Abstract
In medulloblastoma, abnormal expression of pluripotency factors such as LIN28 and OCT4 has been correlated with poor patient survival. The miR-302/367 cluster has also been shown to control self-renewal and pluripotency in human embryonic stem cells and induced pluripotent stem cells, but there is limited, mostly correlational, information about these pluripotency-related miRNA in cancer. We evaluated whether aberrant expression of such miRNA could affect tumor cell behavior and stem-like traits, thereby contributing to the aggressiveness of medulloblastoma cells. Basal expression of primary and mature forms of miR-367 were detected in four human medulloblastoma cell lines and expression of the latter was found to be upregulated upon enforced expression of OCT4A. Transient overexpression of miR-367 significantly enhanced tumor features typically correlated with poor prognosis; namely, cell proliferation, 3-D tumor spheroid cell invasion and the ability to generate neurosphere-like structures enriched in CD133 expressing cells. A concurrent downregulation of the miR-367 cancer-related targets RYR3, ITGAV and RAB23, was also detected in miR-367-overexpressing cells. Overall, these findings support the pro-oncogenic activity of miR-367 in medulloblastoma and reveal a possible mechanism contributing to tumor aggressiveness, which could be further explored to improve patient stratification and treatment of this important type of pediatric brain cancer.
Keywords: Cancer stem cell, medulloblastoma, microRNA, miR-367, pluripotency
Medulloblastoma is the most common malignant brain tumor in children aged 4 or younger and one of the leading causes of morbidity and mortality related to childhood cancer.1,2 This embryonal tumor of the cerebellum, classified by the World Health Organization (WHO) as a grade IV tumor, is highly malignant and often spreads to other brain regions as well as to the spinal cord. The current treatment consisting of surgically removing as much tumor as possible, followed by radiation and chemotherapy, is not effective in approximately one-third of patients, who eventually succumb to the disease.
Although early age and metastatic spread at diagnosis are usually associated with poor survival,3 predictions of prognosis and response to standardized treatments based on clinical parameters have proven difficult. Medulloblastoma cells present a high degree of phenotypic and functional heterogeneity, which may result from stochastic genetic4 and/or epigenetic changes,5 as well as from interaction between cancer cells and the tumor niche.6 Based on molecular and genetic traits, the most recent consensus classification stratifies medulloblastoma in four distinct groups, two of which relate to developmental signaling pathways.7
The existence of highly tumorigenic cancer cells displaying stem cell properties in several types of malignant tumors adds another layer of complexity in understanding the mechanisms underlying inter-tumor and intra-tumor heterogeneity.8 Due to their self-renewal and differentiation capability, cancer stem cells may generate different progenies of cancer and stromal cells along tumor development. The continuous process of subclonal development and divergence fuels heterogenic aberrant tumor cell behavior, with important impacts on the clinical outcome of patients.9 Investigating stemness properties in cancer cells is, therefore, of great relevance to improve medulloblastoma treatment.
As in most embryonal tumors, medulloblastoma cells display primitive cell features, including aberrant function of SHH, NOTCH, WNT and TGFB1 signaling pathways, which are involved in normal neurodevelopment.10 Some of these abnormalities are thought to arise from prenatal mutations more likely occurring in neural stem or progenitor-like cells, favoring an increasing genomic instability in those cells.11 Despite the cell of origin, alterations conferring stemness to cells may increase their fitness and favor tumor development.
Certain early driver alterations may also be related to embryonic stem cell (ESC) traits, because these cells display unlimited growth potential and tumorigenicity.12 At the molecular level, cancer cells and ESC also share many similarities in their gene expression programs.13,14 Indeed, aberrant expression of ESC-related pluripotency factors such as LIN2815 and OCT4 have been reported in medulloblastoma specimens, with OCT4 expression being significantly correlated with shorter survival in either clinically high or average risk patients.16 Although the underlying alteration leading to such aberrant expression of pluripotency-related genes is not clear in medulloblastoma, the very low frequency of driver mutations in these tumors17 and the recent association of differential DNA methylation status with LIN28B misexpression18 indicate the contribution of chromosomal and epigenetic aberrations.
OCT4 is one of the four transcription factors capable of reprogramming somatic cells to pluripotency.19 More recently, overexpression of the miR-302/367 cluster has also been shown to induce pluripotency in somatic cells, without requirement of exogenous transcription factors, and with an efficiency two orders of magnitude higher than the standard OCT4/SOX2/KLF4/MYC-based methods.20 In fact, earlier studies had reported specific miRNA highly expressed by embryonic stem cells (ESC), with a critical role in controlling pluripotency and cell differentiation.21,22
Similar to what has been reported for transcription factors, aberrant expression of miRNA involved in pluripotency may also contribute to stemness traits in cancer cells. Yet, information about pluripotency-related miRNA and cancer aggressiveness is scarce in the literature and, thus far, no such studies have been reported for medulloblastoma. In this work, we found that miR-367 is upregulated by OCT4 in medulloblastoma cells and that transient overexpression of miR-367 enhanced cell proliferation, spheroid cell invasion, as well as generation of neurosphere-like structures in vitro, suggesting the pro-oncogenic activity of this microRNA originally related to pluripotency and ESC self-renewal.
