Abstract
Nasopharyngeal carcinoma (NPC) is highly incident in southern China, where 40% of world's new cases arise each year. Detection of Epstein–Barr virus (EBV) DNA load in nasopharyngeal (NP) brush/swab samples has gradually been established as a method for diagnosis of NPC. However, its applicable value in NPC diagnosis has never been investigated in southern China. It is important to explore whether such a test could be applicable to our local population. A total of 245 consecutive participants undergoing NP brushing examination were recruited to obtain the NP brushing samples in this study. Quantitative PCR assays were used to obtain the EBV DNA load. Mann–Whitney, ANOVA and receiver operating characteristic tests were used to analyze its diagnostic value. NP brushing samples from NPC patients showed extremely high levels of EBV DNA load (mean = 46360 copy/ng DNA) compared to its expression from non-NPC control (mean = 28 copy/ng DNA) and high-risk control (mean = 50 copy/ng DNA) groups. It produced 96% sensitivity and 97% specificity, at the COV = 225 copy/ng DNA. Furthermore, EBV DNA load could reflect disease progress. Our data showed a better performance of EBV DNA load in NP brushing samples compared with an initial biopsy, immunoglobulin A (IgA) antibody titers to viral capsid antigen in serum and EBV DNA load in plasma. Detection of EBV DNA load in NP brushing samples could be an effective supplement for NPC diagnosis. Being minimally invasive and low cost, NP brush sampling combined with EBV DNA detection demonstrates great potential for screening high-risk populations for NPC.
Keywords: Diagnosis, disease progress, Epstein–Barr virus DNA load, nasopharyngeal brush, nasopharyngeal carcinoma
Nasopharyngeal carcinoma (NPC) is a highly invasive and metastatic cancer that is widely prevalent in southern China. Although the overall survival rate is approximately 90% for patients diagnosed at an early clinical stage, unfortunately, most patients are diagnosed with advanced-stage disease at their first visit, and the survival rate decreases to <50%.1 Developing a simple and reliable method facilitating diagnosis of NPC is of great significance to increase patients’ survival in clinical practice.2
Previous studies have demonstrated that genetic susceptibility, endemic environmental factors and Epstein–Barr virus (EBV) infection constitute the three etiological contributors to NPC.3 Although EBV is ubiquitous worldwide, it is so closely linked to NPC that nearly 100% of tumor lesions possess EBV genomes in undifferentiated type NPC (WHO type III), the predominant type of NPC in endemic areas. Moreover, squamous cell (WHO type I) and non-keratinizing (WHO type II) NPC are also frequently associated with EBV in endemic areas.4 The infection of epithelial cells by EBV is a typical characteristic of NPC in high-risk areas.5,6
At present, the gold standard NPC diagnosis depends on a biopsy of the suspected tumor site with the aid of nasopharyngeal (NP) endoscopy, followed by histopathologic assessment, and even an evaluation of EBV involvement, including assessing expressions of the EBV encoded small RNA (EBER), latent membrane protein 1 (LMP1) and EBV nuclear antigen 1 (EBNA1). However, NP biopsy sampling is highly invasive and painful, and can lead to several complications.7 Image examination is another useful method for determining the disease progression. However, imaging techniques such as computed tomography or magnetic resonance (MR) are costly and time-consuming, making them unsuitable for use in repeated examination in suspected individuals.
Driven by the demand for painless and affordable diagnosis and screening of NPC, substantial efforts have been made to develop novel approaches for diagnosis. Considering the strong association between EBV infection and NPC, EBV-related markers have been the primary detection targets. Due to the advantages of blood sampling, antibody titers against EBV serum antigens, including viral capsid antigen (VCA), EBNA1 and early antigen (EA), have frequently been used to assist in the diagnosis of NPC in southern China. However, the results of these serological tests alone have proven to be insufficient to accurately diagnose NPC.8
Since the late 1990s, detection of EBV DNA load in plasma or serum has gradually been established as a method for diagnosis of NPC.9–12 Although EBV DNA testing has been proposed in clinical practice to aid in the diagnosis of NPC, EBV DNA appears to be of limited value in diagnosing NPC patients with early clinical stage disease and local recurrence,13 likely due to the low levels of DNA fragments released from small tumors into the blood.
