Background: The IVVY motif and TRAF-binding sites in the RANK cytoplasmic domain initiate distinct modes of action to promote osteoclastogenesis.
Results: The ability of the IVVY motif to mediate osteoclast lineage commitment for osteoclastogenesis requires TRAF-binding sites.
Conclusion: The RANK IVVY motif cooperates with TRAF-binding sites to induce osteoclastogenesis.
Significance: This reveals a novel mechanism of RANK signaling in osteoclastogenesis.
Keywords: cell signaling, IL-1, osteoclast, TNF receptor-associated factor (TRAF), TNF, IVVY motif, RANK, RANKL
Abstract
Receptor activator of NF-κB (RANK) activation by RANK ligand (RANKL) mediates osteoclastogenesis by recruiting TNF receptor-associated factors (TRAFs) via three cytoplasmic motifs (motif 1, PFQEP369–373; motif 2, PVQEET559–564; and motif 3, PVQEQG604–609) to activate the NF-κB and MAPK signaling pathways. RANK also has a TRAF-independent motif (IVVY535–538), which is dispensable for the activation of TRAF-induced signaling pathways but essential for osteoclast lineage commitment by inducing the expression of nuclear factor of activated T-cells c1 (NFATc1) to regulate osteoclast gene expression. Notably, TNF/IL-1-mediated osteoclastogenesis requires RANK ligand assistance, and the IVVY motif is also critical for TNF/IL-1-mediated osteoclastogenesis by rendering osteoclast genes responsive to these two cytokines. Here we show that the two types of RANK cytoplasmic motifs have to be on the same RANK molecule to mediate osteoclastogenesis, suggesting a functional cooperation between them. Subsequent osteoclastogenesis assays with TNF or IL-1 revealed that, although all three TRAF motifs play roles in TNF/IL-1-mediated osteoclastogenesis, motifs 2 and 3 are more potent than motif 1. Accordingly, inactivation of motifs 2 and 3 blocksTNF/IL-1-mediated osteoclastogenesis. Mechanistically, double mutation of motifs 2 and 3, similar to inactivation of the IVVY motif, abrogates the expression of nuclear factor of activated T-cells c1 and osteoclast genes in assays reflecting RANK-initiated and TNF/IL-1-mediated osteoclastogenesis. In contrast, double inactivation of motifs 2 and 3 did not affect the ability of RANK to activate the NF-κB and MAPK signaling pathways. Collectively, these results indicate that the RANK IVVY motif cooperates with the TRAF-binding motifs to promote osteoclastogenesis, which provides novel insights into the molecular mechanism of RANK signaling in osteoclastogenesis.
Introduction
Osteoclasts, the sole bone-resorbing cells, play a critical role in skeleton development and normal bone homeostasis (1). Furthermore, osteoclasts are implicated in the pathogenesis of various bone diseases, including postmenopausal osteoporosis (2) and bone loss in inflammatory bone conditions such as rheumatoid arthritis and periodontitis (3). Osteoclasts are multinucleated giant cells that are derived from mononuclear cells of the monocyte/macrophage lineage upon stimulation by the monocyte/macrophage colony-stimulating factor (M-CSF)2 and RANKL (4). Although M-CSF stimulates the proliferation and survival of osteoclast precursors (5), RANKL drives osteoclast differentiation and regulates the survival and activation of mature osteoclasts (4).
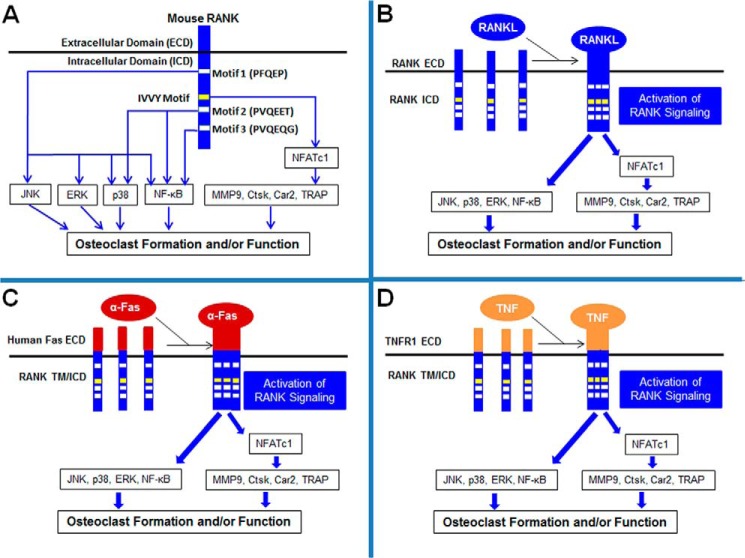
RANKL, a member of the TNF superfamily (6, 7), exerts its effects by activating its receptor, RANK, on the cell surface of osteoclast precursors. RANK belongs to the TNF receptor superfamily (7). RANK contains three functional motifs, PFQEP369–373 (motif 1), PVQEET559–564 (motif 2), and PVQEQG604–609 (motif 3), that recruit TRAF proteins to promote osteoclastogenesis (8–10) (Fig. 1A). However, these three TRAF-binding sites activate distinct sets of signaling pathways during osteoclastogenesis. Specifically, in response to RANK activation from RANKL stimulation, motif 1 activates the NF-κB and three MAPK pathways (JNK, ERK, and p38) in bone marrow macrophages (BMMs), which are primary osteoclast precursors (9). In contrast, motif 2 activates the NF-κB and p38 pathways, and motif 3 only activates the NF-κB pathway in BMMs (9). Collectively, these three RANK TRAF motifs trigger the activation of the NF-κB, JNK, ERK, and p38 pathways (Fig. 1A) which play important roles in osteoclast formation and/or function (10).
FIGURE 1.