Materials and Methods
Cell culture
Human medulloblastoma cell lines D283-Med (ATCC HTB-185), Daoy (ATCC HTB-186) and CHLA-01-Med (ATCC CRL-3021) were purchased directly from ATCC. Cell line USP-13-Med was obtained from HUG-CEL (Human Genome and Stem Cell Center, Universidade de São Paulo). CHLA-01-Med cells were maintained in DMEM–Nutrient Mixture F-12 (DMEM/F12) with 20 ng/mL human recombinant EGF, 20 ng/mL human recombinant basic FGF and B-27 Supplement (Invitrogen, Carlsbad, CA) to a final concentration of 2%. D283-Med, Daoy and USP-13-Med were cultivated in low DMEM supplemented with 10% FBS and 1% 100 μg/mL penicillin/streptomycin solution. The H9 cell line of human ESC (hESC) was kindly provided by LANCE (Laboratório Nacional de Células-tronco Embrionárias, UFRJ) and cultivated under standard conditions, as previously described.14 All cell lines were maintained at 37°C, 5% CO2.
Overexpression of miR-367
For miR-367 transient overexpression, cells at 50% confluence were transfected with mirVANA miRNA 367 mimic, inhibitor (Life Technologies, Carlsbad, CA) or an universal non-specific ncRNA control that does not target any sequence in the human transcriptome (DS-NC1; Integrated DNA Technologies, Coralville, IA), using Lipofectamine RNAiMAX (Life Technologies). Transfection was performed according to the manufacturer's instructions, using a final oligonucleotide concentration of 20 nM. Medium was replaced 24 h after transfection. Transient miR-367 overexpression efficiency was confirmed by analyzing the cellular levels of RYR3 and RAB23, two experimentally validated targets of miR-367.23
Protein analysis
After 48 h of cellular transfection, cells were permeabilized with the FIX & PERM Cell Fixation and Cell Permeabilization Kit (Life Technologies) and incubated with anti-RYR3 antibody (NPB1-70399; Novus Biologicals, Littleton, CO). The amount of RYR3+ cells was estimated by flow cytometry in the Guava easyCyte 5HT Flow Cytometer (Millipore, Billerica, MA). RAB23 protein levels were analyzed by western blotting under standard conditions. After protein transfer, PVDF membranes (GE Healthcare, Little Chalfont, UK) were blocked, incubated first with anti-RAB23 antibody (ABcam Cat. No.169491) and then with a goat anti-mouse HRP-conjugated secondary antibody. Protein bands were revealed using the Immobilon Western Chemiluminescent HRP Substrate (Millipore).
miRNA and gene expression analyses
Total RNA was extracted from tumors and hESC using the RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. MicroRNA were purified using the mirVana miRNA Isolation kit (Life Technologies), following the manufacturer's protocol. RNA concentration was determined by absorbance at 260 nm with NANO DROP (Qiagen). A total of 1 μg of RNA was reverse transcribed with the SuperScript III Reverse Transcriptase Kit (Invitrogen) according to the manufacturer's instructions. Quantitative PCR was performed in a 7500 Real-Time PCR System Thermal Cycler (Applied Biosystems, Carlsbad, CA). Expression of miR-367 and pri-miR-367 were quantitated using individual TaqMan MicroRNA Assays (Applied Biosystems). Small nucleolar RNA, C/D Box 58A (RNU58A), was used as endogenous control. Expression analysis of pluripotency-related genes was performed with Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies), using GAPDH as endogenous control. Reaction specificity was assessed by dissociation curve analysis. Primer sequences: SOX2 for: 5′-GCTACAGCATGATGCAGGACCA-3′, SOX2 rev: 5′-TCTGCGAGCTGGTCATGGAGTT-3′, NANOG for: 5′-CCTGAAGAAAACTATCCATCC-3′, NANOG rev: 5′-CCTTGTCTTCCTTTTTTGCGA-3′, OCT4A for: 5′-TCGCAAGCCCTCATTTCACCA-3′, OCT4A rev: 5′GGACTCCTCCGGGTTTTGCT-3′, GAPDH for: 5′-GCATCCTGGGCTACACTG-3′, GAPDH rev: 5′-CCACCACCCTGTTGCTGCTGTA-3′. RT-PCR quantification was based on linear regression analysis from standard curves with amplification efficiency ranging from 90% to 100%. Reactions were performed in triplicate. RNA from hESC was used as a positive control in all analyses.24
Cell cycle analysis
After 48 h of transfection with miR-367 mimic or non-specific control, cells were washed twice with PBS and fixed in cold 70% ethanol at 4°C for 1 h. Cells were then incubated with Guava Cell Cycle Reagent (Millipore) for 30 min, at room temperature, shielded from light, and analyzed using the Guava easyCyte 5HT Flow Cytometer (Millipore).
Immunofluorescence and mitotic index
After 48 h of transfection with either miR-367 mimic or non-specific control, cells were fixed with 3.7% formaldehyde for 30 min. For the mitotic index, cells were stained with 1 μg/mL DAPI for 2 min, washed in PBS and mounted on slides with VectaShield. Then, 1000 cells/slide were counted on three different slides for each experimental group. For cell morphology analysis, cells were submitted to immunofluorescence. After fixation, cells were treated with PBSAT (0.1% Triton X-100, 1% BSA) for 30 min and then incubated with anti-alpha tubulin (1:200 dilution; Sigma) and anti-pericentrin (1:800 dilution; Abcam) antibodies, for 1 h at room temperature. After washing with PBS three times, cell nuclei were stained with 1 μg/mL DAPI for 2 min and cells were mounted on slides with VectaShield.