In the meantime, NP brushing/swab samples are also used for qualitative and quantitative detection of EBV DNA load, because NP brush sampling can accurately obtain samples from the nasopharynx in a minimally invasive fashion. High sensitivity (87.3–96.4%) and specificity (90.0–98.4%) in NPC diagnosis have been observed in patients from Hong Kong, Taiwan, Canada and Indonesia, respectively.14–18 NP brush/swab sampling combined with EBV DNA detection brings hope of diagnosis for NPC patients with early stage and locally recurrent disease, due to its original lesion at the site of the nasopharynx. However, the highest prevalence of NPC is observed in southern China, contributing 40% of world's new cases every year. In our local high-risk area, the potential value of detecting EBV DNA in NP brushing samples for NPC's diagnosis has yet to be investigated. It is necessary and meaningful to explore whether such test can be applicable to our local population, especially to test its use in diagnosing NPC with early stage as well as locally recurrent disease.
Materials and Methods
Clinical specimens
Between January 2013 and October 2013, 245 participants undergoing NP brush sampling were recruited in this study. A total of 163 NP brushing samples were collected consecutively from participants when they underwent NP biopsy at Sun Yat-Sen University Cancer Center (SYSUCC). Among the total participants, 129 patients exhibited biopsy-proven NPC and 34 patients (non-NPC control) were diagnosed with chronic nasopharyngitis (n = 26) or other non-NPC tumors (n = 8). Meanwhile, NP brushing samples from healthy individuals (n = 82) were collected as high-risk controls from Sihui City, a high-risk area of Guangdong Province, China. For the present study, 3 mL of anticoagulant blood and 3 mL of non-anticoagulant blood were collected from participants at SYSUCC; 3 mL of anticoagulant blood was collected from the high-risk controls in Sihui City. This study was approved by the Human Ethics Committee of the SYSUCC, and all participants provided informed consent.
Sampling procedures
Nasopharyngeal brush sampling was conducted under the guidance of endoscopy by experienced specialists and resident trainees. An endoscope was used to evaluate the entire nasopharynx, and images were captured at the site of suspicious tumors. Before biopsy, an NP brush (Copan Diagnostics, Murrieta, CA, USA) was inserted via the nose until the NP cavity was reached. Subsequently, the brush was rotated several times over the NP epithelium at the site of the suspected lesion and quickly removed. Immediately after sampling, the brush tip (1.5 cm) was cut and placed in 1 mL of RNAlater (Invitrogen, Carlsbad, CA, USA) and stored at −80°C until use. For healthy participants, NP brush sampling was conducted at the lateral pharynx because this was the most common site for disease occurrence.
DNA extraction and quantitative PCR
Total DNA from NP brushing samples were extracted using an automated workstation (Chemagic Star; Hamilton Robotic, Bonaduz, GR, Switzerland) following the protocol recommended by the manufacturer. A final elution volume of 150 μL was obtained after the extraction procedure was complete. DNA concentration was quantified using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Waltham, MA, USA). Our real-time quantitative PCR was based on a previously mature fluorogenic PCR reaction system.10 In this system, amplification primers targeting BamHI-W region of EBV DNA genome (5′-CCCAACACTCCACCACACC-3′, 5′-TCTTAGGAGCTGTCCGAGGG-3′) and a dual-labeled hybridization probe (5′-FAM-CACACACTACACACACCCACCCGTCTC-TAMRA-3′) were included. β-globin gene was used as a reference for quality control. The two primers’ sequences were: 5′-GTGCACCTGACTCCTGAGGAGA-3′ and 5′-CCTTGATACCAACCTGCCCAG-3′ and the probe sequence was: 5′-FAM-AAGGTGAACGTGGATGAAGTTGGTGG-TAMRA-3′. Data on the amplified fragment of EBV DNA (76 bp) and globin DNA (102 bp) in NP brushing samples are shown in Supplementary Figure S1. The standard samples ladders (103, 104, 105, 106 and 107 copies/μL) was used to obtain the standard curve. Each PCR reaction was set up in a reaction volume of 8 μL including 4 μL PCR master mix, 1 μL primers, 0.2 μL probe, 0.8 μL water and 2 μL DNA template. Thermal cycling was initiated with a denaturation step of 5 min at 95°C, and then 45 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 15 s were carried out. The EBV DNA and β-globin levels in brushing samples were expressed as copy/ng DNA.