RANK signaling pathways in osteoclasts and schematic of the chimeric receptor systems for studying RANK signaling pathways in osteoclasts. A, RANK motifs and their modes of action in osteoclast formation and/or function. B, model showing how RANKL activates its receptor, RANK, to transduce signaling pathways for osteoclast formation and/or function. C, schematic of the chimeric receptor containing the human Fas external domain linked to the transmembrane and cytoplasmic domains of mouse RANK, which can be stimulated by α-Fas to activate RANK signaling for osteoclast formation and/or function. D, schematic of the chimeric receptor consisting of the mouse TNFR1 external domain linked to the transmembrane and cytoplasmic domains of mouse RANK, which can be stimulated by TNF to activate RANK signaling for osteoclast formation and/or function. TM, transmembrane domain.
Moreover, our group further identified a TRAF-independent motif (IVVY535–538) in the RANK cytoplasmic domain by carrying out a detailed structure/function study and showed that the IVVY motif plays an essential role in osteoclastogenesis by committing BMMs to the osteoclast lineage (11) (Fig. 1A). The importance of the IVVY motif in osteoclastogenesis is supported further by clinical data revealing that truncating mutations causing the loss of a RANK region containing the IVVY motif has resulted in osteopetrosis in humans (12). Mechanistically, our group and others showed that the IVVY motif, unlike the TRAF motifs, plays a dispensable role in the activation of the NF-κB, JNK, ERK, and p38 pathways (11, 13, 14). However, the IVVY motif is critical for inducing the expression of nuclear factor of activated T-cells c1 (NFATc1) (15, 16) and the subsequent induction of the osteoclast genes encoding matrix metalloproteinase 9 (MMP9), carbonic anhydrase 2 (Car2), cathepsin K (Ctsk), and tartrate-resistant acid phosphatase (TRAP) during osteoclastogenesis (17) (Fig. 1A).
TNF and IL-1 are two proinflammatory cytokines that play crucial roles in immune and inflammatory responses (18, 19) and are also implicated in the bone loss associated with various pathological conditions, such as postmenopausal osteoporosis (20), rheumatoid arthritis (21), and periodontitis (22). Notably, although IL-1 and TNF can activate TRAF-dependent signaling pathways through activation of their receptors (18, 19), they are unable to stimulate osteoclastogenesis independently of RANKL (23–26), indicating that RANK signaling is required for TNF/IL-1-mediated osteoclastogenesis. Importantly, we have shown recently that the RANK IVVY motif is critical for TNF/IL-1-mediated osteoclastogenesis by appointing osteoclast genes to a TNF/IL-1-inducible state (15, 17). We have further demonstrated that although IL-1 alone is unable to induce the expression of NFATc1 and osteoclast genes (Cstk, TRAP, MMP9, and Car2), it can do so with the assistance of RANKL, whose effect is drastically abrogated with the inactivation of the IVVY motif (15). Notably, it has also been reported that inhibitory peptide targeting RANK-IVVY-induced signaling blocks inflammatory bone loss and protects against bone loss in ovariectomy (13). Taken together, these findings establish an essential role for the IVVY motif in osteoclastogenesis by promoting osteoclast lineage priming by inducing the expression of NFATc1 and osteoclast genes despite playing a dispensable role in the activation of most of the known osteoclast signaling pathways.
In this study, we investigated the functional relationship between the RANK IVVY motif and TRAF-functional sites in osteoclastogenesis. Moreover, we examined whether RANK TRAF sites play roles in inducing osteoclast lineage commitment for TNF/IL-1-mediated osteoclastogenesis. This study uncovered a novel RANK signaling mechanism in osteoclastogenesis.
Experimental Procedures
Chemical and Biological Reagents
All chemicals were from Sigma. Synthetic oligonucleotides were purchased from Sigma-Genosys (Woodlands, TX). Recombinant mouse IL-1α (catalog no. 400-ML-005) and recombinant mouse TNF-α (TNF, catalog no. 410-TRNC-050) were purchased from R&D Systems (Minneapolis, MN). Anti-human Fas-activating antibody (α-Fas) was from Millipore (Temecula, CA). Anti-human Fas (catalog no. sc-21730PE) and mouse anti-TNF receptor 1 (TNFRI) (catalog no. sc-12746PE) antibodies conjugated with phycoerythrin were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Recombinant GST-RANKL was prepared as described previously (11). The anti-NFATc1 (catalog no. sc-7294) antibody was from Santa Cruz Biotechnology, Inc., and antibodies against p44/42ERK (catalog no. 9102), phospho-p44/42ERK (catalog no. 9101s), JNK (catalog no. 9252), phospho-JNK (catalog no. 9251s), p38 (catalog no. 9212), phospho-p38 (catalog no. 9211s), Iκβα (catalog no. 9242), and phospho-IκBα (catalog no. 2859s) were obtained from Cell Signaling Technology, Inc. (Beverly, MA). Mouse M-CSF was generated from the M-CSF-producing cell line CMG14-12 (27).
Preparation of the Retrovirus Encoding the Chimeras
The retrovirus 293GPG-packaging cell line was cultured in DMEM containing 10% heat-inactivated FBS supplemented with tetracycline, penicillin/streptomycin, puromycin, and G418 as described previously (28). The chimeric receptor constructs pMX-Neo-hFas-mRANK-IVVY (hFas-mIVVY), pMX-puro-TNFR1-mRANK-M123 (TNFR-mTRAF), pMX-puro-hFas-mRANK-M123 (hFas-mTRAF), pMX-puro-hFas-mRANK-M1 (hFas-M1), pMX-puro-hFas-mRANK-M2 (hFas-M2), pMX-puro-hFas-mRANK-M3 (hFas-M3), pMX-puro-hFas-mRANK-M12 (hFas-M12), pMX-puro-hFas-mRANK-M13 (hFas-M13), and pMX-puro-hFas-mRANK-M23 (hFas-M23) were prepared as indicated in previous studies (9, 11, 29). The pMX-puro-GFP (GFP), pMX-puro-hFas-RANK (hFas-WT), and pMX-puro-TNFR1-RANK (TNFR-WT) constructs were generated as in previous studies (9, 11). 293GPG cells were transiently transfected with specific pMX constructs using Lipofectamine and Plus reagent (Invitrogen), and the virus supernatant was collected after 2, 3, and 4 days of transfection. Harvested retroviruses were used in the subsequent osteoclastogenesis assays.
