT, it is likely that this increased susceptibility of albumin to InpA under reducing conditions was due to structural changes in the substrate rather than to any over-stimulatory effect of DTT on protease activity per se.
Fig. 8.
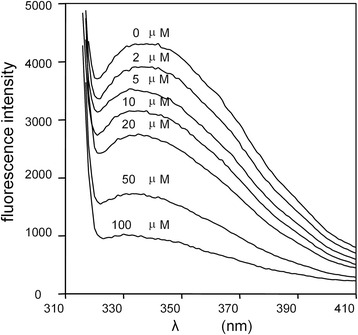
Intrinsic fluorescence spectra of bovine albumin in the presence of DTT. Albumin (16 μM) was treated with increasing concentrations of DTT in 0.14 M NaCl, 0.1 M Tris–HCl buffer, pH 7.5, at 37 °C, and the fluorescence emission monitored after excitation at 295 nm
The low redox potential of diseased periodontal pockets is attributable to the presence of volatile sulphur compounds (VSCs) including H2S and methyl mercaptan, generated by organisms including P. intermedia, P. gingivalis and Fusobacterium nucleatum [34, 35]. P. intermedia is also recovered from periodontal disease sites of patients suffering from oral malodour linked to VSC production [36]. The 17 disulphide bridges of albumin are sensitive to reduction, which stimulates tertiary structural changes [33, 37], leading to increased molecular flexibility and enhanced susceptibility to pepsin [38]. Chemical reduction resulting from bacterial metabolism occurs during batch-wise enrichment culture of sub-gingival plaque in serum, where redox potentials of -70 to −550 mV have been recorded, and correlate with both H2S production and enrichment in numbers of P. intermedia [39]. Since 3 mM DTT was sufficient for optimal InpA activity, and is a concentration which can depress the redox potential to between −60 and −220 mV in E. coli growth medium [40], it is likely that the reducing environment of deep periodontal pockets also results in structural changes in albumin because of disulphide bond disruption [33], rendering albumin highly susceptible to InpA. Predictably therefore, treatment of albumin with 3 mM DTT greatly increased degradation by InpA, especially at pH 7.5. The minimal breakdown at acid pH with DTT present, where InpA activity towards azo-albumin and aquomethaemoglobin is highest [12], suggests that pH-dependent conformational changes [23, 25, 41] may dictate the effectiveness of proteolysis by InpA. It is noteworthy also that accessibility of reducing agents to disulfide bonds in albumin (which would be in the B form) increases above pH 7.0 [42].
In the presence of 3 mM DTT the susceptibility of haemalbumin (1:1 haem: protein ratio) to InpA was also dramatically increased (Fig. 9). This was especially noticeable at pH 7.5; an observation explained by the fact that haemin binding to albumin expands sub-domain IB (normally constrained by the Cys124 and Cys169 S-S bridges [43]) which may render other regions of the peptide chain available for attack. To further assess the ability of InpA to bring about proteolytic haem release from albumin at pH 7.5 haemalbumin conjugated agarose was used as a substrate. After 1 h in the presence of DTT, InpA liberated approximately 3-fold more haem than under non-reducing conditions (data not presented). In this context it is noteworthy that the albumin-haem affinity is lowered under reducing conditions, which in vivo favours haem transfer to haemopexin [44]. Furthermore, we observed that under reducing conditions haemalbumin was intrinsically more unstable than its non-liganded counterpart, particularly at alkaline pH (Fig. 5), which may explain its heightened susceptibility to enzymic digestion. Thus, should the local plaque or periodontal pocket redox potential decline, the additional effect of reducing agents in facilitating haem liberation may supplement InpA-mediated haem release.
Fig. 9.