Viral capsid antigen-IgA antibody titers in blood specimens
Detection of VCA-IgA antibody titers in serum was conducted at tge Department of Clinical Laboratory, SYSUCC. The VCA-IgA titer was measured using a commercial kit (Zhongshan Bio-Tech, Zhongshan City, China) based on standard techniques, which have been described previously.19
Epstein–Barr virus DNA load in blood specimens
Detection of EBV DNA load in plasma was conducted at the Department of Molecular Diagnosis, SUSUCC. DNA from plasma samples was extracted using the Qiamp Blood Kit (Qiagen, Hilden, Germany) following the protocol recommended by the manufacturer. The EBV DNA load was measured by real-time Q-PCR based on a mature system, which has been described previously.20
Statistical analyses
The Mann–Whitney test (two-tailed) was used to compare the nonparametric variables between NPC and control groups. A one-way ANOVA test was used to compare the difference in multiple groups. P < 0.05 was considered significant. Receiver operating characteristic (ROC) curves were used to evaluate the diagnostic value. We used the curves to select the cut-off value (COV) with maximum sensitivity and specificity. The statistical analyses were performed using SPSS 16 software.
Results
Characteristics of the study population
A total of 245 NP brushing samples were collected from different participants. In the NPC group, it was noted that all of the patients were classified as WHO type 3. TNM staging was performed for the patients using the Chinese 2008 staging system. The information for cancer stage was evaluated by clinical doctors based on the comprehensive results of MR, histopathology and clinical symptoms. Some patients were biopsy-diagnosed with NPC in our Cancer Center but subsequently moved to other hospitals for further diagnosis and treatment; for these patients, results such as MR were not collected in our Cancer Center. Therefore, the staging information for 47 patients was lost in this study, and only histopathology information was obtained. The control group consisted of 82 healthy individuals and 34 clinical controls. The detailed characteristics of the study population are presented in Table1.
Table 1.
Characteristics of patient and control groups
| Characteristic | NPC patient | Non-NPC control | High-risk control |
|---|---|---|---|
| Number | 129 | 34 | 82 |
| Age (years) | |||
| Mean (minimum, maximum) | 46 (18, 73) | 48 (25, 67) | 50 (20, 68) |
| Gender | |||
| Male | 95 | 23 | 53 |
| Female | 34 | 11 | 29 |
| VCA-IgA titer | |||
| Negativity | 4 | 21 | 46 |
| 1:10–1:40 | 10 | 5 | 12 |
| ≥1:80 | 110 | 6 | 19 |
| No data | 5 | 2 | 5 |
| EBV DNA load in plasma | |||
| Negativity | 20 | 20 | |
| 1–1000 copy/mL | 4 | 0 | |
| ≥1000 copy/mL | 76 | 3 | |
| No data | 29 | 11 | |
| Cancer stage | |||
| Stage I/II | 11 | ||
| Stage III | 38 | ||
| Stage IV | 33 | ||
| Unknown | 47 | ||
| Recurrence | |||
| Yes | 3 | ||
| No | 126 | ||
| T stage | |||
| T1/T2/T3/T4 | 5/8/45/24 | ||
| N stage | |||
| N0/N1/N2/N3 | 5/35/30/12 | ||
| M stage | |||
| M0/M1 | 78/4 | ||
| Histopathology | |||
| WHO 3 | 129 | ||
EBV, Epstein–Barr virus; NPC, nasopharyngeal carcinoma.