In Vitro Osteoclastogenesis Assays
BMMs were isolated from long bones of 4- to 6-week-old C3H mice (Harlan Industries, Indianapolis, IN) for most of the assays or from TNF receptor-deficient mice (TNFR1−/−R2−/− mice) (The Jackson Laboratory, Bar Harbor, ME) for the assays involving double retroviral infection. BMMs were maintained in α-minimal essential medium (α-MEM) supplemented with 10% heat-inactivated FBS and M-CSF (220 ng/ml) (30) before infection with the retrovirus for 24 h in the presence of M-CSF (220 ng/ml) and 8 μg/ml Polybrene. Cells were further cultured with 220 ng/ml M-CSF and 2 μg/ml puromycin and/or 0.9 mg/ml G418 for selection and expansion. Selected BMMs (5 × 104 cells/well), seeded in a 24-well tissue culture dish, were treated with M-CSF (44 ng/ml) and different dosages of α-Fas with or without IL-1 (5 ng/ml) or TNF (5 ng/ml), as indicated in individual assays. Cultures were then stained for TRAP activity using a Leukacyte acid phosphatase kit (catalog no. 387-A, Sigma). These assays were performed in triplicate and repeated at least twice. The experiments involving mice were performed in accordance with the regulations of the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Flow Cytometric Analysis
1 × 106 BMMs suspended in 200 ml of α-MEM supplemented with 10% heat-inactivated FBS were blocked with 1 μg of 2.4G2 antibody for 30 min at 4 °C as described previously (17). Under dim light, 10 μl of human Fas or 20 μl of mouse TNFRI antibodies conjugated with phycoerythrin was added to each cell suspension, and cells were incubated for another 30 min at 4 °C. Cells were then centrifuged at 2000 rpm for 5 min and washed three times by gently resuspending cells with α-MEM, followed by centrifugation at 2000 rpm for 5 min. Cells were finally suspended in 1 ml of α-MEM for cytometric analysis using a BD LSR II flow cytometer at the University of Alabama at Birmingham Center for AIDS Research.
Semiquantitative RT-PCR Analysis
Total RNA from BMMs was isolated using TRIzol reagent (Invitrogen). 1 μg of total RNA was reverse-transcribed to cDNA with oligo(dT) in a 20-μl volume at 50 °C for 60 min using the SuperScript III RT-PCR system (Invitrogen) as described previously (15). Amplification of the MMP9, Cstk, TRAP, Car2, and GAPDH genes was performed as described previously (17). 20 μl of PCR product was loaded on 2% agarose gel for electrophoretic analysis. These RT-PCR assays were repeated twice independently.
Western Blot Analysis
BMMs were first cultured in serum-free α-MEM for 16 h and then treated with α-Fas as indicated in the individual experiments. Cells were then washed twice with ice-cold PBS and lysed in buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 mm NaF, 1× protease inhibitor mixture 1 (Sigma, catalog no. P-2850), and 1× protease inhibitor mixture 2 (Sigma, catalog no. P-5726). The cell lysate was submitted to Western blot analysis as described previously (15). Membranes were washed extensively, and an enhanced chemiluminescence detection assay was performed using a SuperSignal West Dura kit from Pierce. The experiment was repeated at least twice independently.
Statistical Analysis
Some osteoclastogenesis assays were quantified by counting the multinucleated TRAP-positive cells (>3 nuclei) in a representative area in each of three replicate samples. The results are shown as mean ± S.D. of the number or percentage of TRAP-positive cells. Statistical significance was determined using Student's t test, and p values of less than 0.05 were considered significant (*, p < 0.05; **, p < 0.001).
Results
The IVVY and TRAF Functional Motifs Need to Be on the Same RANK Molecule to Mediate Osteoclastogenesis
RANK is a member of the TNF receptor superfamily, which also includes TNFR and Fas (31). Members of this superfamily of receptors are activated by ligand-induced trimerization or oligomerization (32). Therefore, binding of RANK by RANKL leads to trimerization of RANK, which, in turn, activates the NF-κB, JNK, ERK, and p38 pathways and induces the expression of NFATc1 to regulate the expression of genes involved in osteoclast formation and/or function (Fig. 1B) (33). Previously, we developed two chimeric receptors for performing RANK structural/functional studies in osteoclastogenesis. The first chimera contains the human Fas external domain linked to the transmembrane and cytoplasmic domains of mouse RANK. This chimeric receptor system can be activated specifically to transmit RANK signaling by α-Fas as a surrogate for RANKL (Fig. 1C) (29). The second chimera consists of the mouse TNFR1 external domain linked to the transmembrane and cytoplasmic domains of mouse RANK. These chimeras can be activated to initiate RANK signaling by TNF as a surrogate for RANKL (Fig. 1D) (9).