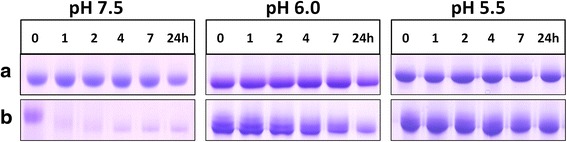
Effect of reducing conditions on breakdown of haemalbumin by InpA at different pH. Haemalbumin (16 μM) with a 1:1 haem to protein ratio was incubated at 37 °C with InpA (100 μg ml−1) in the absence (a panels) and presence of 3 mM DTT (b panels). Samples and gels were run under non-reducing conditions on 10 % gels and stained for protein with CBB. Each gel lane had a nominal loading of 20 μg albumin
It is noteworthy that Grenier et al. [45] demonstrated albumin breakdown by P. gingivalis R- and K-gingipains in the presence of 10 mM DTT. We have observed that albumin and haemalbumin are resistant to active forms of both HRgpA and Kgp under non-reducing conditions (data not presented). These and the above findings highlight the importance of reducing conditions which prevail in the diseased periodontal pocket and which are essential for disrupting both the haem liganded and apoalbumin structures.
InpA has specificity for K, R, A and F residues at the P1 position of hydrolysed peptide bonds [12; J. Potempa, unpublished findings] and this was seen in the large number of peptide fragments generated under both reducing and non-reducing conditions (Fig. 10). In all, 19 InpA scission sites were exclusive to reducing conditions and 12 to non-reducing conditions, with 24 sites common to digestions under both incubation conditions. The formation of more fragments in the presence of DTT is not surprising, as it may reflect the exposure of more susceptible cleavage sites at previously inaccessible residues resulting from disulphide bond disruption.
Fig. 10.

Primary sequence showing all the cleavage sites inflicted during incubation of bovine serum albumin with InpA. Albumin (16 μM) was incubated at 37 °C with InpA (100 μg ml−1) in 10 mM Tris buffer, pH 7.5, in the presence or absence of 2mM DTT, and the incubation mixture sampled over time subjected to MALDI-TOF mass spectrometry. InpA scission sites exclusive to reducing ( ) , non-reducing (
) , non-reducing ( ), and to both incubation conditions (
), and to both incubation conditions ( ) are indicated. See text for details
) are indicated. See text for details
Bleeding, and consequently increased levels of haemoglobin and hence haem, is associated clinically with the inflamed gingival crevice and diseased periodontal pocket, where the host responds to bacterial presence by increasing levels of plasma stress proteins including haemopexin and albumin [6, 46, 47]. In addition, the combined haemolytic and proteolytic activities of other sub-gingival micro-organisms may also transiently elevate local haem levels resulting in greater haem-albumin concentrations. However, we show that InpA not only degrades apoalbumin which normally reduces the potential for haem-induced inflammatory events [48] and limits the bioavailability of haem to micro-organisms, but is more active towards the haem-liganded protein. Depreciation in haem scavenging by albumin at periodontal sites through the action of InpA would hinder local haem clearance and potentially advantage P. intermedia and other haem-requiring organisms. In addition, in regard to the periodontal pocket environment, it is known that in addition to low redox potential, slightly alkaline pH conditions also severely perturb host haem recycling, since they lower the albumin-haem binding affinity [44, 49].
Conclusions
The current findings show that InpA can function as an albuminase and may enable P. intermedia to specifically target albumin present in whole plasma. Although reducing and slightly alkaline conditions may be optimal for InpA breakdown of albumin and haem release, this protease may also be versatile in degrading albumin in a plaque environment where non-reducing conditions prevail, such as a newly inflamed site or at early stages of periodontal pocket formation. In conclusion, by more easily degrading haem-liganded albumin, InpA may contribute to haem acquisition by P. intermedia where the availability of this essential growth factor is otherwise tightly restricted by the host.
Methods
Purification of InpA
InpA was produced as a recombinant protein in Escherichia coli and purified as previously described [12, 13] by affinity chromatography on Fast Flow Ni-NTA (Ni2+-nitrilotriacetate)–Sepharose (Qiagen) followed by anion exchange chromatography (MonoQ, GE Healthcare). The active concentration of InpA was determined by active-site titration using an appropriate dilution of a standardized 1 mM aqueous solution of the protease inhibitor N-(trans-epoxysuccinyl)-L-leucine 4-guanidinobutylamide (E-64) (Sigma-Aldrich; product E3132) needed for total inactivation of the proteinase. Residual enzyme activity was determined by measurement of fluorescence (λex = 380 nm and λem = 460 nm) of AMC (7-amino-4-methylcoumarin) released from t-butoxycarbonyl-Val-Leu-Lys-AMC as described previously [13]. Immediately before use, InpA was activated by incubation for 15 min with 3 mM DTT in 0.1 M Tris–HCl, 0.14 M NaCl, pH 7.5 [50]. When used for protein breakdown assays under non-reducing conditions, the above buffer was exchanged for that without reducing agent by ultrafiltration using 10 kDa cut-off Microcons (Amicon).