High Epstein–Barr virus DNA loads were detected in nasopharyngeal brushing samples from nasopharyngeal carcinoma patients
To demonstrate that NP brush sampling was a reliable approach to directly collect cells from the nasopharynx, β-globin used as the internal reference was first detected. Highly consistent expression of β-globin copies in NP brush samples among NPC patients and controls was observed. The range of β-globin copies in the NPC group was from 3670 to 37 000, with mean = 17 800, while the range was from 4192 to 33 480, with mean = 17 400, in the non-NPC control and from 6000 to 32 940, with mean = 18 670, in the high-risk control. There was no significant difference between NPC patients and controls (P = 0.83; Fig.1a).
Figure 1.
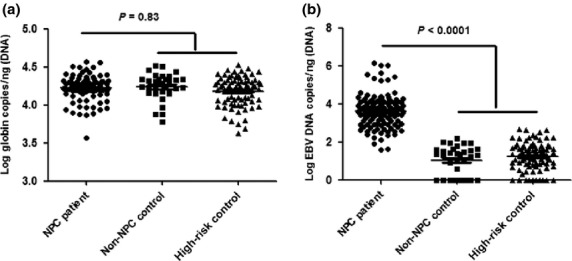
(a) Quantitative PCR test showed a highly consistent expression of β-globin copies in nasopharyngeal (NP) brushing samples between nasopharyngeal carcinoma (NPC) patients and controls, indicating that NP brush sampling was a reliable method. (b) A significant higher Epstein–Barr virus (EBV) DNA load was observed in NPC patients (P < 0.0001), compared to controls. No significant difference was observed between non-NPC control and high-risk control groups.
All NP brushing samples from NPC patients were positive for EBV DNA, with extremely high DNA loads (mean = 46 360, range from 40 to 1 395 000). Although EBV DNA was detectable in 70.6% of the non-NPC control group (mean = 28, range from 0 to 158) and in 87.8% of the high-risk control group (mean = 50, range from 0 to 469), the load value was very low. A significantly higher EBV DNA load was observed in NPC patients (P < 0.0001), compared to controls. In contrast, no significant difference was observed between non-NPC control and high-risk control (Fig.1b).
Epstein–Barr virus DNA load produced high sensitivity and specificity in diagnosing nasopharyngeal carcinoma
To calculate the diagnostic efficiency of EBV DNA load in NP brushing samples, the COV was defined by the mean expression of EBV DNA load in the control group plus the standard deviations as previously described,17 or by the ROC curve. The COV were used to determine the sensitivity, specificity, positivity and negative predictive values, as indicated in Table2. Similar results were obtained using these two methods. However, at COV = 225 copy/ng DNA, a higher sensitivity (96%) and specificity (97%) was obtained, compared to the COV = 260 copy/ng DNA (94% sensitivity and 97% specificity). Under this condition, only 4 controls from high-risk group showed a slightly elevated EBV DNA load above the COV (EBV DNA load = 322, 407, 418 and 469, respectively). However, there were still 5 NPC cases with EBV DNA load below the COV (EBV DNA load = 40, 41, 77, 122 and 164, respectively).
Table 2.
Sensitivity, specificity, PPV, NPV of EBV DNA load at different COV
| COV1 | COV2 | |||
|---|---|---|---|---|
| Below | Above | Below | Above | |
| NPC patients (n = 129) | 8 | 121 | 5 | 124 |
| Controls (n = 116) | 112 | 4 | 112 | 4 |
| Sensitivity (%) | 94 | 96 | ||
| Specificity (%) | 97 | 97 | ||
| PPV (%) | 97 | 97 | ||
| NPV (%) | 93 | 96 | ||
COV1 = mean + 3 standard deviations (260 copy/ng DNA). COV2 = directly determined by ROC (225 copy/ng DNA). EBV, Epstein–Barr virus; NPV, negative predictive value; PPV, positive predictive value.
