Background: NEDD8 conjugates are increased in cells with proteasome function impairment, and neddylation of ubiquitinated proteins on the ubiquitin chain requires ubiquitin- but not NEDD8-activating enzyme.
Results: NEDD8 ultimate buster 1 long (NUB1L) suppresses atypical neddylation and promotes the degradation of misfolded proteins.
Conclusion: NUB1L is an important regulator of protein quality control.
Significance: Selective targeting of atypical neddylation is a novel strategy to enhance protein quality control.
Keywords: cardiomyopathy, posttranslational modification (PTM), proteasome, protein degradation, ubiquitin, NEDD8, NUB1L
Abstract
Neddylation is a posttranslational modification that controls diverse biological processes by covalently conjugating the ubiquitin-like protein NEDD8 to specific targets. Neddylation is commonly mediated by NEDD8-specific enzymes (typical neddylation) and, sometimes, by ubiquitin enzymes (atypical neddylation). Although typical neddylation is known to regulate protein function in many ways, the regulatory mechanisms and biological consequence of atypical neddylation remain largely unexplored. Here we report that NEDD8 conjugates were accumulated in the diseased hearts from mouse models and human patients. Proteotoxic stresses induced typical and atypical neddylation in cardiomyocytes. Loss of NUB1L exaggerated atypical neddylation, whereas NUB1L overexpression repressed atypical neddylation through promoting the degradation of NEDD8. Activation of atypical neddylation accumulated a surrogate misfolded protein, GFPu. In contrast, suppression of atypical neddylation by NUB1L overexpression enhanced GFPu degradation. Moreover, NUB1L depletion accumulated a cardiomyopathy-linked misfolded protein, CryABR120G, whereas NUB1L overexpression promoted its degradation through suppressing neddylation of ubiquitinated proteins in cardiomyocytes. Consequently, NUB1L protected cells from proteotoxic stress-induced cell injury. In summary, these data indicate that NUB1L suppresses atypical neddylation and promotes the degradation of misfolded proteins by the proteasome. Our findings also suggest that induction of NUB1L could potentially become a novel therapeutic strategy for diseases with increased proteotoxic stress.
Introduction
Protein posttranslational modification by ubiquitin (Ub)2 and ubiquitin-like proteins is an important mechanism regulating the function of eukaryotic proteins. Since the discovery of ubiquitin in the 1970s, a dozen ubiquitin-like proteins have been identified that modify protein substrates in a manner similar to ubiquitination (1, 2). Among all ubiquitin-like proteins, neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) shares the highest (80%) homology with Ub and is evolutionarily conserved among most species (3). Similar to Ub, NEDD8 is covalently attached to target proteins by forming an isopeptide bond between the C-terminal glycine of NEDD8 and the ϵ-amino group of lysine residues on the recipient substrate. Typical neddylation requires the NEDD8-specific heterodimeric E1-activating enzyme (NAE), the E2 conjugation enzyme (Ubc12), and a still largely unknown set of E3 ligases. The conjugated NEDD8 can be cleaved off the substrates by deneddylases such as the COP9 signalosome (CSN) and NEDD8 protease 1 (NEDP1) in a process known as deneddylation (4, 5). Unlike ubiquitination, neddylation does not signal modified proteins to the proteasome for degradation but, instead, alters the conformation (6), stability (7), intracellular compartmentalization (8), and binding affinity of the substrate to DNA and protein partners (9, 10). Consequently, by modulating the cellular functions of the substrates, neddylation influences diverse cellular processes, including ubiquitination, cell proliferation, DNA damage response, transcriptional regulation, signaling transduction, and others (4, 11). Not surprisingly, dysregulation of neddylation has been linked to developmental defects (12), cancer (13), heart failure (14, 15), and synaptic dysfunction (16).
Cullin family proteins are the best characterized NEDD8 substrates. However, an increasing number of cellular proteins have been identified recently as NEDD8 targets (11). Interestingly, recent reports indicate that, under various stress conditions, NEDD8 incorporates into the existing Ub chain, leading to mixed ubiquitin- and NEDD8-modified conjugates (17). Such neddylation requires a ubiquitin-conjugation system, such as the ubiquitin-activating enzyme UBE1, but not the canonical NAE. Therefore, this process has been termed atypical neddylation (17, 18). It remains unclear how atypical neddylation is controlled or what its functional consequence is in cells.
The interferon-inducible protein NEDD8 ultimate buster 1 long (NUB1L) has been proposed as a negative regulator of neddylation (19). It possesses a ubiquitin-like domain and three ubiquitin-associating domains. NUB1L specifically binds to NEDD8 and promotes the transfer of NEDD8 to the proteasome for degradation by cooperating with the p97UFD1-NPL4 complex (20). NUB1L overexpression has been shown previously to reduce neddylated proteins in non-cardiac cells (19). Moreover, NUB1L controls the degradation of synphilin 1, tau, and mutant Hungtingtin (21–23), suggesting that NUB1L may play a role in certain neurodegenerative diseases.
Although neddylation does not lead to the proteasomal degradation of NEDD8-modified substrates, accumulating evidence suggests a cross-talk between NEDD8 and ubiquitin-mediated proteolysis. In support of this, typical neddylation of cullin-based or non-cullin Ub ligases controls their activities and the degradation of their substrates (6, 10). Furthermore, neddylation of cellular proteins can actually antagonize their ubiquitination and increase their stability (7, 24). So far, whether atypical neddylation, particularly neddylation of ubiquitinated proteins, influences ubiquitin-mediated proteolysis has not been explored in any cell type, including cardiomyocytes. Here we report that proteotoxic stress activates atypical neddylation in cardiomyocytes, which, in turn, impairs global proteasomal proteolysis. We further demonstrate that NUB1L suppresses atypical neddylation and promotes the proteasomal degradation of a misfolded protein that causes cardiomyopathy, thereby conferring protection to cardiomyocytes against proteotoxic stress. Our studies reveal a previously unrecognized cross-talk between ubiquitin and the NEDD8 pathway. Given that proteotoxicity has been identified as a pathogenic factor in many cardiac disorders, selectively targeting atypical neddylation could be a novel therapeutic strategy to combat cardiac pathogenesis involving prevalent proteotoxic stress.
Experimental Procedures
Animal Models
Transgenic (Tg) mouse models expressing a missense mutant (arginine 120 to glycine) of αB-crystallin (CryABR120G) or a seven-amino acid deletion mutant of desmin (D7-des) haven been described previously (25, 26). These mice were maintained in the FVB/N inbred background for our studies. All animal experiments were approved by the Georgia Regents University Institutional Animal Care and Use Committee.
Human Samples
Formalin-fixed and paraffin-embedded human heart tissue blocks from 10 patients with end-stage heart failure and five non-failing controls (without a history of cardiovascular disorders) were retrieved from the archive of the Department of Pathology and Laboratory Medicine at the University of Rochester Medical Center. The protocol for the use of human heart samples was reviewed and exempted by the Institutional Review Board at the University of Rochester Medical Center.
Cell Culture and Plasmid Transfection
The HEK293 cell line stably expressing a degron signal CL1-fused green fluorescent protein (GFPu) and red fluorescent protein (RFP) was created as described previously (27). Mouse embryonic fibroblasts (MEFs) were isolated from NUB1L knockout or littermate wild-type embryos at embryonic day 14.5 as described previously (28). The primary cultured MEFs were used within five passages. HEK293 and MEFs were maintained in DMEM with 10% FBS. pShuttle-CMV-SF-NEDD8 (Strep- and FLAG-tagged NEDD8) or pShuttle-CMV-SF-NEDD8ΔGG (a conjugation-deficient mutant because of the deletion of the last two glycine residues) was transfected into the cells using X-tremeGene HP DNA transfection reagent (Roche) according to the protocol of the manufacturer. Cells were harvested for analysis 48 h after transfection.
Cardiomyocyte Isolation, Adenovirus Infection, and siRNA Transfection
Neonatal rat ventricular myocytes (NRVMs) were isolated using the neonatal cardiomyocyte isolation system (Worthington) following the protocol of the manufacturer (29).