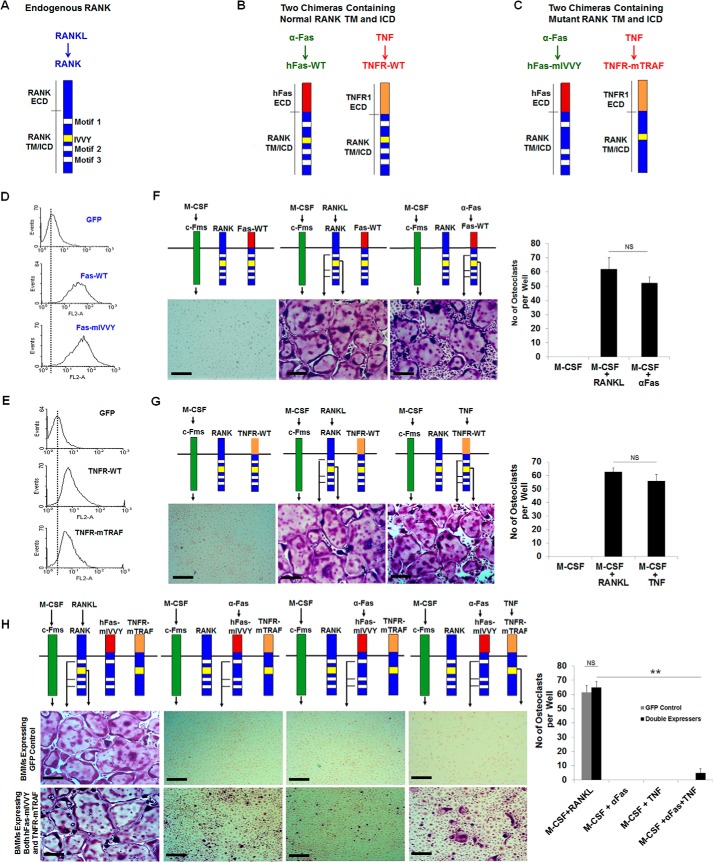
RANK structural/functional studies using the chimeric receptor approach have led to the identification of three TRAF-functional motifs (motif 1, motif 2, and motif 3) and a TRAF-independent IVVY motif in the RANK cytoplasmic domain (9, 11) (Fig. 2A). These two types of RANK motifs initiate distinct modes of action to promote osteoclastogenesis. Specifically, although these three TRAF sites collectively activate the NF-κB and MAPK signaling pathways, the IVVY motif induces the expression of NFATc1 to regulate the expression of osteoclast genes (Fig. 1A). Moreover, mutational inactivation of the IVVY motif does not affect the activation of the NF-κB and MAPK signaling pathways (11). These observations have raised the possibility that these two types of RANK motifs may act independently to initiate distinct modes of action to promote osteoclastogenesis. To address this possibility, we examined whether concurrent but independent activation of the TRAF-functional sites and IVVY motif in the same BMMs using the two different types of chimeric receptors can promote osteoclastogenesis. The two chimeras containing normal RANK transmembrane and intracellular domains, hFas-WT and TNFR-WT, were prepared in our previous studies and used as controls in this study (9, 29) (Fig. 2B). Here we further modified the hFas-WT and TNFR-WT chimeras to generate two chimeras containing mutated RANK intracellular domains: hFas-mIVVY and TNFR-mTRAF, respectively (Fig. 2C). hFas-mIVVY consists of the human Fas extracellular domain linked to the mouse RANK transmembrane and intracellular domains bearing an inactivating mutation in the IVVY motif but with normal TRAF sites. TNFR-mTRAF comprises the mouse TNFR1 extracellular domain linked to the mouse RANK transmembrane and intracellular domains bearing inactivating mutations in all three TRAF sites but with a normal IVVY motif (Fig. 2C). To select for BMMs expressing both hFas-mIVVY and TNFR-mTRAF, we engineered hFas-mIVVY and its corresponding control hFas-WT retroviral constructs with the G418-resistant gene, but TNFR-mTRAF and its corresponding control TNFR-WT retroviral constructs contain the puromycin-resistant gene.
FIGURE 2.
The IVVY motif and the TRAF sites need to be on the same RANK molecule to promote osteoclastogenesis. A, schematic of RANK. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. B, schematic of hFas-WT and TNFR-WT. C, schematic of hFas-mIVVY and TNFR-mTRAF. D, flow cytometric analysis of BMMs from TNFR1−/−R2−/− mice expressing GFP, hFas-WT, or hFas-mIVVY and TNFR-mTRAF. E, flow cytometric analysis of BMMs from TNFR1−/−R2−/− mice expressing GFP, TNFR-WT, or hFas-mIVVY and TNFR-mTRAF. F, BMMs expressing hFas-WT were treated with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and α-Fas (100 ng/ml) for 5 days. G, BMMs expressing TNFR-WT were treated with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (100 ng/ml) for 5 days. H, BMMs expressing GFP control or hFas-mIVVY and TNFR-mTRAF were treated with M-CSF (44 ng/ml) and RANKL (100 ng/ml), M-CSF (44 ng/ml) and α-Fas (100 ng/ml), M-CSF (44 ng/ml) and TNF (100 ng/ml), or M-CSF (44 ng/ml) and TNF (100 ng/ml) plus α-Fas (100 ng/ml) for 5 days. The cultures were stained for TRAP activity on day 5. Quantification of the osteoclast formation assays in F–H is shown in the right panels as the mean number of multinucleated TRAF-positive cells (>3 nuclei) per well. Data are mean ± S.D. **, p < 0.001; NS, not significant. Scale bars = 200 μm.
BMMs were doubly infected with retroviruses encoding hFas-mIVVY as well as TNFR-mTRAF and cultured with G418 and puromycin to select cells expressing both chimeras. Given that TNFR-mTRAF consists of the mouse TNFR1 external domain and is activated by TNF, BMMs from TNFR1−/−R2−/− mice were used to avoid cross-signaling through endogenous TNF receptors, as in our previous studies (9, 34). As controls, TNFR1−/−R2−/− BMMs were infected with retroviruses encoding GFP, hFas-WT, or TNFR-WT and selected with a single antibiotic. Cell surface expression of hFas-mIVVY and TNFR-mTRAF on doubly infected BMMs and that of hFas-WT or TNFR-WT on singly infected BMMs was determined by flow analysis (Fig. 2, D and E). Cell surface expression levels of hFas-mIVVY (Fig. 2D) and TNFR-mTRAF (Fig. 2E) on doubly infected BMMs were comparable with those of hFas-WT or TNFR-WT, respectively, on singly infected BMMs. Moreover, BMMs expressing hFas-WT generated numerous osteoclasts when treated with M-CSF and α-Fas (Fig. 2F), and TNFR-WT-expressing cells formed many osteoclasts in response to M-CSF and TNF stimulation (Fig. 2G), indicating that the expression levels of hFas-mIVVY and TNFR-mTRAF on doubly infected BMMs were sufficient to be functional.