Epstein–Barr virus DNA load in different subgroup
Further analysis revealed that the EBV DNA load in NP brushing samples of non-NPC control was significantly lower than that in stage I/II (Fig.2a), T1/T2 (Fig.2b), N0 (Fig.2c) and M0 (Fig.2d) diseases (P < 0.0001). Stage I/II and T1/2 disease also had a significantly lower level of EBV DNA load in NP brushing samples compared with advanced disease (P < 0.001). In stage I/II disease, 82% of cases (9 out of 11) had EBV DNA loads above COV, while 80% of cases (4 out of 5) in the T1 subgroup and 88% of cases (7 out of 8) in the T2 subgroup had EBV DNA loads above the COV. No significant difference was observed between node negative and positive cases and between metastasis negative and positive cases.
Figure 2.
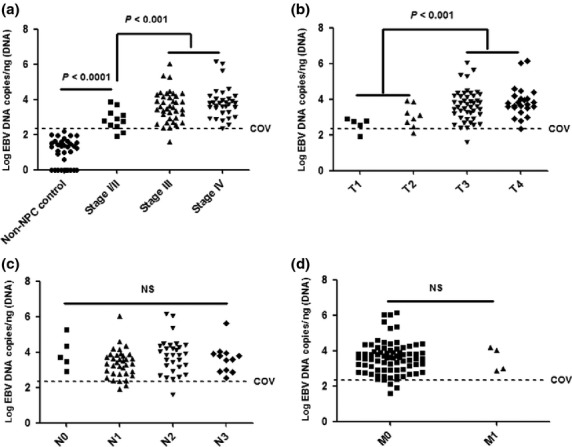
Epstein–Barr virus (EBV) DNA load in nasopharyngeal (NP) brushing samples of non-nasopharyngeal carcinoma (NPC) control was significantly lower than that in stage I/II (a), T1/T2 (b), N0 (c) and M0 (d) diseases (P < 0.0001). Stage I/II (A) and T1/2 (b) disease also had a significantly lower level of EBV DNA load in NP brushing samples compared with advanced disease (P < 0.001). No significant difference was observed between node negative and positive cases (c) and between metastasis negative and positive cases (d).
Diagnosis by Epstein–Barr virus DNA load versus initial biopsy
Among the 129 biopsy-proven NPC patients, 11 patients (8.5% false negativity) needed to undergo repeated sampling before obtaining a correct pathological diagnosis. Two patients even underwent more than two biopsies. In contrast, the EBV DNA loads in the only NP brushing samples from these 11 patients were all above the COV, directly allowing for a correct diagnosis (Table3). However, it was noted that the mean value of EBV DNA loads (mean = 2346 copy) was far less than its mean expression in the whole case group (mean = 46 360 copy).
Table 3.
Diagnosis by EBV DNA load versus pathological diagnosis based on biopsy
| Sample ID | EBV DNA load | Numbers of biopsy sampling |
|---|---|---|
| 187 | 818 | 2 |
| 207 | 6089 | 2 |
| 216 | 334 | 2 |
| 232 | 3062 | 3 |
| 245 | 2927 | 2 |
| 254 | 499 | 2 |
| 281 | 3953 | 2 |
| 347 | 245 | 2 |
| 397 | 2807 | 2 |
| 460 | 2320 | 5 |
| 469 | 2749 | 2 |
EBV, Epstein–Barr virus.
Diagnosis by Epstein–Barr virus DNA load versus other molecular markers
In this study, EBV DNA load in plasma showed both higher false negativity (24%) and false positivity (13%) at COV = 1000 copy/mL than EBV DNA load in NP brushing samples (4% false negativity and 3% false positivity). Similarly, VCA-IgA titer exhibited 11% false negativity and 23% false positivity at COV ≥ 1:80, also both higher than EBV DNA load in NP brushing samples (Table4).
Table 4.