Gingipain purification
Soluble HRgpA and Kgp gingipains were purified from the culture medium of P. gingivalis strain HG66 as described previously using gel filtration and arginine–Sepharose chromatography [51, 52]. Before use, both HRgpA and Kgp were also activated for 15 min in 0.1 M Tris–HCl, 0.14 M NaCl, pH 7.5, plus 3 mM DTT. However, when employed to determine protein degradation in non-reducing conditions, this buffer was replaced for that without DTT by ultrafiltration as above.
Effect of thiol reducing agents on InpA activity
The fluorescent substrate Z-Arg-Arg-7-amido-4-methylcoumarin (Z-Arg-Arg-AMC; 250 μM) was used to determine the enzyme Vmax as previously described [51], the hydrolysis of AMC being recorded by measuring fluorescence increase (λex = 360 nm; λem = 460nm) in a micro-titre fluorescence plate reader (SpectraMax M5, Molecular Devices) in kinetic mode at an interval of 9 s for 5–10 min.
Temperature-dependent activity profile and thermal stability of InpA
The temperature-dependent activity profile was determined by pre-incubating the enzyme for 10 min in 0.1M Tris–HCl buffer, pH 7.5, containing 2 mM DTT, at temperatures in the range 4 to 60 °C, and then assaying activity at these temperatures using azocoll as substrate. To determine the thermal stability of InpA, the enzyme was pre-incubated for 30 min at temperatures in the above range, and in the above buffer, followed by assay with Z-Arg-Arg-AMC at 37 °C.
Incubations of InpA with albumin
InpA was incubated with apo-and haemalbumin at 37 °C at 1:4 enzyme to substrate ratio, at pH 5.5 (in 0.06 M potassium hydrogen phthalate), pH 6.0 (0.2 M phosphate buffer, pH 6.0) or pH 7.5 (0.14 M NaCl, 0.1M Tris–HCl). Incubations were terminated by incubation for 30 min at 20 °C after addition of 0.5 mM E-64 to inhibit InpA activity, and then sampled for SDS-PAGE on 10 % gels under non-reducing conditions. For some experiments, to examine the effects of reducing conditions, buffers were supplemented with 3 mM DTT. For MALDI-TOF-MS analysis incubations were done in 10 mM Tris–HCl buffer (pH 7.5) with or without 3 mM DTT.
Haemalbumin
Haemalbumin was prepared by incubating a 100 μM stock solution of bovine albumin (Sigma-Aldrich; product A-8763) at 37 °C in 0.14 M NaCl, 0.1 M Tris–HCl, pH 7.5, with iron(III) protoporphyrin IX at a 1:1 protein to haem molar ratio [31].
Haem release from immobilised haemalbumin (agarose)
Bovine albumin-agarose beads (Sigma-Aldrich; product A3790) were washed in 0.14 M NaCl, 0.1M Tris–HCl, pH 7.5, until no unconjugated free albumin could be detected by UV absorbance at 280 nm in the eluate. The albumin was then complexed with haem (albumin to haem molar ratio 1:1) by gentle agitation at 4 °C for 18 h, and the beads (45 μM with respect to albumin) were incubated with gentle agitation with or without 2 μM InpA and with or without 3 mM DTT in 0.14 M NaCl, 0.1M Tris–HCl, pH 7.5 at 37 °C. Aliquots of the whole incubation mixture were periodically removed, having first been thoroughly mixed to ensure homogeneity. The samples were then centrifuged (5,000 g for 1 min at 20 °C) and the supernatants were removed and assayed for haem release by the pyridine haemochromogen method [53].
Fluorescence monitoring of structural changes to albumin
Fluorescence emission of 16 μM albumin in the 310–410 nm range was measured at pH 7.5 in the presence of DTT (up to 100 μM), employing a Flexstation 3 Benchtop (Molecular Devices) multi-mode microplate reader using λex = 295 nm [32, 33].