Adenoviruses expressing SF-NEDD8 (Ad-SF-NEDD8) or FLAG-tagged NUB1L (Ad-NUB1L) were created using the AdEasy adenoviral vector system (Stratagene). Adenoviruses expressing GFPu (Ad-GFPu), RFP (Ad-RFP), HA-tagged wild-type CryAB (Ad-CryAB), or HA-tagged CryABR120G (Ad-HA-CryABR120G) were used. Infection of NRVMs was generally performed 24∼48 h after plating in serum-free DMEM for 3 h. The adenovirus-containing medium were replaced with fresh medium containing 2% serum. Ad-NUB1L was applied to NRVMs at a multiplicity of infection (MOI) of 20 unless specified otherwise. Control cells were infected with Ad expressing β-galactosidase (Ad-β-gal) at a comparable MOI.
All gene knockdown assays were performed using siRNA (Qiagen) as described previously (29). The siRNAs used for NUB1L were 5′-CCGGAAACGATGCCTTACTTA-3′ and 5′-TCCGAGGCTCCGGAAAGATAA-3′. siRNA against luciferase (5′-AACGTACGCGGAATACTTCGA-3′) was used as a negative control in all knockdown experiments. Briefly, NRVMs were transfected with siRNA (100 pmol/2 × 106 cells) using Lipofectamine 2000 (Life Technologies) following the protocol of the manufacturer 24∼48 h after plating. Six hours after transfection, the siRNA-containing medium was replaced with fresh medium containing 2% FBS. A second round of siRNA transfection was performed 3 days after the first transfection to achieve a sustained silencing effect.
Simulated Ischemia (sI) and sI/Reperfusion
Simulated ischemia or ischemia reperfusion was achieved using a modified Krebs buffer/anaerobic environment protocol as described previously (30). Briefly, NRVMs were incubated with ischemic buffer containing 1.3 mm CaCl2, 5 mm KCl, 0.3 mm KH2PO4, 0.5 mm MgCl2, 0.4 mm MgSO4, 128 mm NaCl, 4 mm NaHCO3, and 10 mm HEPES (pH 6.8) and placed in a chamber mimicking hypoxic conditions (95% N2 and 5% CO2) for 12 h. Following ischemia, the cells were reoxygenated (perfused) for another 12 h in DMEM with 2% serum under normal culture conditions.
Protein Extraction, Western Blot Analysis, and Protein Dot Blot Analysis
Protein extraction from ventricular myocardium tissues or cultured cells, protein concentration determination with BCA reagents (Pierce), SDS-PAGE, immunoblotting, and densitometry were performed as described previously (14).
The soluble and insoluble fractions of cell lysates were prepared as described previously (29). Briefly, cultured cardiomyocytes were lysed in cold PBS (pH 7.4) containing 1% Triton X-100, 2.5 mm EDTA, and complete protease inhibitor mixture on ice for 30 min and vortexed for 30 s every 10 min. The cell extracts were centrifuged at 12,000 × g for 15 min, and the supernatants were collected as soluble fractions. The pellets were dissolved in 1× SDS sample buffer (50 mm Tris-HCl (pH 6.8) containing 2% SDS, 10% glycerol, and complete protease inhibitor mixture), sonicated on ice, boiled for 5 min, and centrifuged at 10,000 × g for 10 min at 4 °C. The supernatants were collected as insoluble fractions.
For the protein dot blot, equal amounts of soluble or insoluble protein fractions were loaded onto a PVDF membrane using a Bio-Dot apparatus (Bio-Rad). The membrane was then rinsed with cold PBS and subjected to Western blot analysis.
RNA Preparation and Real-time PCR
Isolation of total RNA and reverse transcription into single-stranded cDNA was performed as described previously (29). Gene expression levels were measured in duplicate per sample by real-time PCR (StepOnePlus real-time PCR system, Life Technologies) using the SYBR Green assay with gene-specific primers at a final concentration of 200 nm. The following primers were used: GFPu forward, GGGCACAAGCTGGAGTACAACT; GFPu reverse, ATGTTGTGGCGGATCTTGAAG; HA-CryABR120G forward, TTCTTCGGAGAGCACCTGTT; and HA-CryABR120G reverse, CCCCAGAACCTTGACTTTGA. Relative gene expression was calculated using the 2−ΔΔct method against rat housekeeping gene acidic ribosomal phosphoprotein P0 (RPLP0). Each experiment was repeated at least three times independently.
Proteasomal Peptidase Activity Assay
This was performed as described previously with minor modifications (31). Briefly, cultured NRVMs were lysed on ice in cytosolic extraction buffer (50 mm Tris-HCl (pH 7.5), 250 mm sucrose, 5 mm MgCl2, 0.5 mm EDTA, and 1 mm DTT). Protein concentrations were determined with BCA reagents (Pierce) and diluted to equal concentration in proteasome assay buffer (50 mm Tris-HCl (pH 7.5), 40 mm KCl, 5 mm MgCl2, and 1 mm DTT). Peptidase activities were determined in the presence of ATP for chymotrypsin-like (28 μm) and caspase-like activity (14 μm). The synthetic fluorogenic substrates Suc-LLVY-AMC (18 μm, Enzo Life Sciences) and Suc-LLE-AMC (45 μm, Millipore) were used for measuring chymotrypsin-like and caspase-like activities, respectively. Peptide cleavage reactions were carried out in the absence or presence of the proteasome inhibitor MG-132 (Enzo Life Sciences). The portion of activities that was inhibited by MG132 was considered to be the proteasome activity. The fluorescence was read in a Bio-Tek Synergy H4 plate reader at an excitation wavelength of 380 nm and an emission wavelength of 460 nm.
Cycloheximide (CHX) Chase Assay
NRVMs subject to various experimental treatments were incubated in serum-free DMEM containing CHX (100 μm, Sigma) to suppress protein synthesis. The cells were collected at different consecutive time points after CHX administration, and whole-cell lysates were analyzed by Western blot for the protein of interest.
Immunohistochemistry Microscopy
For immunohistochemical staining of NEDD8, 4-μm sections were deparaffinized and rehydrated. To recover antigen, these sections were incubated in preheated sodium citrate buffer (pH 6.0, 99 °C) for 20 min with the PT Link system (Dako). Endogenous peroxidase activity was blocked with 3% H2O2. After preincubation with 10% non-immune goat serum (Invitrogen) to prevent nonspecific binding, tissue sections were incubated with rabbit anti-NEDD8 antibody (1:400, Cell Signal Technology) at 4 °C overnight. The signal was amplified with the Histostain-SP kit (Life Technologies) and detected with 3,3′-diaminobenzidine tetrahydrochloride substrate (Dako). Special attention was paid to avoid color overdevelopment. Hematoxylin was used as a counterstain. Negative controls were incubated with appropriate non-immune serum instead of primary antibody under the same conditions.
Non-denaturing and Denaturing Immunoprecipitation
NRVMs were lysed in radioimmunoprecipitation assay buffer supplemented with complete protease inhibitor mixture (Roche) and a deubiquitin enzyme inhibitor, PR-619 (30 μm, Lifesensors), followed by centrifugation at 12,000 × g for 10 min. The supernatant was then quantified for protein concentration with BCA reagents, and 1 mg of the supernatant, in a total volume of 1 ml, was used for the subsequent immunoprecipitation. The tissue lysates were first precleared by incubation with appropriate IgG for 2 h at 4 °C. The precleared lysates were then incubated with mouse anti-HA antibody-conjugated agarose beads to pull down HA-tagged CryABR120G or tandem ubiquitin binding entity-conjugated agarose beads (Lifesensors) to pull down polyubiquitinated proteins at 4 °C overnight. Protein A/G-agarose beads (Santa Cruz Biotechnology) along with unconjugated mouse IgG (4 μg of IgG/1 mg of protein, Santa Cruz Biotechnology) or control agarose (Lifesensors) served as immunoprecipitation controls. The immunoprecipitates were finally eluted using 2× sample loading buffer (160 mm Tris-HCl, 4% SDS, and 20% glycerol), resolved by SDS-PAGE, and subjected to immunoblot analyses.
Lactate Dehydrogenase Activity Assay
Lactate dehydrogenase activity in the collected culture medium was measured using a cytotoxicity detection kit (Roche) and following the instructions of the manufacturer.
Measurement of Cellular ATP Levels
Cell viability was determined by measuring cellular ATP levels using the CellTiter-Glo luminescent cell viability kit (Promega) and following the instructions of the manufacturer. Luminescence was recorded with a Centro XS3 LB 960 microplate luminometer (Berthold).
Statistical Analysis
Results are shown as mean ± S.E. Paired data were evaluated using two-tailed Student's t test. For multiple comparisons, one-way analysis of variance or, when appropriate, two-way analysis of variance, followed by a post hoc test was performed. p < 0.05 was considered statistically significant.