Diagnosis by EBV DNA load versus other two molecular markers
| COV | NPC patients | Controls | |||||
|---|---|---|---|---|---|---|---|
| Positivity | Negativity | Data lost | Positivity | Negativity | Data lost | ||
| EBV DNA load in NP brushing samples | 225/ng | 124 (96%) | 5 (4%) | 0 | 4 (3%) | 112 (97%) | 0 |
| EBV DNA load in plasma | 1000/mL | 76 (76%) | 24 (24%) | 29 | 3 (13%) | 20 (87%) | 93 |
| VCA-IgA | 1:80 | 110 (89%) | 14 (11%) | 5 | 25 (23%) | 84 (77%) | 7 |
COV, cut-off value; EBV, Epstein–Barr virus; NP, nasopharyngeal; NPC, non-nasopharyngeal carcinoma.
Discussion
Convenient diagnosis of NPC is important for increasing patients’ survival. Blood sampling combined with VCA-IgA titer or EBV DNA load detection demonstrates great potential for use in assisting in NPC diagnosis and screening. Detecting VCA-IgA titer in serum or EBV DNA load in plasma has been widely studied and even used in our clinical practice. However, quantitative detection of EBV DNA load combined with NP brush sampling has never been investigated in our population in southern china. Because the highest prevalence of NPC is observed in southern China, it is important to explore and expand its application in our high-risk area. Our study showed that detection of EBV DNA load combined with NP brush sampling could be a valuable supplement in the diagnosis of NPC.
Nasopharyngeal brush or swab sampling combined with EBV DNA detection has been conducted in patients in Hong Kong, Taiwan, Canada and Indonesia. The method used in detecting EBV DNA has developed from insensitive, qualitative PCR14,16 to sensitive, quantitative real-time PCR.15,17,18 Different DNA fragments have been detected in the various studies. For example, some studies have targeted a 213-bp region of EBNA-1.17,18 Another study targets a 75-bp fragment of the EBV BamHI-W region.15 Different normalization methods have also been applied. For example, one study reports EBV DNA load as EBV copies per β-actin15; other studies report the EBV DNA load as EBV copies per brush17,18 or as EBV copies per μg DNA.21 Although there are no standardized approaches, similar sensitivity (91–96.4%) and specificity (90–98%) are obtained. High sensitivity and specificity were also observed in the present study using both the EBV DNA load per ng DNA (96% sensitivity and 97% specificity) and EBV DNA load per β-globin DNA (95% sensitivity and 96% specificity) (Suppl. Table S1). There were only minor differences between these two normalized methods (Suppl. Table S2). This was the first study to quantitatively evaluate the value of EBV DNA load in NP brushing samples using people from our local high-risk area. In contrast, Tong and colleagues found that T1 tumors had a significantly lower EBV DNA level than cases with T2/T3/T4 disease (P = 0.038), while no difference was found between stage I/II disease and stage III/IV disease; they used a small sample size (n = 28).15 Another study also only observed a tendency of increasing EBV DNA level between early and advanced tumor stage, but there was no significant difference.18 In our study, not only did the T1/T2 subgroup have a significantly lower EBV DNA level than was the case for the T3/T4 stage (P < 0.001), but also stage I/II tumors had a significantly lower EBV DNA level than was the case for stage III/IV tumors (P < 0.001) (Fig.2). EBV DNA level exhibited a greater potential in diagnosing NPC using this method in our population.
The nasopharynx is an anatomic area that is difficult to access. Therefore, pathological diagnosis based on biopsy may lead to false negativity. In this study, 11 out of 129 NPC patients had to receive repeated biopsy sampling in order to obtain a correct pathological confirmation, with an 8.5% false negativity for the first biopsy sampling. It is noteworthy that patients who failed pathological diagnosis in the initial biopsy did not exhibit T1/T2 disease. Although EBV DNA loads in the only NP brushing samples were all above the COV (Table3), the mean value of EBV DNA loads (mean = 2346 copy) was far less than its mean expression in the whole case group (mean = 46360 copy). This suggested that tumor cells in this condition were located in a difficult anatomic area for both biopsy and NP brush sampling. However, it seemed NP brush sampling was easier to obtain tumor cells under this condition compared to biopsy sampling. Therefore, it was suggested that detecting EBV DNA in NP brushing samples could be an effective supplement in NPC diagnosis.