Next we determined whether BMMs expressing hFas-mIVVY and TNFR-mTRAF could form osteoclasts when stimulated with α-Fas to activate hFas-mIVVY and TNF to activate TNFR-mTRAF. As positive controls, BMMs expressing GFP or both hFas-mIVVY and TNFR-mTRAF formed numerous osteoclasts when treated with M-CSF and RANKL (Fig. 2H). However, these cells failed to form osteoclasts when stimulated with M-CSF plus α-Fas or M-CSF plus TNF, replicating our previous findings that both the IVVY and TRAF motifs are essential for osteoclastogenesis (9, 11). Interestingly, when BMMs expressing hFas-mIVVY and TNFR-mTRAF were treated with M-CSF and TNF plus α-Fas, the cells formed only few small osteoclasts (Fig. 2H). These results indicate that the two types of RANK motifs have to be on the same RANK molecule to efficiently promote osteoclastogenesis.
The RANK TRAF Sites Are Involved in TNF-/IL-1-mediated Osteoclastogenesis
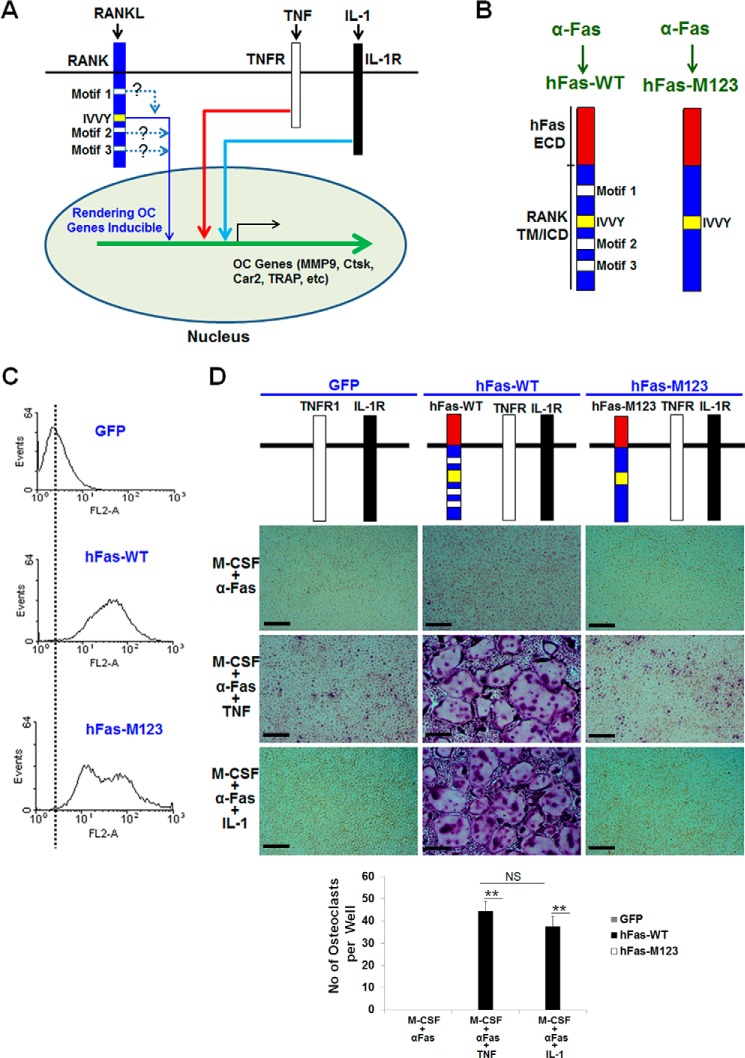
Although TNF or IL-1 alone cannot mediate osteoclastogenesis, they can stimulate osteoclastogenesis with permissive levels of RANKL, indicating that TNF/IL-1-mediated osteoclastogenesis requires prior commitment of osteoclast precursors to the osteoclast lineage (23–26). The permissive levels of RANKL required for TNF/IL-1-mediated osteoclastogenesis is about one-tenth of the optimal RANKL dosage (100 ng/ml) utilized for standard in vitro osteoclastogenesis assays involving M-CSF and RANKL treatment (23–26). Mechanistically, we have reported that RANKL-evoked TNF/IL-1-mediated osteoclastogenesis involves the activation of the IVVY motif, which, in turn, renders osteoclast genes in a TNF/IL-1 inducible state (15, 17) (Fig. 3A). To further address the potential cooperation between the RANK IVVY motif and the TRAF sites in osteoclastogenesis, we extended our study to investigate whether the three TRAF-functional sites are also involved in committing BMMs to the osteoclast lineage for TNF/IL-1-mediated osteoclastogenesis (Fig. 3A). We reasoned that if the TRAF sites functionally collaborate with the IVVY motif, then mutational inactivation of these sites should block TNF/IL-1-mediated osteoclastogenesis by abrogating the ability of the IVVY motif to initiate its mode of action, namely, rendering osteoclast genes responsive to TNF or IL-1, and, thereby, blunt the osteoclast lineage. To this end, we used hFas-WT and also prepared another chimeric receptor, hFas-M123, which consists of the human Fas external domain linked to RANK transmembrane and cytoplasmic domains bearing inactivating mutations in all three TRAF sites but with a normal IVVY motif (Fig. 3B). Importantly, the use of hFas-WT and hFas-M123 eliminates the need for TNFR1−/−R2−/− BMMs because wild-type BMMs expressing endogenous TNFR and IL-1R are required for assays aimed at studying the roles of TNF and IL-1 in osteoclastogenesis. BMMs expressing comparable cell surface levels of hFas-WT or hFas-M123 as assessed by flow cytometry analysis (Fig. 3C) were treated with M-CSF and 10 ng/ml of α-Fas or M-CSF and 10 ng/ml of α-Fas plus TNF or IL-1 to promote osteoclastogenesis (Fig. 3D). 10 ng/ml α-Fas was used for these assays because the concentration is about one-tenth of the α-Fas dose, which is capable of promoting optimal osteoclastogenesis from BMMs expressing hFas-WT (29). Although BMMs expressing hFas-WT failed to form osteoclasts when treated with M-CSF and 10 ng/ml of α-Fas, these cells gave rise to many osteoclasts when stimulated with M-CSF and 10 ng/ml of α-Fas plus TNF or IL-1 (Fig. 3D). However, BMMs expressing hFas-M123 were unable to generate osteoclasts when treated with M-CSF and 10 ng/ml of α-Fas plus TNF or IL-1. These results indicate that one or more of the three RANK TRAF-functional sites are also required for TNF/IL-1-mediated osteoclastogenesis.