SDS-PAGE
SDS-PAGE was carried out as previously described on either 10 or 12 % polyacrylamide gels [54]. Where appropriate, samples were solubilised at 100 °C for 5 min in reducing application buffer. For visualisation of protein-bound haem, samples were solubilised at 37 °C for 1 h in non-reducing application buffer and the gels were stained with tetramethylbenzidine-H2O2 (TMB) and then counter stained with Coomassie brilliant blue G250 (CBB) [13]. Concentrations of protein for SDS-PAGE were determined using the Bradford method.
Densitometry
Densitometry was carried out on CBB- and TMB-H2O2-stained bands using UVIband gel analysis software (UVItech Ltd., Cambridge, UK) after digital image capture using a UMax Powerlook 1000 flatbed transmission scanner. Digital pixel volumes of CBB- and TMB-H2O2-stained bands were used as indirect measures of protein and haem loss from a sample (calculated as a percentage of the time zero sample).
Determination of pH activity profile of InpA using azoalbumin
The activity profile of InpA over the pH range 3 to 10 was determined with azoalbumin, using 0.1 M sodium acetate for pH 3–5.5, 0.1 M sodium phosphate for pH 5.5-7.0, and 0.1 M Tris–HCl for pH 7–10. Azoalbumin (3 % w/v) was incubated with enzyme (final protein concentration 50 μg ml−1) in the above buffers together with freshly added DTT (3 mM) at 37 °C with shaking for 1 h. Subsequently, the protease activity was inhibited by mixing with two volumes of 10 % (w/v) ice-cold TCA. After 10 min, the insoluble protein was pelleted by centrifugation (10,000 × g for 10 min), and released azo dye was measured at 440 nm.
MALDI-TOF-mass spectrometry
This was carried out using a Micromass M@LDI mass spectrometer, using an α-cyano-4-hydroxy-cinnamic acid matrix (Sigma-Aldrich). The spectra were recorded in the positive ion mode and the mass range scanned was 800 to 4000 Daltons. The spectrometer was calibrated using a mixture of authentic peptide samples. The observed peptide masses were compared to those predicted by theoretical cleavage using the PeptideMass program (ExPASy).
Acknowledgements
We thank Mr Mark Prescott and Dr Deborah Ward of the Proteomics Laboratory, School of Biological Sciences, University of Liverpool, who performed the mass spectrometry. JWS is grateful for financial support (to DPB) from the University of Liverpool, School of Dentistry, and the British Council [British-Polish Young Scientist Programme; project WAR/342/146). JP is supported by grants from the EC [FP7-HEALTH-F3-2012-306029 “TRIGGER”, and FP7-PEOPLE-2011-ITN-290246 “RAPID”], the National Institutes of Health/NIDCR, USA [grant DE 09761], National Science Center, Poland [UMO-2012/04/A/NZ1/00051], and Polish Ministry of Science and Higher Education. The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of structural funds from the European Union [POIG.02.01.00-12-064/08].
Abbreviations
- InpA
Interpain A
- CBB
Coomassie brilliant blue
- DTT
Dithiothreitol
- SDS-PAGE
Sodium dodecyl sulphate polyacrylamide electrophoresis
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JP and JWS conceived the study, coordinated the experimental work and, along with DPB and SPM, wrote the manuscript. Both DPB and SPM carried out the experimental work. All authors read and approved the final manuscript.
Contributor Information
Dominic P. Byrne, Email: bs0u4193@liv.ac.uk
Surya P. Manandhar, Email: Surya.manandhar@csulb.edu
Jan Potempa, Email: jan.potempa@louisville.edu.