EdU incorporation assay
Cells in the S phase of cell cycle were determined by EdU (5-ethynyl-2′-deoxyuridine) incorporation assay (Click-It EdU Alexa Fluor 488 Imaging Kit, Life Technologies), according to the manufacturer's protocol. EdU (10 μM) incorporation time was 30 min for Daoy cells and 45 min for CHLA-01-Med, D283-Med and USP-13-Med cells.
Neurosphere formation assay
Cells were transfected with miR-367 or non-specific control and plated 24 h later into 96-well low-attachment plates (Corning, New York, NY), at a density of 1 × 103 cells/mL in DMEM/F12 medium containing 20 ng/mL EGF, 20 ng/mL bFGF, 1× B-27 and 1× N2 (Invitrogen). Well-developed neurospheres (D283-Med and Daoy: ≥30 μm in diameter; CHLA-01-Med: ≥50 μm in diameter; USP-13-Med: ≥100 μm in diameter) were measured at day 4. D283-Med, Daoy and USP-13-Med neurospheres were dissociated with 0.05% trypsin/1 mM EDTA (Tryple, Life Technologies) and CHLA-01-Med neurospheres were dissociated with Cell Strainer Cap (Corning) to reach single cell suspension. For CHLA-01-Med, Daoy and D283-Med cells, 1 × 105 cells were labeled with CD133/1 PE antibody (AC133; Miltenyi Biotec, Cologne, Germany) for 40 min at 4°C. Cells were analyzed in the flow cytometer FACS Aria II with DIVA software (BD Biosciences, Franklin Lakes, NJ). USP-13-Med cells were labeled as described above, fixed (PFA 4% for 1 h) and permeabilized (0.1% Triton X-100 for 10 min) before analysis in the Guava easyCyte 5HT Flow Cytometer (Millipore). Cells displaying fluorescence higher than the basal fluorescence of unstained cells were deemed CD133-positive.
Apoptosis assay
CHLA-01-Med, USP-13-Med, D283-Med and Daoy cells were transfected with miR-367 or non-specific control and 24 h later treated with 9.2, 126.4, 50 and 3.7 μM of cisplatin, respectively, for 48 h. These concentrations correspond to the LD50 previously determined for each native cell line by dose-response curves. Apoptotic cells were evaluated using the Guava Nexin Annexin V Assay kit (Millipore), following the manufacturer's instructions, and samples were analyzed using the Guava easyCyte 5HT Flow Cytometer (Millipore).
Invasion assay
Tumor cell invasion activity was accessed using the Cultrex 96 Well 3-D Spheroid BME Cell Invasion Assay (Amsbio, Abingdon, UK). After 24 h of transfection with miR-367, miR-367 inhibitor or non-specific control, cells were plated into 96-well low-attachment round plate (Corning) at a density of 4 × 103 cells/mL, along with a specialized Spheroid Formation ECM to drive cell aggregation and spheroid formation. Spheroids were then embedded in an invasion matrix composed of basement membrane proteins that form a hydrogel network on which invasive cells can travel. Invading cells form spindle-like protrusions out from the spheroid core. The amount of these spindle-like protrusions and their length are proportional to the degree of cell invasion, which was visualized under a digital inverted microscope (EVOS Cell Imaging System; Life Technologies) and quantitated with the ImageJ software, according to the manufacturer's instructions. Daoy, D283-Med and CHLA-01-Med cells were not included in this analysis because they did not display intrinsic invasive properties under such experimental conditions (Suppl. Fig. S1).
Statistical analysis
All experiments were performed in triplicate and three independent experiments were carried out. Data were analyzed by anova and the Bonferroni post hoc test. Significance was established at the P ≤ 0.05 level. Results are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Results
Expression of miR-367 is upregulated by OCT4A in medulloblastoma cells
All medulloblastoma cell lines displayed detectable expression of the primary and mature forms of miR-367, although at significantly lower levels when compared with hESC (Fig.1a,b). Given the role of this microRNA in pluripotency and the aberrant OCT4 expression reported in aggressive medulloblastoma, a possible connection between miR-367 and OCT4 expression was evaluated. Medulloblastoma cells stably overexpressing OCT4A, encoding an OCT4 isoform with a well-known role in pluripotency, were obtained, reaching transcript levels similar to those found in hESC (Suppl. Fig. S2). Indeed, basal expression of miR-367 was significantly upregulated in the four medulloblastoma cell lines after enforced OCT4A expression (Fig.1c). Conversely, transient overexpression of miR-367 in medulloblastoma cells did not significantly increase OCT4A expression, nor the expression of other pluripotency-related genes encoding protein partners of OCT4A. Significant expression variation due to miR-367 was cell line-dependent (Fig.1d–f).
Figure 1.