FIGURE 3.
Mutation of the RANK TRAF-functional motifs blocks TNF/IL-1-mediated osteoclastogenesis. A, roles of the RANK motifs in TNF/IL-1-mediated osteoclastogenesis. OC, osteoclast. B, schematic of hFas-WT and hFas-M123. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. C, flow cytometric analysis of BMMs expressing GFP, hFas-WT, or hFas-M123. D, BMMs expressing GFP, hFas-WT, or hFas-M123 were treated with M-CSF (44 ng/ml) and α-Fas (10 ng/ml), M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus TNF (5 ng/ml), or M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus IL-1 (5 ng/ml) for 5 days. BMMs expressing GFP or hFas-WT were used as negative and positive controls, respectively. The cultures were stained for TRAP activity at the end of the assays. Quantification of the osteoclast formation assays is shown in the bottom panel as the mean number of multinucleated TRAF-positive cells (>3 nuclei) per well. Data are mean ± S.D. **, p < 0.001; NS, not significant. Scale bars = 200 μm.
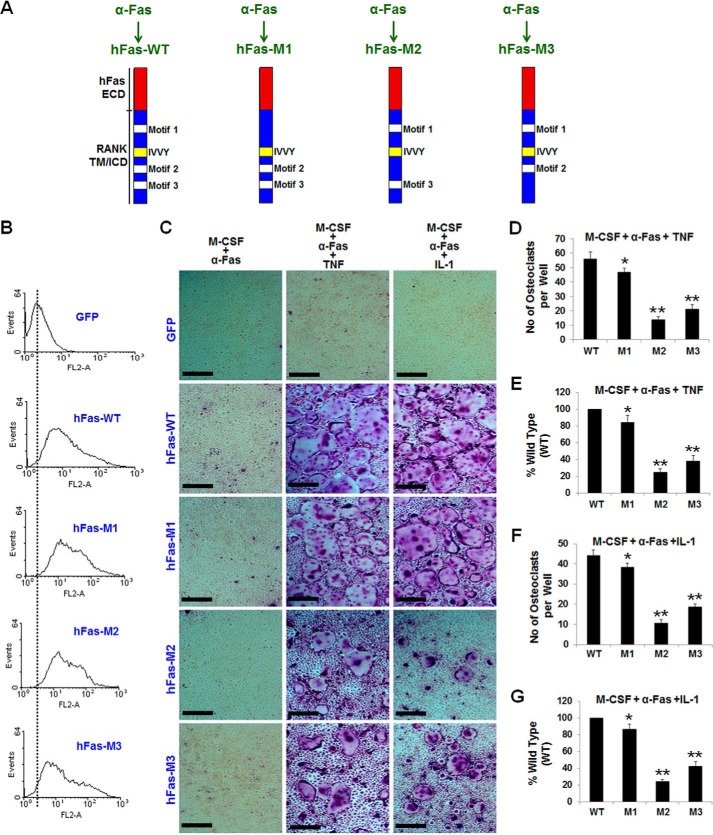
We next sought to delineate which one of the three RANK TRAF-functional sites is involved in TNF/IL-1-mediated osteoclastogenesis. To do so, we prepared three more chimeras that contain the human Fas external domain linked to mouse RANK transmembrane and intracellular domains bearing inactivating mutations in motif 1 (hFas-M1), motif 2 (hFas-M2), or motif 3 (hFas-M3) (Fig. 4A). Flow cytometry analysis showed that BMMs expressing hFas-WT, hFas-M1, hFas-M2, or hFas-M3 exhibited comparable cell surface levels (Fig. 4B). Although BMMs expressing WT formed numerous osteoclasts when treated with M-CSF and 10 ng/ml α-Fas plus TNF or IL-1 (Fig. 4C), cells expressing hFas-M1, hFas-M2, or hFas-M3 formed 15.7%, 75%, or 62% fewer osteoclasts, respectively, compared with hFas-WT expressers treated with M-CSF and 10 ng/ml α-Fas plus TNF (Fig. 4, C–E) and 13.4%, 76%, or 57.7% fewer osteoclasts, respectively, compared with hFas-WT expressers treated with M-CSF and 10 ng/ml α-Fas plus IL-1 (Fig. 4, C, F, and G). These data demonstrate that, although all three RANK TRAF-functional sites play a role in TNF/IL-1-mediated osteoclastogenesis, motif 2 and motif 3 are more potent than motif 1 in mediating TNF/IL-1-induced osteoclastogenesis.
FIGURE 4.
The RANK functional TRAF motifs exert different effects on TNF/IL-1-mediated osteoclastogenesis. A, schematic of hFas-WT, hFas-M1, hFas-M2, and hFas-M3. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. B, flow cytometric analysis of BMMs expressing GFP, hFas-WT, hFas-M1, hFas-M2, or hFas-M3. C, BMMs expressing GFP, hFas-WT, hFas-M1, hFas-M2, or hFas-M3 were treated with M-CSF (44 ng/ml) and α-Fas (10 ng/ml), M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus TNF (5 ng/ml), or M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus IL-1 (5 ng/ml) for 5 days. BMMs expressing GFP or hFas-WT were used as negative and positive controls, respectively. The cultures were stained for TRAP activity on day 5. Scale bars = 200 μm. D and E, quantification of the osteoclastogenesis assays induced by TNF is shown in mean number (D) or mean percentage (E) of multinucleated TRAP-positive cells (<3 nuclei) per well. F and G, quantification of the osteoclastogenesis induced by IL-1 is shown in mean number (F) or mean percentage (G) of multinucleated TRAP-positive cells (>3 nuclei) per well. Data are mean ± S.D. *, p < 0.05; **, p < 0.001.