John W. Smalley, Email: josmall@liv.ac.uk
References
- 1.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 2.Smalley JW, Silver J, Birss AJ, Withnall R, Titler PJ. The haem pigment of the oral anaerobes Prevotella nigrescens and Prevotella intermedia is composed of iron (III) protoporphyrin IX in the monomeric form. Microbiology. 2003;149:1711–1718. doi: 10.1099/mic.0.26258-0. [DOI] [PubMed] [Google Scholar]
- 3.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the μ-oxo bishaem of iron protoporphrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 4.Beaven GH, Chen SH, D’Albis A, Gratzer WD. A spectroscopic study of the haemin-human-serum-albumin system. Eur J Biochem. 1974;72:539–546. doi: 10.1111/j.1432-1033.1974.tb03295.x. [DOI] [PubMed] [Google Scholar]
- 5.Hrkal Z, Vodrážka Z, Kalousek I. Transfer of haem from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur J Biochem. 1974;43:73–78. doi: 10.1111/j.1432-1033.1974.tb03386.x. [DOI] [PubMed] [Google Scholar]
- 6.Curtis MA, Sterne JAC, Price SJ, Griffiths GS, Coulthurst SK, Wilton JMA, et al. The protein composition of gingival crevicular fluid sampled from male adolescents with no destructive periodontitis: baseline data of a longitudinal study. J Periodont Res. 1990;25:6–16. doi: 10.1111/j.1600-0765.1990.tb01202.x. [DOI] [PubMed] [Google Scholar]
- 7.Makela M, Soderling E, Paunio K, Talonpoika J, Hyyppa A. Protein composition of crevicular fluid before and after treatment. Scand J Dent Res. 1991;99:413–423. doi: 10.1111/j.1600-0722.1991.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 8.Hanioka T, Matsuse R, Shigemoto Y, Ojima O, Shizukuishi S. Relationship between periodontal status and combination of biochemical assays of gingival crevicular fluid. J Periodont Res. 2005;40:331–338. doi: 10.1111/j.1600-0765.2005.00807.x. [DOI] [PubMed] [Google Scholar]
- 9.Hrkal Z, Kalousek I, Vodrazka Z. Haem binding to albumin and equilibria in the albumin-ferrihaemoglobin and albumin-haemopexin system. Int J Biochem. 1980;12:619–624. doi: 10.1016/0020-711X(80)90014-2. [DOI] [PubMed] [Google Scholar]
- 10.Muller-Eberhard U, Morgan WT. Porphyrin binding proteins in serum. Ann N Y Acad Sci. 1975;244:624–649. doi: 10.1111/j.1749-6632.1975.tb41558.x. [DOI] [PubMed] [Google Scholar]
- 11.Walji F, Rosen A, Hider RC. The existence of conformationally labile (preformed) drug binding sites in human serum albumin as evidenced by optical rotation measurement. J Pharm Pharmacol. 1993;45:551–558. doi: 10.1111/j.2042-7158.1993.tb05597.x. [DOI] [PubMed] [Google Scholar]
- 12.Byrne DP, Wawrzonek K, Jaworska A, Birss AJ, Potempa J, Smalley JW. Role of the cysteine protease interpain A of Prevotella intermedia in breakdown and release of haem from haemoglobin. Biochem J. 2009;425:257–264. doi: 10.1042/BJ20090343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallorquí-Fernández N, Manandhar SP, Mallorquí-Fernández G, Usón I, Wawrzonek K, Kantyka T, et al. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J Biol Chem. 2008;283:2871–2882. doi: 10.1074/jbc.M708481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potempa J, Golonka E, Flipek R, Shaw LN. Fighting an enemy within: cytoplasmic inhibition of bacterial cysteine proteases. Mol Microbiol. 2005;57:605–610. doi: 10.1111/j.1365-2958.2005.04714.x. [DOI] [PubMed] [Google Scholar]
- 16.Byrne DP, Potempa J, Olczak T, Smalley JW. Evidence of mutualism between two periodontal pathogens: co-operative haem acquisition by the HmuY haemophore of Porphyromonas gingivalis and the cysteine protease interpain A (InpA) of Prevotella intermedia. Mol Oral Microbiol. 2013; doi: 10.1111/omi.12018. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 17.Carlsson J, Hofling JG, Sundquist GK. Degradation of albumin, haemopexin, haptoglobin and transferrin by black-pigmented Bacteroides species. J Med Microbiol. 1984;18:39–46. doi: 10.1099/00222615-18-1-39. [DOI] [PubMed] [Google Scholar]