Results
Proteotoxic Stress Induces Typical and Atypical Neddylation in Cardiomyocytes
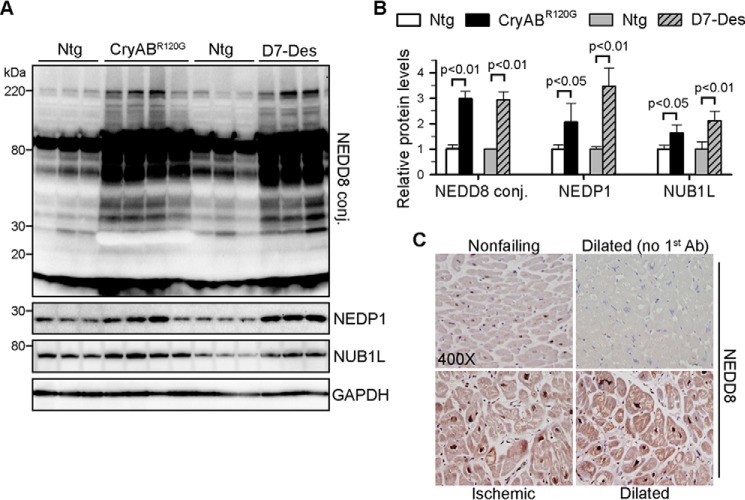
To study the role of neddylation in ubiquitin-mediated proteolysis, we first asked whether neddylation is dysregulated in hearts exposed to proteotoxic stress. It has been established previously that proteotoxic stress is prevalent in Tg mouse hearts overexpressing the misfolded proteins CryABR120G or D7-des in which these misfolded proteins form protein aggregates in cardiomyocytes (25, 26). In adult CryABR120G and D7-des Tg mouse hearts, we observed ∼3-fold increases of neddylated proteins compared with littermate non-Tg mice. This was accompanied by up-regulation of the negative regulator of neddylation NUB1L and the deneddylase NEDP1 (Fig. 1, A and B), suggesting that the increase in neddylated proteins likely results from activation of neddylation rather than defective deneddylation. Furthermore, proteotoxic stress is also prevalent in human failing hearts, as evidenced by the accumulation of ubiquitinated proteins and protein aggregates (32, 33). In immunostained myocardium sections of human patients suffering from ischemic or dilated cardiomyopathy, we observed increased NEDD8 immunoreactivity compared with those from control hearts without diagnosed cardiac abnormalities (Fig. 1C).
FIGURE 1.
Increased NEDD8 conjugates in diseased hearts. A and B, representative Western blots (A) of the indicated proteins and densitometric analysis (B) of 2-month-old CryABR120G and D7-Des Tg mouse hearts. n = 4–6 for each group. Conj, conjugate. C, NEDD8 immunohistochemistry of myocardium sections of explanted failed human hearts with ischemic or dilated cardiomyopathy and nonfailing hearts from donors without cardiovascular disorders. Normal rabbit serum was used as the primary antibody for a negative control. Shown are representative images from five sets of staining.
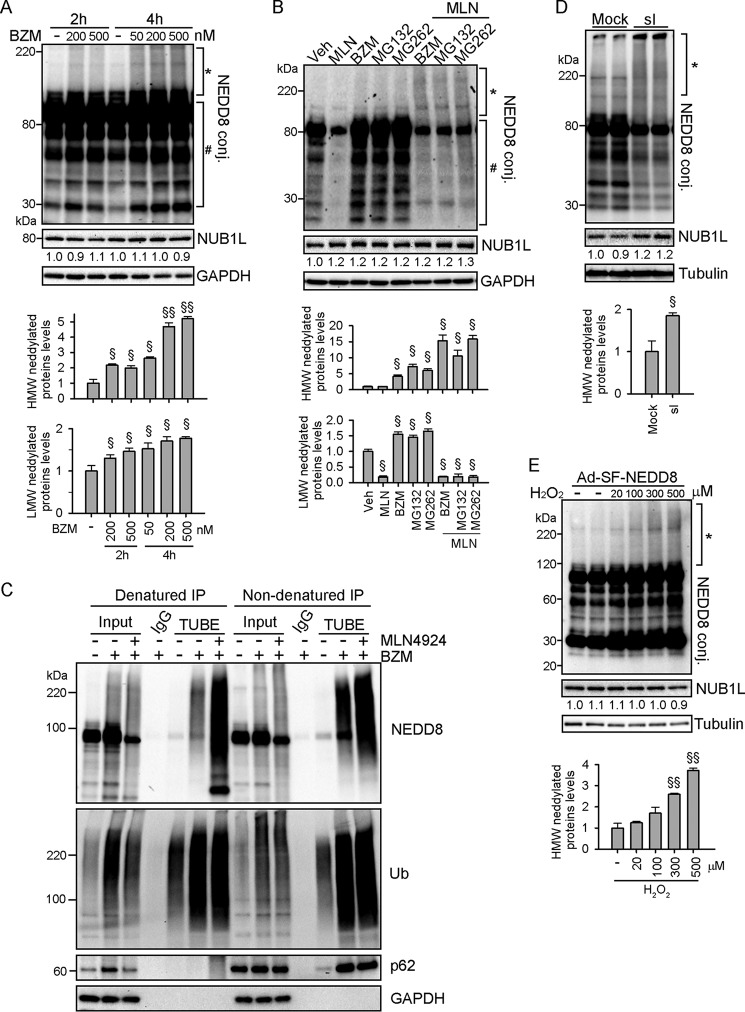
Various stresses induce UBE1-mediated mixed ubiquitin- and NEDD8- protein modification (atypical neddylation) in non-cardiac cells (17, 18). In cultured NRVMs, the proteasome inhibitor bortezomib dose- and time-dependently increased both low and high molecular weight NEDD8 conjugates (Fig. 2A). Similarly, other proteasome inhibitors such as MG132 and MG262 also substantially increased neddylated proteins in NRVMs (Fig. 2B). The NAE inhibitor MLN4924 (13) effectively attenuated the increase of NEDD8 conjugates with low molecular weight but not of those with high molecular weight upon proteasome inhibition (Fig. 2B), indicating that proteasome inhibition also activates typical neddylation in cardiomyocytes. To determine whether the high molecular weight neddylated proteins are Ub- and NEDD8-mixed modified proteins, we used tandem ubiquitin-binding entities (TUBEs) (34) to pull down polyubiquitinated proteins under denaturing conditions, which excluded non-covalently associated proteins such as p62 (Fig. 2C). As expected, we detected a remarkable increase of NEDD8 species with high molecular weight in the precipitates from NRVMs subject to proteasome inhibition, which was not attenuated but, instead, augmented by MLN4924 (Fig. 2C), confirming that the neddylation of ubiquitinated proteins does not require NAE. We next exposed NRVMs to sI or oxidative stress by H2O2 treatment. Each of these stimuli has been reported to elevate proteotoxic stress in cardiac cells (35, 36). We observed dramatically increased levels of high molecular weight neddylated proteins in stressed cardiomyocytes, which, presumably, are ubiquitin- and NEDD8-mixed modified (Fig. 2, D and E). The NUB1L protein levels were not affected by the tested stresses (Fig. 2, A, B, D, and E), suggesting that the increase of neddylated proteins was not due to loss of NUB1L. Together, these data suggest that proteotoxic stress activates typical and atypical neddylation in cardiomyocytes.
FIGURE 2.
Activation of typical and atypical neddylation by multiple stresses in cardiomyocytes. A, NRVMs were treated with the proteasome inhibitor bortezomib (BZM) for the indicated times. HMW, high molecular weight; conj, conjugate. B, NRVMs were treated with the proteasome inhibitors BZM (100 nm), MG132 (5 μm), and MG262 (1 μm) for 6 h. The NAE inhibitor MLN4924 (MLN) was added 1 h after the administration of proteasome inhibitors. Veh, vehicle. C, NRMMs were treated with vehicle or BZM (100 nm) in the absence or presence of MLN4924 (1 μm) for 6 h. Polyubiquitinated proteins were purified from cell extracts using TUBEs under denaturing or non-denaturing conditions and immunoblotted with the indicated antibodies. D, NRMMs were subjected to sI for 12 h. E, NRMMs were infected with Ad-SF-NEDD8 for 24 h and treated with H2O2 for another 8 h. A–E, Shown are representative Western blots and the densitometric analysis from three independent experiments. The asterisks and # denote high and low molecular weight NEDD8 conjugates respectively.