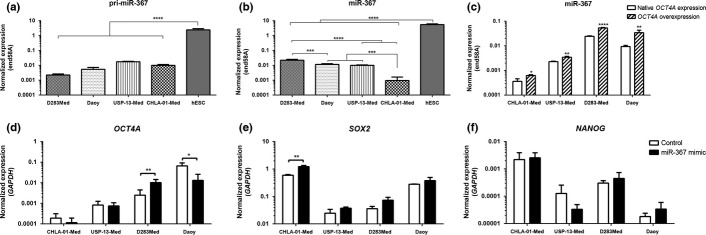
Expression profile of miR-367 and pluripotency factors in medulloblastoma cells. Expression of (a) pri-miR-367 and (b) mature miR-367 were detected in in four human medulloblastoma cell lines by real-time PCR, using RNU58A as endogenous control. Expression levels of non-coding RNA in tumor cells were compared with the levels in native pluripotent cells (hESC). (c) Upregulation of miR-367 in medulloblastoma cells stably overexpressing OCT4A. Expression of genes encoding the pluripotency factors (d) OCT4A, (e) SOX2 and (f) NANOG, 48 h post-transfection with either miR-367 mimic or non-specific control. Expression of these protein-coding genes was accessed by real-time PCR, using GAPDH as endogenous control. Significance level: *P < 0.05, **P < 0.01, ****P < 0.0001.
Overexpression of miR-367 increases medulloblastoma cell proliferation
Overexpression of miR-367 significantly increased the amount of viable cells in CHLA-01-Med and USP-13-Med cell line cultures up to 48 h after transfection. A similar tendency was observed for D283-Med and Daoy cells (Suppl. Fig. S3). Accordingly, cell cycle analysis of CHLA-01-Med, USP-13-Med and D283-Med, but not Daoy cells overexpressing miR-367 indicated a higher percentage of cells at S+G2/M phases, and lower percentage of cells at G0/G1, compared with control cells (Fig.2a).
Figure 2.
Overexpression of miR-367 affects cell cycle and proliferation of medulloblastoma cells. (a) Cell cycle analysis of CHLA-01-Med, USP-13-Med, D283Med and Daoy cells by flow cytometry. Cell proliferation was investigated in CHLA-01-Med, USP-13-Med, D283Med and Daoy cells by the mitotic index (b) and (c) EdU incorporation. (d) Representative immunofluorescence images of tumor cells marked with antibody against microtubules (red) and DAPI DNA staining (blue). (e) Quantification of apoptotic tumor cells induced by cisplatin. Data is expressed as percentage of total cell population analyzed. Significance level: *P < 0.05, **P < 0.01.
In agreement with this result, immunofluorescence analysis of CHLA-01-Med and USP-13-Med cell populations revealed a significant increase in the mitotic index and EdU incorporation in subsets of cells overexpressing miR-367, when compared with control cells displaying basal levels of miR-367 expression (Fig.2b,c). Again, a corresponding similar tendency was observed for D283-Med and Daoy cells. Interestingly, the immunofluorescence analysis also revealed morphological changes in USP-13-Med cells overexpressing miR-367. Control cell cultures were mainly comprised of fusiform cells with tapered and very thin ends, displaying a maximum length of 500 μm. After transfection with miR-367 mimic, these fusiform cells with long extensions were rarely observed, and most cells had a maximum length of 200 μm. Morphological analysis of CHLA-01-Med cells was difficult to perform because they naturally grow in suspension, forming tight cell clusters (Fig.2d).
In contrast, miR-367 overexpression did not significantly affect medulloblastoma cell apoptosis induced by treatment with cisplatin, except in Daoy cells (Fig.2e). Altogether, these results support that the increment previously observed in the population of viable cells due to miR-367 overexpression is likely due to a positive effect of miR-367 on cell proliferation, rather than on resistance to apoptosis.
Overexpression of miR-367 accentuates stem-like traits in medulloblastoma cells
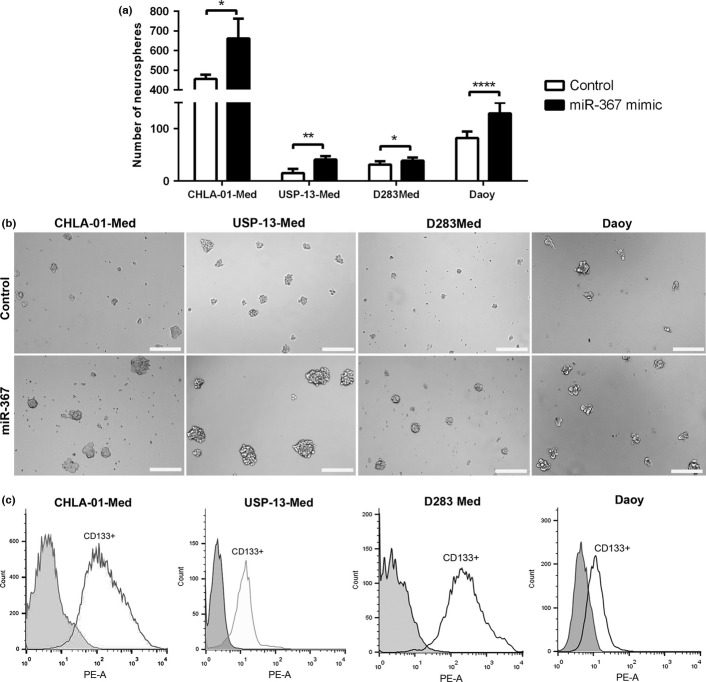
Medulloblastoma cells overexpressing miR-367 were more capable of generating neurosphere-like structures in vitro than control cells. The amount of neurospheres formed after 4 days in neural stem cell media was significantly higher in all medulloblastoma cell line cultures subjected to miR-367 mimic transfection, when compared with cultures of control cells (Fig.3a). Notably, neurospheres in cultures of CHLA-01-Med, USP-13-Med and D283-Med cells overexpressing miR-367 were not only more abundant but also more developed than their control counterparts, displaying a mean diameter of approximately 100 μm. Control neurospheres presented an average diameter of approximately 50 μm (Fig.3b). Despite being more numerous, neurospheres in cultures of Daoy cells overexpressing miR-367 were not oversized, displaying a general diameter comparable with that of neurospheres in control cultures. These neurospheres from all cell lines were highly enriched in cells expressing the neural stem cell marker CD133 (Fig.3c).