- 18.Jansen H-J, van der Hoeven JS, Göertz JHC, Bakkern JAJM. Breakdown of various serum proteins by periodontal bacteria. Microbial Ecol Health Dis. 1994;7:299–305. doi: 10.3109/08910609409141369. [DOI] [Google Scholar]
- 19.Ichiro Y. Prevotella intermedia can utilize materials in the oral cavity. J Jap Ass Periodont. 2000;42:175–181. doi: 10.2329/perio.42.175. [DOI] [Google Scholar]
- 20.Kenny EB, Ash MM. Oxidation reduction potential of developing plaque, periodontal pockets and gingival sulci. J Periodontol. 1969;40:630–633. doi: 10.1902/jop.1969.40.11.630. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi N, Yamada T. Glucose metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol Immunol. 2000;15:188–195. doi: 10.1034/j.1399-302x.2000.150307.x. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 2003;18:109–113. doi: 10.1034/j.1399-302X.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 23.Peters T. Serum albumin. Adv Prot Chem. 1985;37:161–245. doi: 10.1016/S0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 24.Carter DC, Ho JX. Structure of serum albumin. Adv Prot Chem. 1994;45:153–196. doi: 10.1016/S0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 25.Ross PD, Finlayson JS, Shrake A. Thermal stability of human albumin measured by differential scanning calorimetry. II. Effects of isomers of N-acetyltryptophanate and tryptophanate, pH, reheating and dimerization. Vox Sang. 1984;47:19–27. doi: 10.1111/j.1423-0410.1984.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 26.Harmsen BJM, DeBruin SH, Janssen LH, DeMiranda JFR, Van Os GAJ. pK change of imidazole groups in bovine serum albumin due to the conformational change at neutral pH. Biochem. 1971;10:3217–3221. doi: 10.1021/bi00793a009. [DOI] [PubMed] [Google Scholar]
- 27.Markus G, McClintock DE, Castellani BA. Tryptic hydrolysis of human serum albumin. J Biol Chem. 1997;242:4395–4401. [PubMed] [Google Scholar]
- 28.Bickel M, Cimasoni G. The pH of human crevicular fluid measured by a new microanalytical technique. J Periodont Res. 1985;20:35–40. doi: 10.1111/j.1600-0765.1985.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 29.Eggert FM, Drewell L, Bigelow JA, Speck JE, Goldner M. The pH of gingival crevices and periodontal pockets in children, teenagers and adults. Arch Oral Biol. 1991;36:233–238. doi: 10.1016/0003-9969(91)90091-8. [DOI] [PubMed] [Google Scholar]
- 30.Shin W-S, Yamashita H, Hirose M. Multiple effects of haem binding on protease susceptibility of bovine serum albumin and a novel isolation procedure for its large fragment. Biochem J. 1994;304:81–86. doi: 10.1042/bj3040081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalley JW, Birss AJ. Albumin and hemalbumin degradation by Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:254–258. doi: 10.1111/j.1399-302X.1997.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 32.Steiner RF, Edelhoch H. The ultraviolet fluorescence of proteins 1. The influence of pH and temperature. Biochem Biophys Acta. 1963;66:341–355. doi: 10.1016/0006-3002(63)91203-4. [DOI] [PubMed] [Google Scholar]
- 33.Kang YN, Kim H, Shin WS, Woo G, Moon TW. Effect of disulfide bond reduction on bovine serum albumin-stabilized emulsion gel formed by microbial transglutaminase. J Food Sci. 2003;68:2215–2220. doi: 10.1111/j.1365-2621.2003.tb05749.x. [DOI] [Google Scholar]
- 34.Morita M, Wang HL. Association between oral malodour and adult periodontitis: a review. J Clin Periodontol. 2001;28:813–819. doi: 10.1034/j.1600-051x.2001.028009813.x. [DOI] [PubMed] [Google Scholar]
- 35.Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulphide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302X.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 36.Krespi YP, Shrime MG, Kacker A. The relationship between oral malodour and volatile sulphur compound-producing bacteria. Otolaryngol Head Neck Surg. 2006;135:671–676. doi: 10.1016/j.otohns.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Naoko O, Yotsuyanagi T, Chen D, Ono R, Ito S, Ikeda K. Disulfide bond cleavage of human serum albumin and alterations of its secondary structure by cis-diamminedichloroplatinum(II) Int J Pharm. 1992;85:39–44. doi: 10.1016/0378-5173(92)90131-K. [DOI] [Google Scholar]