NUB1L Suppresses Atypical Neddylation
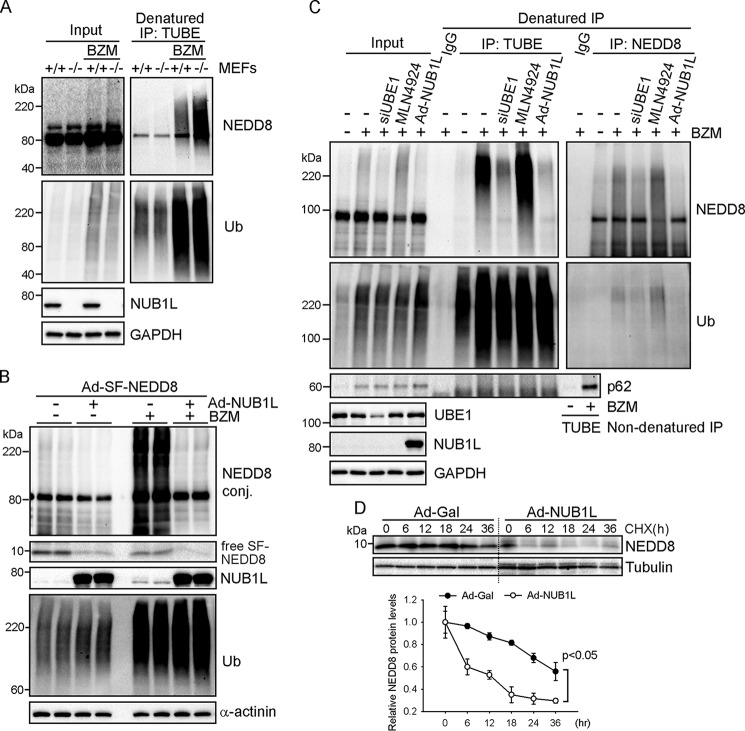
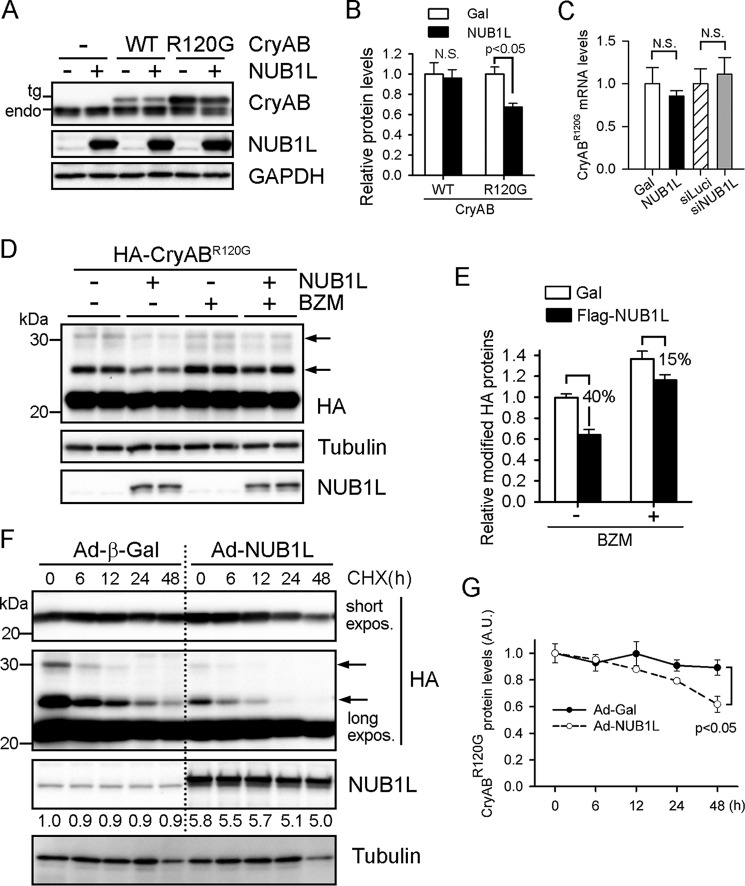
NUB1L overexpression has been shown to reduce neddylated proteins in non-cardiac cells (19, 20), but it is not known whether endogenous NUB1L controls neddylation in cardiac and other cells. We created NUB1L wild-type and knockout MEFs. Loss of NUB1L did not alter the levels of total neddylated proteins at baseline and upon proteasome inhibition (Fig. 3A). However, by pulling down polyubiquitinated proteins using TUBEs under denaturing conditions, we found a remarkable increase in Ub-NEDD8 mixed modified proteins in NUB1L knockout MEFs treated with bortezomib compared with those in NUB1L wild-type MEFs (Fig. 3A). In cultured cardiomyocytes, NUB1L overexpression mildly decreased neddylated proteins at baseline and robustly reduced protein neddylation induced by bortezomib treatment, whereas NUB1L expression had no effect on the levels of ubiquitinated proteins (Fig. 3B). Denatured immunoprecipitation followed by Western blot analysis confirmed that silencing of UBE1, but not MLN4924 treatment, largely reduced bortezomib-induced Ub-NEDD8 mixed modification (Fig. 3C). Although NUB1L overexpression did not alter UBE1 protein levels, it greatly diminished Ub-NEDD8 mixed modification upon bortezomib treatment (Fig. 3C). Reciprocally, neddylated proteins were pulled down using specific NEDD8 antibodies and probed for ubiquitin, and we also found a reduction of Ub-NEDD8 mixed modified proteins in NUB1L-overexpressing cardiomyocytes (Fig. 3C). These data demonstrate that NUB1L potently suppresses atypical neddylation but not typical neddylation. Supporting a role of NUB1L in NEDD8 degradation (20), NUB1L expression decreased free NEDD8 proteins (Fig. 3B). A cycloheximide-based chase assay further revealed that NUB1L overexpression significantly shortened the half-life of NEDD8 from >36 h to <12 h (Fig. 3D). Therefore, we conclude that NUB1L suppresses stress-induced atypical neddylation by promoting NEDD8 degradation.
FIGURE 3.
NUB1L suppresses neddylation of ubiquitinated proteins. A, extracts from NUB1L wild-type (+/+) or deficient (−/−) MEFs treated with BZM (100 nm) for 6 h were precipitated with TUBEs to pull down polyubiquitinated proteins under denaturing conditions and analyzed by Western blots. IP, immunoprecipitation. B and C, NRVMs expressing Strep- and FLAG-tagged NEDD8 (SF-NEDD8) were infected with Ad expressing FLAG-tagged NUB1L (Ad-NUB1L) or β-gal (Ad-Gal) or transfected with siRNA against UBE1 (siUBE1). BZM (100 nm) or MLN4924 (1 μm) was added 6 h before cell harvest. B, representative Western blots of total cell lysates. Conj, conjugate. C, polyubiquitinated or neddylated proteins were pulled down with TUBEs or NEDD8 antibody from cell extracts under denaturing conditions and analyzed by Western blots. D, CHX chase assay. Representative Western blots (top panel) and densitometric analysis (bottom panel) are shown. The NEDD8 densitometries were relative to its level at 0 h.
Activation of Atypical Neddylation Increases GFPu Protein Levels
The functional consequence of atypical neddylation is not known. Given that polyubiquitination often causes protein degradation by the proteasome (37), we sought to determine whether the addition of NEDD8 into the ubiquitin chain impacts proteasomal function. Ubiquitin-proteasome system (UPS) function was assessed using a sensitive GFPu/RFP reporter system in which GFPu protein levels inversely reflect proteasomal function because GFPu, but not RFP, is degraded readily by the proteasome because of the fusion of a CL1 degron signal to GFP (27).
It has been shown that forced expression of NEDD8 suffices to induce UBE1-dependent atypical neddylation (38). In HEK293 cells stably expressing GFPu and RFP, NEDD8 overexpression dramatically increased neddylated proteins and accumulated GFPu without altering RFP protein levels in a dose-dependent manner (Fig. 4, A and B). In contrast, expression of its conjugation-deficient mutant SF-NEDD8ΔGG (deletion of C-terminal diglycine) at equivalent or higher levels did not increase neddylated proteins or accumulate GFPu. Similarly, in Ad-GFPu- and Ad-RFP-coinfected cultured cardiomyocytes, NEDD8 overexpression also led to a dose-dependent increase of GFPu with no effect on RFP (Fig. 4, C and D). Our data suggest that activation of atypical neddylation impairs the proteolytic function of the UPS.