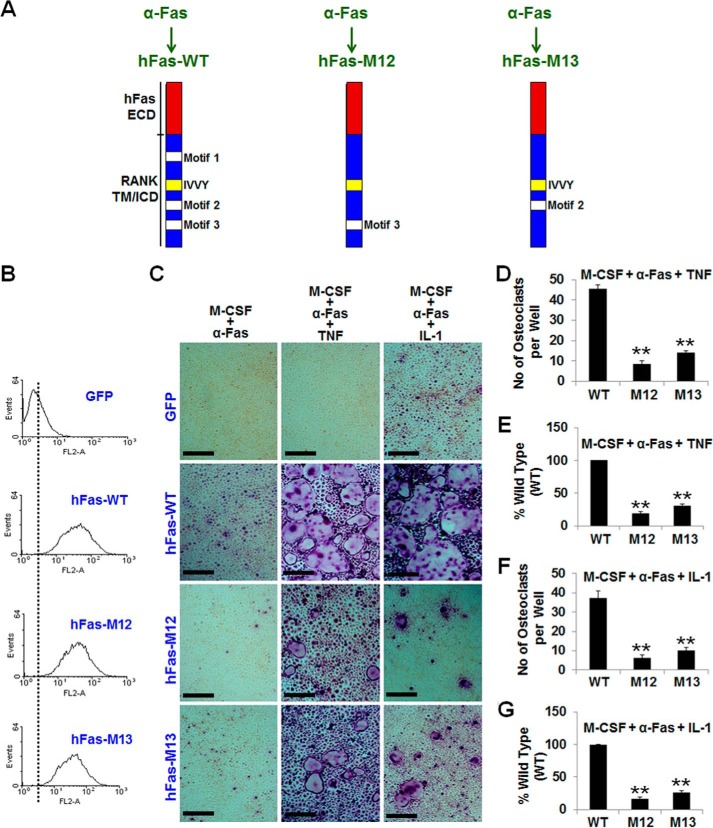
Double Mutation of Motif 1 with Motif 2 or Motif 3 Results in a Partial Reduction in TNF/IL-1-mediated Osteoclastogenesis
To further examine the contribution of the three TRAF-functional sites to TNF/IL-1-mediated osteoclastogenesis, we investigated the impact of mutating two of these three motifs in this osteoclastogenic setting. We engineered hFas-M12 and hFas-M13, which consist of the human Fas extracellular domain linked to mouse RANK transmembrane and intracellular domains bearing a double inactivation of motif 1 and motif 2 (hFas-M12) or motif 1 and motif 3 (hFas-M13) (Fig. 5A). Flow cytometry analysis showed that BMMs expressing hFas-WT, hFas-M12, or hFas-M13 exhibited comparable cell surface levels of these chimeras (Fig. 5B). As expected, the hFas-WT expressers formed many osteoclasts when treated with M-CSF and α-Fas (10 ng/ml) plus TNF or IL-1 (Fig. 5C). However, BMMs expressing hFas-M12 or hFas-M13 gave rise to 80.9% or 69.1% fewer osteoclasts, respectively, compared with hFas-WT expressers treated with M-CSF and α-Fas (10 ng/ml) plus TNF (Fig. 5, D and E) and 83.2% or 73.3% fewer osteoclasts, respectively, compared with hFas-WT expressers stimulated by M-CSF and α-Fas (10 ng/ml) plus IL-1 (Fig. 5, F and G). These data reveal that double mutation of motif 1 with motif 2 or motif 3 markedly inhibited TNF/IL-1-mediated osteoclastogenesis.
FIGURE 5.
Double mutation of motif 1 with motif 2 or motif 3 results in a partial reduction in TNF/IL-1-mediated osteoclastogenesis. A, schematic of hFas-WT, hFas-M12, and hFas-M13. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. B, flow cytometric analysis of BMMs expressing GFP, hFas-WT, hFas-M12, or hFas-M13. C, BMMs expressing GFP, hFas-WT, hFas-M12, or hFas-M13 were treated with M-CSF (44 ng/ml) and α-Fas (10 ng/ml), M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus TNF (5 ng/ml), or M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus IL-1 (5 ng/ml) for 5 days. BMMs expressing GFP or hFas-WT were used as negative and positive controls, respectively. The cultures were stained for TRAP activity on day 5. Scale bars = 200 μm. D and E, quantification of the osteoclastogenesis assays induced by TNF is shown in mean number (D) or mean percentage (E) of multinucleated TRAP-positive cells (<3 nuclei) per well. F and G, quantification of the osteoclastogenesis assays induced by IL-1 is shown in mean number (F) or mean percentage (G) of multinucleated TRAP-positive cells (>3 nuclei) per well. Data are mean ± S.D. *, p < 0.05; **, p < 0.001.