Detection of EBV DNA load in plasma has been a breakthrough in the diagnosis of NPC. However, the method demonstrates limited application in early-stage or recurrent NPC.13 Up to 50% of patients with early stage or recurrent NPC show undetectable plasma EBV DNA, likely due to the relatively low levels of DNA fragments released from small-sized tumors into the blood. Although there were few patients with early stage (n = 11) and recurrent NPC (n = 3) in this study, our data still suggested the potential value of detecting EBV DNA in NP brushing samples. More than 80% of patients with early stage and all 3 with recurrent NPC had EBV DNA load above COV. Although titer of VCA-IgA is widely used in clinic and for screening high-risk populations, it is insufficient for accurately diagnosing NPC. In this study, up to 25 controls exhibited VCA-IgA titers ≥1:80 but were finally diagnosed with chronic nasopharyngitis. In contrast, some patients with advanced-stage NPC exhibited completely negative VCA-IgA titers (Table4). These observations confirmed prior findings that VCA-IgA may not provide strong diagnostic information.
The NP brushing procedure also exhibited 4% false negativity and 3% false positivity in this study (Table4). There were 5 false negative NPC patients with T3 disease stage. The most likely reason was that NP brush sampling was outside the tumor field. Although repeated brush sampling is more feasible because of its noninvasive and painless characteristics, optimization of the sampling method is still needed. For example, a novel NP brush more suitable for the physiological structure of the nasopharynx should be developed. It is expected that after optimization of the sampling method, the sensitivity should reach 100%. However, we cannot exclude the possibility of a deeply located tumor, for which a deep biopsy may be able to yield sufficient tumors cells for a diagnosis. Four false positive cases were from the high-risk populations, with slightly high EBV DNA loads (EBV DNA load = 322, 407, 418 and 469 copy, respectively). Contamination during the experiment might be one of the explanations for these results. However, the possibility of obscured malignancy cannot be excluded. Regular follow up will be conducted for these participants to monitor them for any clinical evidence of NPC.
In conclusion, quantification of EBV DNA load in NP brushing samples could be used as an effective supplement for NPC diagnosis in southern China. Due to the minimal invasiveness of this sampling approach, it has great potential for use in screening such high-risk populations.
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars (81325018), the Key Project for International Cooperation and Exchange of the National Natural Science Foundation of China (81220108022), the National Basic Research Program of China (2011CB504303) and the Science and Technology Planning Project of Guangdong Province, China (2011B031800218).
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Typical fragment of EBV DNA (76 bp) and globin DNA (102 bp) in nasopharyngeal (NP) brushing samples from 10 NPC patients and 10 non-nasopharyngeal carcinoma (NPC) controls are shown.
Table S1. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) of Epstein–Barr virus (EBV) DNA load at cut-off value (COV) determined by different normalization methods
Table S2. Different diagnosis between the two normalized methods. There were minor differences and only two participants had a different diagnosis.