FIGURE 4.
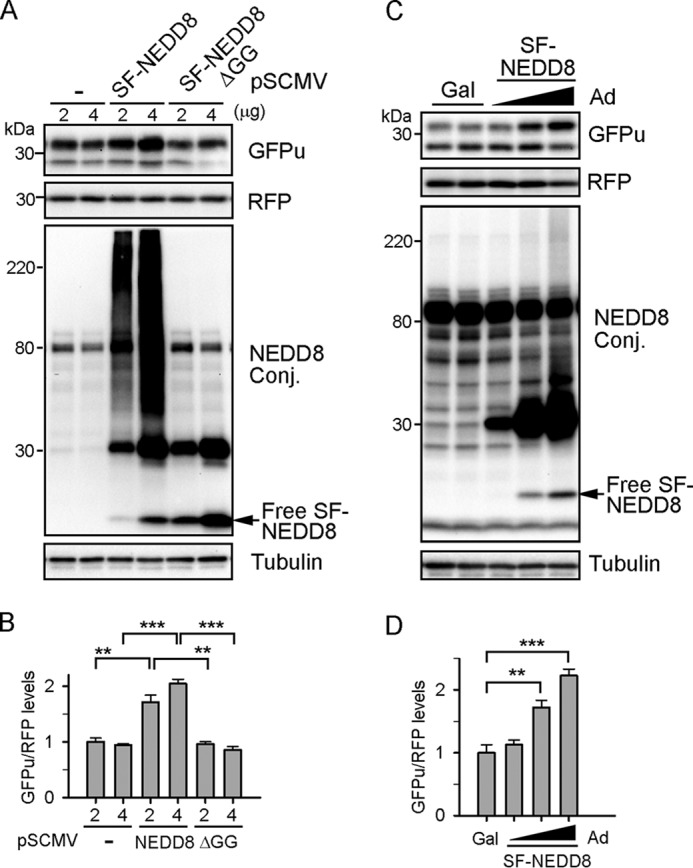
Activation of atypical neddylation accumulates a proteasome surrogate substrate GFPu. A and B, HEK293 cells stably expressing GFPu and RFP were transfected with the SF-NEDD8 or SF-NEDD8ΔGG expression vectors for 48 h. Conj, conjugate. C and D, NRVMs were infected with Ad-GFPu and Ad-RFP before infection with Ad-SF-NEDD8 for another 48 h. Representative immunoblots of the indicated proteins (A and C) and quantitative analyses (B and D) are shown. **, p < 0.01; ***, p < 0.001.
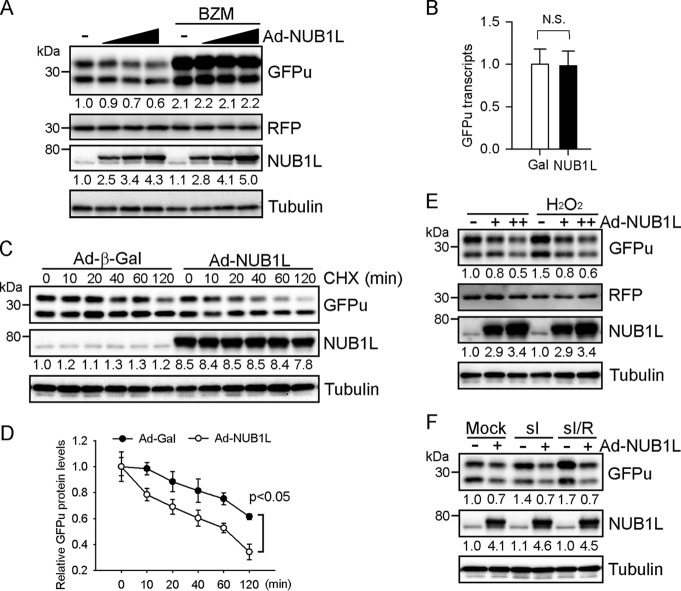
NUB1L Enhances GFPu Degradation in Cardiomyocytes
We next asked whether suppression of atypical neddylation improves proteasomal proteolysis. NUB1L overexpression dose-dependently decreased GFPu protein levels without altering those of RFP in cardiomyocytes (Fig. 5A). The decrease in GFPu proteins levels was not accompanied by changes in its mRNA levels (Fig. 5B) and was effectively abolished by proteasome inhibition (Fig. 5A). We also performed a cycloheximide chase assay and found that NUB1L expression reduced the half-life of GFPu from >120 min to ∼60 min (Fig. 5, C and D). Moreover, in line with the reduced levels of neddylated proteins, NUB1L overexpression abrogated H2O2-, sI-, and sI/R-induced accumulation of GFPu proteins (Fig. 5, E and F). These data indicate that suppression of neddylation by NUB1L overexpression enhances proteasomal proteolysis under basal and stress conditions.
FIGURE 5.
NUB1L overexpression enhances the degradation of GFPu. NRVMs were first infected with Ad expressing GFPu (Ad-GFPu) and RFP (Ad-RFP) for 24 h and subsequently with Ad-NUB1L or Ad-Gal. A, representative Western blots of total cell extracts are shown. Ad-NUB1L was infected at 5, 10, and 20 MOI and Ad-Gal at 20 MOI. BZM (100 nm) was added 6 h prior to cell harvesting. B, quantitative real-time PCR analysis of GFPu transcripts. N.S., not significant. C and D, CHX chase assay. Representative Western blots (C) and the quantification (D) are shown. GFPu protein levels at 0 h were set as 1. E and F, cells were treated with H2O2 (100 μm) for 8 h (E), sI for 6 h, or sI followed by reperfusion for another 6 h (sI/R) (F) prior to cell harvesting. Ad-NUB1L was infected at 10 and 20 (E) or 10 MOI (F). Representative Western blots of total cell extracts are shown. The fast-migrating band of GFPu is its N-terminal truncated form. The densitometries of GFPu normalized to RFP or NUB1L are denoted in A, C, E, and F.
NUB1L Regulates the Clearance of a Bona Fide Misfolded Protein, CryABR120G
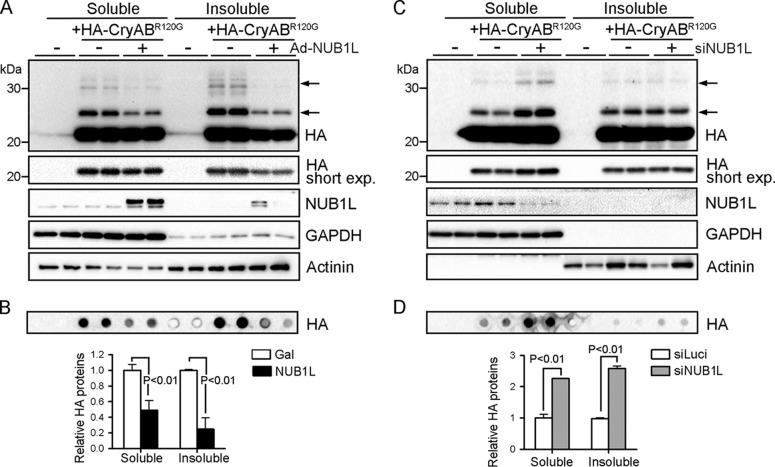
The results above demonstrate the ability of NUB1L to promote the degradation of an artificial proteasome surrogate substrate. We next wanted to determine the effect of NUB1L on the clearance of a bona fide misfolded protein. As mentioned above, CryABR120G expression causes desmin-related cardiomyopathy in mice and humans (25). HA-tagged CryABR120G was expressed in cultured NRMMs via adenoviral delivery. NUB1L overexpression significantly reduced the steady-state protein levels of CryABR120G and its modified species with slower mobility in both Triton X-100-soluble and -insoluble fractions (Fig. 6A). The quantitative protein dot blot analysis revealed ∼50 and ∼80% reductions of CryABR120G in soluble and insoluble fractions, respectively, by NUB1L overexpression (Fig. 6B). In contrast, silencing of NUB1L robustly built up free and modified forms of HA-tagged CryABR120G (Fig. 6C). The protein dot blot analysis showed a more than 2-fold increase of CryABR120G in the soluble and insoluble fractions of NUB1L-silenced cardiomyocytes (Fig. 6D).
FIGURE 6.