Figure 3.
Overexpression of miR-367 induces generation of medulloblastoma neurospheres. (a) Amount of neurospheres generated from medulloblastoma cells transfected with either miR-367 mimic or non-specific control, after 4 days of culture in neural stem cell media. (b) Representative images of CHLA-01-Med, USP-13-Med, D283-Med and Daoy neurospheres. (c) Proportion of CD133+ cells in medulloblastoma neurospheres, assessed by flow cytometry. CHLA-01-Med, USP-13-Med, D283Med and Daoy neurospheres were enriched in cells expressing CD133, with 91.7%, 90.3%, 87.4% and 48.2% of CD133+ cells, respectively. Scale bar: 200 μm. Significance level: *P < 0.05, **P < 0.01, ****P < 0.0001.
Stem cells, either normal or neoplastic, also exhibit a high ability to invade tissues. When grown as cell spheroids surrounded by a biological matrix, a condition that better mimics tumor behavior in vivo, overexpression of miR-367 significantly enhanced invasion of medulloblastoma cells. Invasive cells formed spindle-like protrusions in the invasion matrix, which were more abundant and bigger in length after miR-367 overexpression (Fig.4a). Consequently, the overall area of these spindle-like protrusions was significantly greater in spheroids of cells overexpressing miR-367, compared with spheroids of control cells. Such enhancement on cell invasion was reversed by transfection with miR-367 inhibitor (Fig.4b).
Figure 4.
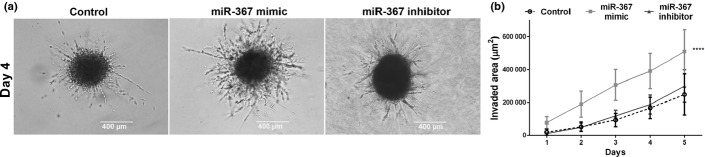
Overexpression of miR-367 in medulloblastoma cells enhances 3-D cell invasion. (a) Representative images of medulloblastoma spheroids displaying arboreal protrusions formed by cells invading the surrounding hydrogel matrix. Scale bar: 400 μm. (b) Kinetics of 3-D cell invasion of USP-13-Med spheroids overexpressing miR-367 (light grey line) and respective controls (miR-367 inhibitor and non-specific control). Significance level: ****P < 0.0001.
Overexpression of miR-367 downregulates cancer-related targets in medulloblastoma cells
The Ryanodine receptor 3 (RYR3) and Ras-related protein 23 (RAB23) transcripts are examples of experimentally validated targets of miR-367.23 Other putative miR-367 targets predicted by in silico analysis, but not experimentally validated, include the Integrin alpha-V (ITGAV) transcript. The respective encoded proteins are known to be involved in cancer. Consistently, both RYR3 and RAB23 protein levels were significantly reduced in medulloblastoma cells after 48 h of initial transfection with miR-367 mimic (Fig.5). Likewise, expression of ITGAV was also found significantly inhibited in medulloblastoma cells overexpressing miR-367 (Suppl. Fig. S4), reflecting a downregulation not necessarily resulting from direct miR-367 targeting.
Figure 5.
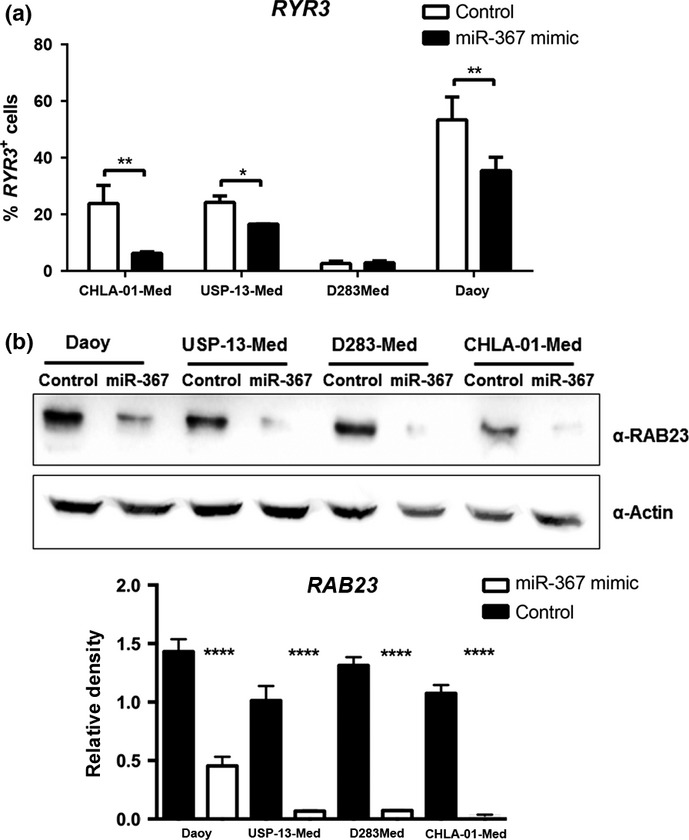
Downregulation of miR-367 cancer-related targets in medulloblastoma cells. (a) Relative amount of cells with positive expression of RYR3, assessed by flow cytometry 48 h post-transfection with either miR-367 mimic or non-specific control. D283-Med cells were negative for RYR3 expression. (b) RAB23 protein levels in medulloblastoma cells, assessed by western blotting 48 h post-transfection with either miR-367 mimic or non-specific control. Respective blot quantification is presented as a bar graph. Significance level: *P < 0.05, **P < 0.01, ****P < 0.0001.