References
- Razak AR, Siu LL, Liu FF, Ito E, O'Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46:1967–78. doi: 10.1016/j.ejca.2010.04.004. [DOI] [PubMed] [Google Scholar]
- He ML, Luo MX, Lin MC, Kung HF. MicroRNAs: potential diagnostic markers and therapeutic targets for EBV-associated nasopharyngeal carcinoma. Biochim Biophys Acta. 2012;1825:1–10. doi: 10.1016/j.bbcan.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Raab-Traub N. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein-Barr virus-infected neoplasia. Am J Pathol. 1995;146:1355–67. [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G, Young LS. Epstein-Barr virus persistence and virus-associated tumours. Lancet. 1994;343:333–5. doi: 10.1016/s0140-6736(94)91167-3. [DOI] [PubMed] [Google Scholar]
- Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–8. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- Low WK. The contact bleeding sign of nasopharyngeal carcinoma. Head Neck. 1997;19:617–9. doi: 10.1002/(sici)1097-0347(199710)19:7<617::aid-hed9>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Chan KC, Lo YM. Circulating EBV DNA as a tumor marker for nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:489–96. doi: 10.1016/s1044579x02000913. [DOI] [PubMed] [Google Scholar]
- Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4:665–9. [PubMed] [Google Scholar]
- Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–91. [PubMed] [Google Scholar]
- Shotelersuk K, Khorprasert C, Sakdikul S, Pornthanakasem W, Voravud N, Mutirangura A. Epstein-Barr virus DNA in serum/plasma as a tumor marker for nasopharyngeal cancer. Clin Cancer Res. 2000;6:1046–51. [PubMed] [Google Scholar]
- Hsiao JR, Jin YT, Tsai ST. Detection of cell free Epstein-Barr virus DNA in sera from patients with nasopharyngeal carcinoma. Cancer. 2002;94:723–9. doi: 10.1002/cncr.10251. [DOI] [PubMed] [Google Scholar]
- Yip TT, Ngan RK, Fong AH, Law SC. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014;50:527–38. doi: 10.1016/j.oraloncology.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Tune CE, Liavaag PG, Freeman JL, et al. Nasopharyngeal brush biopsies and detection of nasopharyngeal cancer in a high-risk population. J Natl Cancer Inst. 1999;91:796–800. doi: 10.1093/jnci/91.9.796. [DOI] [PubMed] [Google Scholar]
- Tong JH, Tsang RK, Lo KW, et al. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res. 2002;8:2612–9. [PubMed] [Google Scholar]
- Hao SP, Tsang NM, Chang KP. Screening nasopharyngeal carcinoma by detection of the latent membrane protein 1 (LMP-1) gene with nasopharyngeal swabs. Cancer. 2003;97:1909–13. doi: 10.1002/cncr.11312. [DOI] [PubMed] [Google Scholar]
- Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int J Cancer. 2006;119:608–14. doi: 10.1002/ijc.21914. [DOI] [PubMed] [Google Scholar]
- Adham M, Greijer AE, Verkuijlen SA, et al. Epstein-Barr virus DNA load in nasopharyngeal brushings and whole blood in nasopharyngeal carcinoma patients before and after treatment. Clin Cancer Res. 2013;19:2175–86. doi: 10.1158/1078-0432.CCR-12-2897. [DOI] [PubMed] [Google Scholar]
- Xu FH, Xiong D, Xu YF, et al. An epidemiological and molecular study of the relationship between smoking, risk of nasopharyngeal carcinoma, and Epstein-Barr virus activation. J Natl Cancer Inst. 2012;104:1396–410. doi: 10.1093/jnci/djs320. [DOI] [PubMed] [Google Scholar]
- Shao JY, Li YH, Gao HY, et al. Comparision of plasma Epstein-Barr Virus (EBV) DNA levels and serum EBV immunoglobulin A/Virus capsid angigen antibody titers in patients with nasopharyngeal carcinoma. Cancer. 2004;100:1162–70. doi: 10.1002/cncr.20099. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jiang WH, Ren CP, et al. Frequent hypermethylation of RASSF1A and TSLC 1, and high viral load of Epstein-Barr Virus DNA in nasopharyngeal carcinoma and matched tumor-adjacent tissues. Neoplasia. 2005;7:809–15. doi: 10.1593/neo.05217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Typical fragment of EBV DNA (76 bp) and globin DNA (102 bp) in nasopharyngeal (NP) brushing samples from 10 NPC patients and 10 non-nasopharyngeal carcinoma (NPC) controls are shown.
Table S1. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) of Epstein–Barr virus (EBV) DNA load at cut-off value (COV) determined by different normalization methods
Table S2. Different diagnosis between the two normalized methods. There were minor differences and only two participants had a different diagnosis.