NUB1L reduces misfolded protein CryABR120G in cardiomyocytes. NRVMs were infected with Ad expressing HA-tagged CryABR120G (Ad-HA-CryABR120G) for 24 h and subsequently infected with Ad-NUB1L (20 MOI) or transfected with siRNA against NUB1L (siNUB1L) for another 72 h. A—D, the Triton X-100 (1%) soluble and insoluble fractions of cell extracts were subjected to Western blot analysis (A and C) and protein dot blot to quantify the abundance of CryABR120G (B and D). Arrows point to the modified forms of CryABR120G. Exp, exposure.
Furthermore, NUB1L expression had no discernible effect on the abundance of endogenous and transgenic wild-type CryAB (Fig. 7, A and B), suggesting its specific effect on misfolded proteins. Quantitative real-time PCR analyses revealed that neither overexpression nor silencing of NUB1L altered the transcripts of CryABR120G (Fig. 7C). CryABR120G is known to be degraded through the UPS (36). Indeed, proteasome inhibition significantly restored NUB1L-diminished CryABR120G proteins (15% reduction with bortezomib treatment versus 40% reduction without bortezomib treatment) (Fig. 7, D and E). The cycloheximide-based chase analysis showed that NUB1L expression significantly reduced the half-life of CryABR120G (Fig. 7, F and G), proving that NUB1L boosts the degradation of CryABR120G. These lines of evidence suggest that NUB1L is necessary for, and sufficient to enhance, the clearance of this cardiomyopathy-linked misfolded protein.
FIGURE 7.
NUB1L promotes the proteasomal degradation of CryABR120G. NRVMs were infected with Ad-HA-CryABR120G or WT CryAB for 24 h and subsequently infected with Ad-NUB1L (20 MOI). A and B, Western blots of total cell extracts (A) and quantification analysis (B). NUB1L overexpression reduced CryABR120G but not WT-CryAB protein levels. Endo, endogenous; N.S., not significant. C, quantitative PCR analysis of CryABR120G transcripts. D and E, proteasome inhibition attenuates the NUB1L-induced reduction of CryABR120G. Representative Western blots (D) of total cell extracts and densitometric analysis (E) are shown. BZM was added 24 h before cell harvesting. F and G, CHX chase assay. Representative Western blots of total cell extracts (F) and pooled densitometry data (G) of CryABR120G are shown. The changes of the native form and modified forms (arrows) of CryABR120G are shown in the short and long exposure (expos) blots, respectively. The abundance of NUB1L is denoted under the blot. A.U., arbitrary unit.
NUB1L Enhances the Proteasomal Degradation of Ubiquitinated Misfolded Proteins
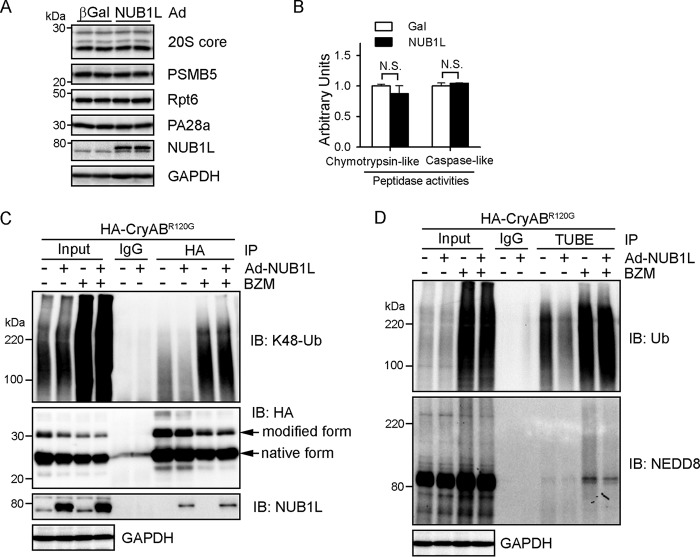
We then asked how NUB1L facilitates the degradation of CryABR120G. Proteasomal degradation of a target protein generally involves two steps: the polyubiquitination of the target protein and its subsequent degradation by the proteasome (37). We first assessed whether NUB1L directly impacts proteasome activities. Overexpression of NUB1L in cardiomyocytes did not alter the protein levels of representative subunits of the 20S (20S core and PSMB5), 19S (Rpt6), and 11S (PA28α) proteasomes (Fig. 8A). We next measured the proteasomal chymotrypsin-like and caspase-like peptidase activities in cardiomyocyte lysates. Neither of these activities was affected by NUB1L overexpression (Fig. 8B), suggesting an intact proteasomal activity in NUB1L overexpressed cardiomyocytes.
FIGURE 8.
NUB1L overexpression accelerates the turnover of ubiquitinated misfolded proteins. A, the effects of NUB1L on proteasome abundance. Western blots of the indicated proteasome subunits in total cell extracts are shown. B, the effects of NUB1L on proteasome peptidase activities. n = 6 biological repeats. N.S., not significant. C and D, NRVMs were infected with Ad-NUB1L or Ad-Gal for 72 h. Ad-HA-CryABR120G was infected 24 h prior to Ad-NUB1L infection, and BZM was added 24 h before cell harvesting. C, NUB1L overexpression reduced Lys-48-linked ubiquitinated CryABR120G, and proteasome inhibition attenuated the reduction. CryABR120G was immunoprecipitated (IP) with anti-HA antibodies from cell lysate and blotted (IB) with the indicated antibodies. Data are representative of three independent repeats. D, polyubiquitinated proteins were precipitated from cell lysates using TUBEs under denaturing conditions and blotted with the indicated antibodies. Data are representative of three independent repeats.
We then tested whether NUB1L overexpression alters the ubiquitination of misfolded proteins. CryABR120G was pulled down and probed with anti-K48 Ub antibody, which specifically detects lysine 48-linked poly-Ub chains, the major signals of proteasomal degradation. NUB1L overexpression substantially reduced Lys-48-linked polyubiquitinated CryABR120G, and the reduction was abolished in the presence of a proteasome inhibitor (Fig. 8C), indicating a quicker turnover of polyubiquitinated CryABR120G. We next purified polyubiquitinated proteins using TUBEs from Ad-CryABR120G-infected cardiomyocytes under denaturing conditions. Consistently, NUB1L overexpression effectively diminished polyubiquitinated proteins, and the decrease was abolished by proteasome inhibition (Fig. 8D). NUB1L expression largely reduced the species modified by the Ub-NEDD8 mixed chain, even in the presence of a proteasome inhibitor (Fig. 5E). We assumed that the Ub-NEDD8 mixed modified (i.e. atypically neddylated) proteins were less efficiently degraded by the proteasome compared with those modified homogeneously by ubiquitin. We therefore propose that NUB1L enhances the proteasomal degradation of misfolded proteins by suppressing the Ub-NEDD8 mixed modification.
NUB1L Protects against Proteotoxic Stress
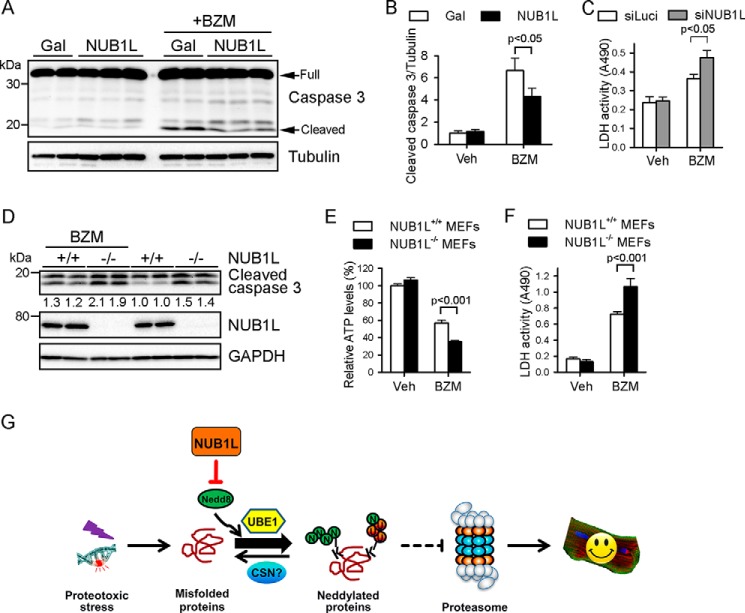
Considerable evidence suggests that misfolded proteins accumulate because of insufficient proteasomal function under stress conditions and are toxic to the cells (39). For this reason, we next sought to examine the role of NUB1L in proteotoxic stress-induced cell injury. As expected, NUB1L overexpression blunted low-dose bortezomib-induced apoptosis in cardiomyocytes, as measured by the cleavage (activation) of caspase 3 (Fig. 9, A and B). In contrast, silencing of NUB1L sensitized cardiomyocytes to bortezomib-induced cell death, as evidenced by increased leakage of lactate dehydrogenase in culture medium (Fig. 9C). In MEFs, proteasome inhibition increased the cleavage of caspase 3 and the release of lactate dehydrogenase and reduced cell viability, which were aggravated significantly by loss of NUB1L (Fig. 9, D–F). These findings together indicate a protective role of NUB1L against proteotoxic stress.