Discussion
The miR-367 is primarily known as an ESC-specific microRNA regulating self-renewal and pluripotency.20,24 Some studies suggest that miR-302/367-mediated reprogramming of somatic cells to pluripotency poses minimum oncogenic risks due to self-regulating anti-tumor mechanisms.24 In agreement with this suggestion, overexpression of the miR-302/367cluster in two human cervical cancer cell lines inhibited cell proliferation and tumor formation.25 Conversely, ESC-specific miRNA, including miR-302/367, have also been shown to positively regulate cell proliferation by promoting G1/S transition in ESC,26,27 indicating that miR-302/367 effects may vary according to the cellular context.
The fact that overexpression of miR-302, but not miR-367, is required to fully reprogram somatic cells to a competent pluripotent state13 indicates functional differences between these two related miRNA, strengthening this point. Thus far, little is known about the specific function of each individual member of this miRNA cluster.
High miR-367 expression has been associated with poor prognosis of cancer patients with Wilm's tumor,28 ependymomas29 and nonsmall cell lung cancer.30 More recently, high levels of circulating miR-367 have been reported in the serum of patients with testicular germ cell cancers, in particular non-seminomas, with advanced local stage and metastasis.31 Despite this clinical evidence, there are few functional studies addressing the direct involvement of miR-367 in tumor aggressiveness.
The positive effects on cell proliferation and neurosphere formation found after overexpression of miR-367 in medulloblastoma cells are in agreement with the previous clinical findings and indicate a possible pro-oncogenic role for this miRNA. Noteworthy, a previous study of embryonal brain tumors, including medulloblastoma specimens, reported that the ability of cancer cells to generate neurospheres ex vivo was significantly correlated with poor disease-free survival and poor overall survival.32 Moreover, the sphere formation assay has been validated as an alternative to xenograft models in determining brain tumor stem-like cell potency and frequency.33 The enrichment of medulloblastoma cell population with self-renewal capacity and stem cell-like phenotype after miR-367 overexpression is of particular interest, because cancer cells displaying similar stem cell properties are known to be highly tumorigenic and to be directly involved in tumor progression.34 Interestingly, a large-scale analysis of miRNA expression in human glioma specimens revealed an expression pattern resembling the miRNA expression profile of stem cells and neuroprogenitors, in which miR-367 was found upregulated.35
The increased medulloblastoma cell invasiveness due to miR-367 overexpression is also in concert with the previous findings, because part of the stem-like cancer cell association with poor prognosis is attributed to the higher invasive growth and metastatic potential of these cells.36–38 Although rare in medulloblastoma, the presence of metastasis at diagnosis, particularly in the neural axis, is a classical hallmark of poor survival.3 However, a very recent study reported that overexpression of miR-367 in two gastric cancer cell lines, HS746T and SGC-7901, had an opposite effect, inhibiting cellular migration and invasion.39 Nonetheless, the same study also found that miR-367 is downregulated in gastric tumor tissues, suggesting a distinct involvement of miR-367 in this particular type of cancer.
Several oncogenes have known effects on both cell proliferation and migration/invasion, which may be stimulated according to the extracellular signals predominating in the tumor microenvironment. Cell migration is highly stimulated by the extracellular matrix surrounding cells, an effect that is well known for neural crest cells during embryonic neurogenesis.40 This cellular environment was mimicked in the 3-D invasion assay used in the present study. Interestingly, by-products of the extracellular matrix degradation can also stimulate proliferation of progenitor cells,41 an effect that might contribute to metastatic colonization in the context of aggressive cancers.
We have previously reported that expression of the pluripotency factor OCT4 is correlated with poor prognosis in medulloblastoma,16 a phenomenon that was later reported to occur in other types of malignant tumors.42–46 Here, we found that ectopic OCT4A expression in medulloblastoma cells upregulates miR-367. It is known that miR-302/367 interact with specific transcription factors (OCT4, SOX2 and NANOG) leading to an auto-regulatory positive loop that induces and sustain pluripotency.47,48 Although upregulation of either OCT4 or SOX2 was observed after miR-367 overexpression in medulloblastoma cells, this effect was cell line-dependent, suggesting that the common effects on cell behavior towards a more aggressive phenotype were not associated with a gain in pluripotency.