- 38.Guo X, Yao H, Chen Z. Effect of heat, rutin and disulfide bond reduction on in vitro pepsin digestibility of Chinese tartary buckwheat protein factions. Food Chem. 2007;102:118–122. doi: 10.1016/j.foodchem.2006.04.039. [DOI] [Google Scholar]
- 39.Ter Steeg PF, Van der Hoeven JS, De Jong MH, Van Munster PJJ, Jansen MJH. Enrichment of subgingival microflora on human serum leading to accumulation of Bacteroides species. Peptostreptococci and Fusobacteria Antonie van Leeuwenhoek. 1987;53:261–271. doi: 10.1007/BF00393933. [DOI] [PubMed] [Google Scholar]
- 40.Kirakosyan G, Bagramyan K, Trchounian A. Redox sensing by Escherichia coli: effects of dithiothreitol, a redox reagent reducing disulphides, on bacterial growth. Biochem Biophys Res Commun. 2004;325:803–806. doi: 10.1016/j.bbrc.2004.10.119. [DOI] [PubMed] [Google Scholar]
- 41.Sogami M, Foster JF. Isomerization reaction of charcoal-defatted bovine plasma albumin. The N-F transition and acid expansion. Biochem. 1968;7:2172–2182. doi: 10.1021/bi00846a020. [DOI] [PubMed] [Google Scholar]
- 42.Katchalski E, Benjamin GS, Gross V. The availability of the disulfide bonds of human and bovine serum albumin and of bovine gamma-globulin to reduction by thioglycolic acid. J Am Chem Soc. 1957;79:4096–4099. doi: 10.1021/ja01572a034. [DOI] [Google Scholar]
- 43.Zunszain P, Ghuman J, Komatsu T, Tsuchida E, Curry S. Crystal structure analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct Biol. 2003;3:6. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan WT, Leu HH, Sutor RP, Muller-Eberhard U. Transfer of haem from haem-albumin to hemopexin. Biochim Biophys Acta. 1976;444:435–445. doi: 10.1016/0304-4165(76)90387-1. [DOI] [PubMed] [Google Scholar]
- 45.Grenier D, Imbeault S, Plamondon P, Grenier G, Nakayama K, Mayrand D. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect Immun. 2001;69:5166–5172. doi: 10.1128/IAI.69.8.5166-5172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller-Eberhard U, Javid J, Liem HH, Hanstein A, Hanna M. Plasma concentrations of hemopexin, haptoglobin and haem in patients with various hemolytic diseases. Blood. 1968;32:811–815. [PubMed] [Google Scholar]
- 47.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 48.Wagener FADTG, Eggert A, Boerman OC, Oyen WJG, Verhofstad A, Abraham NG, et al. Haem is a potent inducer of inflammation in mice and is counteracted by haem oxygenase. Blood. 2001;96:1802–1811. doi: 10.1182/blood.V98.6.1802. [DOI] [PubMed] [Google Scholar]
- 49.Reddi E, Ricchelli F, Jori G. Interactions of human serum albumin with haematoporphyrins and its Zn(2) + − and Fe(3) + −derivatives. Int J Pept Prot Res. 1981;18:402–408. doi: 10.1111/j.1399-3011.1981.tb02998.x. [DOI] [PubMed] [Google Scholar]
- 50.Potempa J, Manandhar SP. Interpain A. In: Rawlings ND ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. 3. Oxford: Elsevier; 2013. pp. 2163–2168. [Google Scholar]
- 51.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis: Isolation and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 52.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen IB, Enghild JJ, et al. Comparative properties of two cysteine proteinases (gingipain Rs), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 53.Gallagher WA, Elliott WB. The formation of pyridine haemochromogen. Biochem J. 1969;97:187–193. doi: 10.1042/bj0970187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smalley JW, Byrne DP, Birss AJ, Wojtowicz H, Sroka A, Potempa J, et al. HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis. PLoS One. 2011;6(2):e17182. doi: 10.1371/journal.pone.0017182. [DOI] [PMC free article] [PubMed] [Google Scholar]