FIGURE 9.
NUB1L ameliorates proteotoxic stress-induced cytotoxicity. A–C, NRVMs were infected with the indicated adenovirus (A and B) or transfected with the indicated siRNA (C) for 48 h before being subjected to BZM (100 nm) treatment for another 24 h. Representative Western blots of total cell extracts (A) and the densitometric analysis (B) are shown. n = 4–6/group. Veh, vehicle. C, culture medium was collected for the lactate dehydrogenase (LDH) activity assay. D–F, NUB1L wild-type (+/+) or deficient (−/−) MEFs were treated with BZM (100 nm) for 24 h. D, representative Western blots of total cell lysates and the densitometries of cleaved caspase 3 relative to those of GAPDH. E, cell viability assay by measuring cellular ATP levels. F, LDH activity assay. For cell viability and LDH release assays (C, E, and F), data are representative of three independent experiments. n = 6–8 samples/group for each replication. G, model of the function of NUB1L in proteasomal proteolysis.
Discussion
In this study, we reveal that neddylation is dysregulated in the diseased hearts of mouse models of desmin-related cardiomyopathy and human patients. We demonstrate that proteotoxic stress induces excessive neddylation in cardiomyocytes, particularly UBE1-mediated atypical neddylation, which, in turn, directly or indirectly impairs proteasome function. NUB1L suppresses the neddylation of ubiquitinated proteins through promoting the degradation of NEDD8 and improves the proteasomal degradation of misfolded proteins such as CryABR120G. Therefore, NUB1L protects the cells against proteotoxic stress by ensuring proteasome-mediated protein quality control (Fig. 9G).
Implications of Aberrant Neddylation in Cardiac Pathogenesis
Germline deletion of NEDD8 E1 (NAE1 or UBA3), E2 (Ubc12), or any subunit of the deneddylase CSN all lead to embryonic lethality in mice (11), which highlights the importance of the NEDD8 pathway in embryonic development but precludes the investigation of its roles in the heart. Our study reveals a remarkable increase of neddylated proteins in the diseased hearts of mouse models of desmin-related cardiomyopathy and human patients (Fig. 1, A–C), which, to our knowledge, is the first report to tie aberrant neddylation to cardiac diseases. We have shown previously that cardiac-specific knockout of CSN8 impairs deneddylation and causes dilated cardiomyopathy in both postnatal and adult mouse hearts (14, 15). We speculate that the overwhelming neddylation seen in diseased hearts is likely a pathogenic factor contributing to cardiac dysfunction. Neddylation inhibition by NAE1 deficiency in neurons results in loss of synapses, impaired neurotransmission, and cognitive deficits (16). In the future, more in-depth studies are imperative to determine the pathophysiological significance of neddylation in the heart.
NUB1L Enhances Proteasomal Proteolysis through Suppression of Atypical Neddylation
Our data uncovered that proteotoxic stress, primarily resulting from proteasome dysfunction, is a critical trigger of excessive neddylation in cardiomyocytes (Fig. 2). In cardiomyocytes, proteasome inhibition activates both NAE-mediated (typical) and UBE1-mediated (atypical) neddylation (Fig. 2, B and C). A substantial change in the Ub-to-NEDD8 ratio under stress condition has been proposed as a trigger to stimulate atypical neddylation (17, 18). Given the similarity of NEDD8 and Ub in amino acid sequence and quaternary structure, it has been postulated that NEDD8 may substitute for Ub and cap the extension of Ub chains, therefore preserving the free Ub pool and avoiding otherwise catastrophic effects on cells (17, 40).
The functional consequence of atypical neddylation, especially neddylation of ubiquitinated proteins, is currently unknown. Using a UPS functional reporter, we revealed that activation of atypical neddylation by multiple means (overexpression of NEDD8, H2O2, or sI treatment) impairs UPS function (Figs. 4 and 5, E and F). Moreover, suppression of atypical neddylation by NUB1L enhances UPS function at baseline and under stress condition (Fig. 5). We further showed that NUB1L promotes the proteasomal degradation of a misfolded protein (Figs. 6–8). These lines of compelling evidence are strongly in agreement with the notion that neddylation of ubiquitinated proteins impairs their proteasomal proteolysis.
How does atypical neddylation impair proteasomal proteolysis? We and others have shown that the increase in neddylated proteins by proteasome inhibition is due to the activation of neddylation (Figs. 2B and 3B) (17, 18), suggesting that the neddylated proteins are not readily degraded by the proteasome. The NEDD8 signals detected in the precipitates from TUBEs, which enrich essentially Lys-48-linked polyubiquitinated proteins under proteasome inhibition condition, suggest that NEDD8 likely incorporates into the Lys-48-linked polyubiquitin chains. NEDD8 and Ub are recognized by Ub shuttle proteins with equivalent affinity in vitro and can form heterodimers in cultured cells (40). This Ub-NEDD8 heterodimer can bind to, and be processed by, the proteasome in vitro (40). Moreover, neddylated proteins have been reported to be copurified with the proteasome complex in mammalian cells (41). Therefore, under proteotoxic stress conditions, massive levels of neddylated proteins, which are resistant to proteasome degradation, may compete for the Ub shuttle proteins and proteasome machineries, leading to inefficient degradation of polyubiquitinated proteins. These possibilities will be a matter of future investigation.
Supporting this model, the deneddylase CSN associates with the proteasome in cells and is able to process Ub-NEDD8 heterodimers in vitro (40). Furthermore, loss of CSN deneddylation activity leads to the accumulation of neddylated proteins and impairs UPS proteolysis in mouse hearts (14, 15). Therefore, CSN could be a proteasome-resident deneddylase to process neddylated proteins and preserve UPS function (14, 15). By suppressing atypical neddylation under proteotoxic stress, NUB1L alleviates the competition and promotes the turnover of polyubiquitinated proteins (Fig. 8, C and D), thereby improving UPS function. Supporting the role of NUB1L in proteasome proteolysis, NUB1L promotes the polyubiquitination and clearance of mutant Huntingtin, tau, and synphilin 1 and reduces the formation of inclusions (21–23). In addition, NUB1L knockdown accumulates Cyclin E and p27, both proteins known to be degraded by the proteasome (42).
The maintenance of protein homeostasis by the UPS is crucial to the survival of cells, especially those with limited renewal capacity, such as cardiomyocytes. Inadequate protein quality control is increasingly recognized as an important pathogenic factor to a large subset of cardiac diseases (39, 43), and measures to enhance protein quality control are promising new strategies to treat cardiac diseases (36, 44). Our results indicate that atypical neddylation may contribute to inefficient proteolysis upon proteotoxic stress. It should be pointed out that targeting UBE1 to inhibit atypical neddylation may not be desirable to the cells because of the essential role of UBE1 in ubiquitin activation (37). Here we identify NUB1L as a potent suppressor of atypical neddylation and a novel regulator of protein quality control. Selective targeting of atypical neddylation, such as by up-regulation of NUB1L, could provide a new avenue to treat cardiac diseases associated with defective protein quality control.
Author Contributions
J. L. and H. S. designed the experiments. J. L., W. M., and H. L. performed and analyzed the majority of experiments. N. H. and F. L. performed and analyzed the immunohistochemistry experiments. X. W. and I. K. provided critical reagents and useful discussions. J. L. and H. S. wrote the manuscript.