Overexpression of miR-367 in medulloblastoma cells, as expected, downregulated RYR3, which is one of the few experimentally verified targets of miR-367.23 Many studies have proposed anti-tumoral effects for RyRs receptors.49 Using next-generation sequencing, three independent studies identified a total of 2102 unique genes as somatically mutated in medulloblastoma patient samples. Of these, only 15 were reported as mutated in all three studies. This restricted list of commonly mutated genes includes RYR3.50 RYR3 inactivation due to aberrant DNA methylation was also reported in acute lymphoblastic leukemia.51 In animal models of tumorigenesis, RyRs activation by caffeine inhibited breast tumor metastasis52 as well as the appearance of UV-induced skin cancer.53
Overexpression of miR-367 in medulloblastoma cells also resulted in decreased transcript levels of RAB23, a recently experimentally validated target of miR-367.39 Members of the RAB family have been reported either as oncoproteins or tumor suppressors.54 RAB23 overexpression enhanced migration and invasion of human breast cancer cells.55 In gliomas, the RAB family was associated with poor clinical outcome.56 Nevertheless, RAB23 is deregulated in many cancers and has been identified as an antagonist of the SHH pathway.57 The SHH pathway is crucial during neurodevelopment and hyperactivation of this signaling pathway characterizes a particular subgroup of medulloblastoma.1 Although downregulation of RYR3 and RAB23 targets supported the effects of miR-367 overexpression observed in medulloblastoma cells, the downstream mechanisms underlying these miR-367-directed events remain to be elucidated.
In sum, ectopic expression of ESC-related miR-367 in medulloblastoma cells significantly enhanced their proliferation, spheroid invasive growth and ability to generate neurospheres enriched in cells with a stem-like phenotype, consistent with oncogenic activity. Upregulation of miR-367 occurred upon activation of another ESC-related factor, OCT4A, which had been previously associated with poor survival of medulloblastoma patients. Overall, these findings reveal a possible mechanism contributing to the aggressiveness of this important type of pediatric brain cancer, which could be further explored therapeutically.
Acknowledgments
The authors thank Dr Felippe Lourenço Claro and Nikon do Brasil for helping with the immunofluorescence imaging, and Dr Marcia Santos for helping with the protein assays. This work was supported by funds from FAPESP-CEPID, CNPq, INCT-CETGEN and FINEP-CTC. CK, PBGS, COR and BAC were recipients of FAPESP.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1. 3-D invasion assay with Daoy, D283-Med and CHLA-01-Med cells.
Fig. S2. Overexpression of OCT4A in medulloblastoma cell lines.
Fig. S3. Amount of viable medulloblastoma cells after miR-367 overexpression.
Fig. S4. Downregulation of ITGAV after miR-367 overexpression in medulloblastoma cells.
References
- Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12:871–84. doi: 10.1586/ern.12.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber NU, Mynarek M, von Hoff K, Friedrich C, Resch A, Rutkowski S. Recent developments and current concepts in medulloblastoma. Cancer Treat Rev. 2014;40:356–65. doi: 10.1016/j.ctrv.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–9. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoranjan B, Venugopal C, McFarlane N, et al. Medulloblastoma stem cells: modeling tumor heterogeneity. Cancer Lett. 2013;338:23–31. doi: 10.1016/j.canlet.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodini CO, Suzuki DE, Nakahata AM, et al. Aberrant signaling pathways in medulloblastomas: a stem cell connection. Arq Neuropsiquiatr. 2010;68:947–52. doi: 10.1590/s0004-282x2010000600021. [DOI] [PubMed] [Google Scholar]
- Marshall GM, Carter DR, Cheung BB, et al. The prenatal origins of cancer. Nat Rev Cancer. 2014;14:277–89. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki DE, Nakahata AM, Okamoto OK. Knockdown of E2F2 inhibits tumorigenicity, but preserves stemness of human embryonic stem cells. Stem Cells Dev. 2014;23:1266–74. doi: 10.1089/scd.2013.0592. [DOI] [PubMed] [Google Scholar]
- Picard D, Miller S, Hawkins CE, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 2012;13:838–48. doi: 10.1016/S1470-2045(12)70257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodini CO, Suzuki DE, Saba-Silva N, et al. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J Neurooncol. 2012;106:72–9. doi: 10.1007/s11060-011-0647-9. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovestadt V, Jones DTW, Picelli S, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu Y, Song F, et al. Functional SNP in the microRNA-367 binding site in the 3′UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci U S A. 2011;108:13653–8. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Deng JH, Deng Q, Ying S-Y. A novel role of miR-302/367 in reprogramming. Biochem Biophys Res Commun. 2012;417:11–6. doi: 10.1016/j.bbrc.2011.11.058. [DOI] [PubMed] [Google Scholar]