Acknowledgments
We thank Dr. John A. Johnson (Georgia Regents University) for insightful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL124248 (to H. S.), R01HL124251 (to I. K.), R01HL085629 and R01HL072166 (to X. W.), and R01HL111480 (to F. L.). This work was also supported by American Heart Association Grants 11SDG6960011 (to H. S.) and 14SDG18970040 (to I. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- Ub
- ubiquitin
- NAE
- NEDD8-specific heterodimeric E1-activating enzyme
- CSN
- COP9 signalosome
- Tg
- transgenic
- RFP
- red fluorescent protein
- MEF
- mouse embryonic fibroblast
- NRVM
- Neonatal rat ventricular myocyte
- MOI
- multiplicity of infection
- Ad
- adenovirus(es)
- sI
- simulated ischemia
- CHX
- cycloheximide
- TUBE
- tandem ubiquitin-binding entity
- UPS
- ubiquitin-proteasome system
- BZM
- bortezomib.
References
- 1. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herrmann J., Lerman L. O., Lerman A. (2007) Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100, 1276–1291 [DOI] [PubMed] [Google Scholar]
- 3. Kumar S., Yoshida Y., Noda M. (1993) Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun. 195, 393–399 [DOI] [PubMed] [Google Scholar]
- 4. Enchev R. I., Schulman B. A., Peter M. (2015) Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watson I. R., Irwin M. S., Ohh M. (2011) NEDD8 pathways in cancer, sine quibus non. Cancer Cell 19, 168–176 [DOI] [PubMed] [Google Scholar]
- 6. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 7. Zuo W., Huang F., Chiang Y. J., Li M., Du J., Ding Y., Zhang T., Lee H. W., Jeong L. S., Chen Y., Deng H., Feng X. H., Luo S., Gao C., Chen Y. G. (2013) c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol. Cell 49, 499–510 [DOI] [PubMed] [Google Scholar]
- 8. Watson I. R., Blanch A., Lin D. C., Ohh M., Irwin M. S. (2006) Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 281, 34096–34103 [DOI] [PubMed] [Google Scholar]
- 9. Xirodimas D. P., Saville M. K., Bourdon J. C., Hay R. T., Lane D. P. (2004) Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118, 83–97 [DOI] [PubMed] [Google Scholar]
- 10. Xie P., Zhang M., He S., Lu K., Chen Y., Xing G., Lu Y., Liu P., Li Y., Wang S., Chai N., Wu J., Deng H., Wang H. R., Cao Y., Zhao F., Cui Y., Wang J., He F., Zhang L. (2014) The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat. Commun. 5, 3733. [DOI] [PubMed] [Google Scholar]
- 11. Kandala S., Kim I. M., Su H. (2014) Neddylation and deneddylation in cardiac biology. Am. J. Cardiovasc. Dis. 4, 140–158 [PMC free article] [PubMed] [Google Scholar]
- 12. Tateishi K., Omata M., Tanaka K., Chiba T. (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 14. Su H., Li J., Menon S., Liu J., Kumarapeli A. R., Wei N., Wang X. (2011) Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ. Res. 108, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Su H., Li J., Osinska H., Li F., Robbins J., Liu J., Wei N., Wang X. (2013) The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ. Heart Fail. 6, 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogl A. M., Brockmann M. M., Giusti S. A., Maccarrone G., Vercelli C. A., Bauder C. A., Richter J. S., Roselli F., Hafner A. S., Dedic N., Wotjak C. T., Vogt-Weisenhorn D. M., Choquet D., Turck C. W., Stein V., Deussing J. M., Refojo D. (2015) Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat. Neurosci. 18, 239–251 [DOI] [PubMed] [Google Scholar]
- 17. Leidecker O., Matic I., Mahata B., Pion E., Xirodimas D. P. (2012) The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 11, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 18. Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L. R., Kurz T. (2012) Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka T., Kawashima H., Yeh E. T., Kamitani T. (2003) Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem. 278, 32905–32913 [DOI] [PubMed] [Google Scholar]
- 20. Liu S., Yang H., Zhao J., Zhang Y. H., Song A. X., Hu H. Y. (2013) NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex. J. Biol. Chem. 288, 31339–31349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanji K., Tanaka T., Mori F., Kito K., Takahashi H., Wakabayashi K., Kamitani T. (2006) NUB1 suppresses the formation of Lewy body-like inclusions by proteasomal degradation of synphilin-1. Am. J. Pathol. 169, 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richet E., Pooler A. M., Rodriguez T., Novoselov S. S., Schmidtke G., Groettrup M., Hanger D. P., Cheetham M. E., van der Spuy J. (2012) NUB1 modulation of GSK3β reduces Tau aggregation. Hum. Mol. Genet. 21, 5254–5267 [DOI] [PubMed] [Google Scholar]
- 23. Lu B., Al-Ramahi I., Valencia A., Wang Q., Berenshteyn F., Yang H., Gallego-Flores T., Ichcho S., Lacoste A., Hild M., Difiglia M., Botas J., Palacino J. (2013) Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat. Neurosci. 16, 562–570 [DOI] [PubMed] [Google Scholar]
- 24. Embade N., Fernández-Ramos D., Varela-Rey M., Beraza N., Sini M., Gutiérrez de Juan V., Woodhoo A., Martínez-López N., Rodríguez-Iruretagoyena B., Bustamante F. J., de la Hoz A. B., Carracedo A., Xirodimas D. P., Rodríguez M. S., Lu S. C., Mato J. M., Martínez-Chantar M. L. (2012) Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 55, 1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., Osinska H., Klevitsky R., Gerdes A. M., Nieman M., Lorenz J., Hewett T., Robbins J. (2001) Expression of R120G-αB-crystallin causes aberrant desmin and αB-crystallin aggregation and cardiomyopathy in mice. Circ. Res. 89, 84–91 [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Osinska H., Dorn G. W. 2nd, Nieman M., Lorenz J. N., Gerdes A. M., Witt S., Kimball T., Gulick J., Robbins J. (2001) Mouse model of desmin-related cardiomyopathy. Circulation 103, 2402–2407 [DOI] [PubMed] [Google Scholar]
- 27. Su H., Huang W., Wang X. (2009) The COP9 signalosome negatively regulates proteasome proteolytic function and is essential to transcription. Int. J. Biochem. Cell Biol. 41, 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu J. (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. Chapter 28, Unit 28.1 [DOI] [PubMed] [Google Scholar]
- 29. Zheng Q., Su H., Ranek M. J., Wang X. (2011) Autophagy and p62 in cardiac proteinopathy. Circ. Res. 109, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fahmi A., Smart N., Punn A., Jabr R., Marber M., Heads R. (2013) p42/p44-MAPK and PI3K are sufficient for IL-6 family cytokines/gp130 to signal to hypertrophy and survival in cardiomyocytes in the absence of JAK/STAT activation. Cell Signal. 25, 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Powell S. R., Davies K. J., Divald A. (2007) Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. J. Mol. Cell Cardiol. 42, 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanbe A., Osinska H., Saffitz J. E., Glabe C. G., Kayed R., Maloyan A., Robbins J. (2004) Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10132–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weekes J., Morrison K., Mullen A., Wait R., Barton P., Dunn M. J. (2003) Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics 3, 208–216 [DOI] [PubMed] [Google Scholar]
- 34. Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M. S. (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J., Powell S. R., Wang X. (2011) Enhancement of proteasome function by PA28α: overexpression protects against oxidative stress. FASEB J. 25, 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J., Horak K. M., Su H., Sanbe A., Robbins J., Wang X. (2011) Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J. Clin. Invest. 121, 3689–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 38. Hjerpe R., Thomas Y., Kurz T. (2012) NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 421, 27–29 [DOI] [PubMed] [Google Scholar]
- 39. Sandri M., Robbins J. (2014) Proteotoxicity: an underappreciated pathology in cardiac disease. J. Mol. Cell Cardiol. 71, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh R. K., Zerath S., Kleifeld O., Scheffner M., Glickman M. H., Fushman D. (2012) Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol. Cell Proteomics 11, 1595–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kamitani T., Kito K., Fukuda-Kamitani T., Yeh E. T. (2001) Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 276, 46655–46660 [DOI] [PubMed] [Google Scholar]
- 42. Hosono T., Tanaka T., Tanji K., Nakatani T., Kamitani T. (2010) NUB1, an interferon-inducible protein, mediates anti-proliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br. J. Cancer 102, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X., Robbins J. (2006) Heart failure and protein quality control. Circ. Res. 99, 1315–1328 [DOI] [PubMed] [Google Scholar]
- 44. Maejima Y., Kyoi S., Zhai P., Liu T., Li H., Ivessa A., Sciarretta S., Del Re D. P., Zablocki D. K., Hsu C. P., Lim D. S., Isobe M., Sadoshima J. (2013) Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 19, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]