- Cai N, Wang Y-D, Zheng P-S. The microRNA-302-367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA. 2013;19:85–95. doi: 10.1261/rna.035295.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Choi E, Hwang K-C. MicroRNAs as novel regulators of atem cell fate. World J Stem Cells. 2013;5:172–87. doi: 10.4252/wjsc.v5.i4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Bryan K, Williams R, et al. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PLoS One. 2013;8(1):e53417. doi: 10.1371/journal.pone.0053417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF, Bischof JM, Vanin EF, et al. Identification of microRNAs as potential prognostic markers in ependymoma. PLoS One. 2011;6:e25114. doi: 10.1371/journal.pone.0025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campayo M, Navarro A, Viñolas N, et al. Low miR-145 and high miR-367 are associated with unfavourable prognosis in resected nonsmall cell lung cancer. Eur Respir J. 2013;41:1172–8. doi: 10.1183/09031936.00048712. [DOI] [PubMed] [Google Scholar]
- Syring I, Bartels J, Holdenrieder S, Kristiansen G, Müller SC, Ellinger J. Circulating serum microRNA (miR-367-3p, miR-371a-3p, miR-372-3p, miR-373-3p) as biomarkers for patients with testicular germ cell cancers. J Urol. 2015;193:331–7. doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Panosyan EH, Laks DR, Masterman-Smith M, et al. Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatr Blood Cancer. 2010;55:644–51. doi: 10.1002/pbc.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal C, Li N, Wang X, et al. Bmi1 marks intermediate precursors during differentiation of human brain tumor initiating cells. Stem Cell Res. 2012;8:141–53. doi: 10.1016/j.scr.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- Lavon I, Zrihan D, Granit A, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12:422–33. doi: 10.1093/neuonc/nop061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Tirino V, Desiderio V, Paino F, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27:13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Lu P, Zhang H, et al. Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 2014;16:408. doi: 10.1186/s13058-014-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Z, Dedong H, Xiangjie F, Hongwei X, Qinghui Y. The microRNA-367 inhibits the invasion and metastasis of gastric cancer by directly repressing Rab23. Genet Test Mol Biomarkers. 2015;19:69–74. doi: 10.1089/gtmb.2014.0210. [DOI] [PubMed] [Google Scholar]
- Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mech Dev. 2000;95:3–21. doi: 10.1016/s0925-4773(00)00365-8. [DOI] [PubMed] [Google Scholar]
- Reing JE, Zhang L, Myers-Irvin J, et al. Degradation products of extracellular matrix affect cell migration and proliferation. Tissue Eng Part A. 2009;15:605–14. doi: 10.1089/ten.tea.2007.0425. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Arao T, Togashi Y, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2013;34:199–208. doi: 10.1038/onc.2013.547. [DOI] [PubMed] [Google Scholar]
- Kim S-H, Singh SV. Mammary cancer chemoprevention by withaferin A is accompanied by in vivo suppression of self-renewal of cancer stem cells. Cancer Prev Res (Phila) 2014;7:738–47. doi: 10.1158/1940-6207.CAPR-13-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher BA. Targets in small cell lung cancer. Biochem Pharmacol. 2014;87:211–9. doi: 10.1016/j.bcp.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Tsai L-L, Hu F-W, Lee S-S, Yu C-H, Yu C-C, Chang Y-C. Oct4 mediates tumor initiating properties in oral squamous cell carcinomas through the regulation of epithelial-mesenchymal transition. PLoS One. 2014;9:e87207. doi: 10.1371/journal.pone.0087207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wu X, Wang J, Huang J. Cancer stem cell theory: therapeutic implications for nanomedicine. Int J Nanomedicine. 2013;8:899–908. doi: 10.2147/IJN.S38641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-L, Chang DC, Lin C-H, Ying S-Y, Leu D, Wu DTS. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–65. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-danso F, Trivedi CM, Juhr D, et al. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Jones DTW, Kool M, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–34. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund J, Milani L, Lundmark A, Lönnerholm G, Syvänen A-C. DNA methylation analysis of bone marrow cells at diagnosis of acute lymphoblastic leukemia and at remission. PLoS One. 2012;7:e34513. doi: 10.1371/journal.pone.0034513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rouse J, Lukes L, et al. Caffeine suppresses metastasis in a transgenic mouse model: a prototype molecule for prophylaxis of metastasis. Clin Exp Metastasis. 2004;21:719–35. doi: 10.1007/s10585-004-8251-4. [DOI] [PubMed] [Google Scholar]
- Lu Y-P, Lou Y-R, Xie J-G, et al. Caffeine and caffeine sodium benzoate have a sunscreen effect, enhance UVB-induced apoptosis, and inhibit UVB-induced skin carcinogenesis in SKH-1 mice. Carcinogenesis. 2007;28:199–206. doi: 10.1093/carcin/bgl112. [DOI] [PubMed] [Google Scholar]
- Jordens I, Marsman M, Kuijl C, Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–7. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Y, Jian Q, Li J, Chi S. Role of Rab23 in invasion and migration of human breast cancer Bcap-37 cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29:813–7. [PubMed] [Google Scholar]
- Liu Q, Tang H, Liu X, et al. miR-200b as a prognostic factor targets multiple members of RAB family in glioma. Med Oncol (Internet) 2014;31:859. doi: 10.1007/s12032-014-0859-x. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–8. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. 3-D invasion assay with Daoy, D283-Med and CHLA-01-Med cells.
Fig. S2. Overexpression of OCT4A in medulloblastoma cell lines.
Fig. S3. Amount of viable medulloblastoma cells after miR-367 overexpression.
Fig. S4. Downregulation of ITGAV after miR-367 overexpression in medulloblastoma cells.