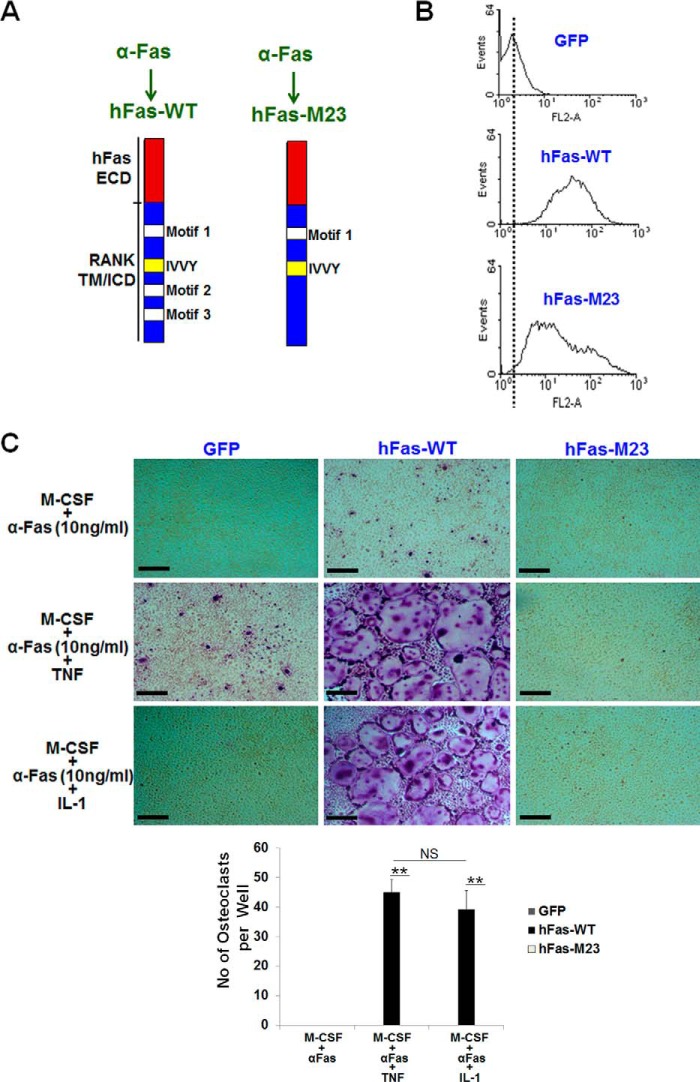
Mutation of Motif 2 and Motif 3 Blocks TNF/IL-1-mediated Osteoclastogenesis
Given that inactivation of motif 2 or motif 3 alone resulted in more than a 70% or 55% reduction, respectively, in TNF/IL-1-mediated osteoclastogenesis compared with the hFas-WT control (Fig. 4), we reasoned that mutation of both motif 2 and motif 3 might lead to a drastic reduction in TNF/IL-1-mediated osteoclastogenesis. To address this possibility, we generated a chimeric receptor consisting of the human Fas extracellular domain linked to mouse RANK transmembrane and intracellular domains bearing a double mutation in both motif 2 and motif 3 (hFas-M23) (Fig. 6A). Flow cytometry analysis showed that BMMs expressing hFas-WT or hFas-M23 exhibited comparable cell surface levels of the chimeras (Fig. 6B). BMMs expressing hFas-WT formed many osteoclasts when treated with M-CSF and α-Fas (10 ng/ml) plus TNF or IL-1, but the hFas-M23 expressers formed no osteoclasts under these treatment conditions (Fig. 6C), indicating that double inactivation of motif 2 and motif 3 leads to complete inhibition of TNF/IL-1-mediated osteoclastogenesis.
FIGURE 6.
Mutation of motif 2 and motif 3 blocks TNF/IL-1-mediated osteoclastogenesis. A, schematic of hFas-WT and hFas-M23. ECD, extracellular domain; TM, transmembrane domain; ICD, intracellular domain. B, flow cytometric analysis of BMMs expressing GFP, hFas-WT, or hFas-M23. C, BMMs expressing GFP, hFas-WT, or hFas-M23 were treated with M-CSF (44 ng/ml) and α-Fas (10 ng/ml), M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus TNF (5 ng/ml), or M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus IL-1 (5 ng/ml) for 5 days. BMMs expressing GFP or hFas-WT were used as negative and positive controls, respectively. The cultures were then stained for TRAP activity on day 5. Quantification of the osteoclast formation assays is shown under the images as mean number of multinucleated TRAF-positive cells (>3 nuclei) per well. Data are mean ± S.D. **, p < 0.001; NS, not significant. Scale bars = 200 μm.
Motif 2 and Motif 3 Cooperate with the IVVY Motif to Induce NFATc1 Expression to Activate Gene Expression to Promote Osteoclastogenesis
Having demonstrated that double mutation of motif 2 and motif 3 blocks TNF/IL-1-mediated osteoclastogenesis, we then focused on motif 2 and motif 3 to decipher the mechanism by which the TRAF sites cooperate with the IVVY motif to induce osteoclastogenesis. Toward this end, we first examined whether dual mutations of motif 2 and motif 3 affect the ability of RANK to activate the NF-κB, JNK, p38, and ERK pathways using the hFas-WT and hFas-M23 chimeras (Fig. 6A). As shown in Fig. 7, the capacity of hFas-M23 to activate the NF-κB, JNK, p38, and ERK pathways is comparable with that of hFas-WT, indicating that double inactivation of motif 2 and motif 3 does not affect the ability of RANK to activate the NF-κB, JNK, p38, and ERK signaling pathways. This is consistent with our previous findings that motif 1 alone is sufficient to activate these signaling pathways (9) (Fig. 1A).
FIGURE 7.
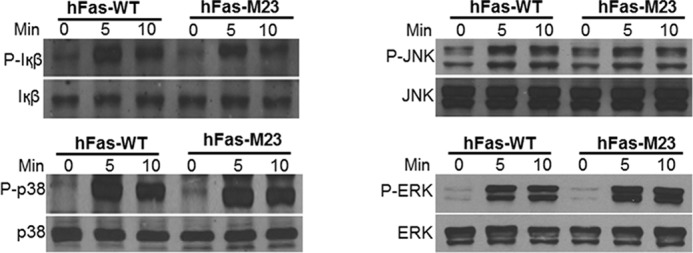
Double inactivation of motif 2 and motif 3 does not affect the ability of RANK to activate the NF-κB, JNK, p38, and ERK pathways. BMMs expressing hFas-WT or hFas-M23 were treated with α-Fas (100 ng/ml) for 0, 5, or 10 min as indicated. The activation of the NF-κB, JNK, p38, and ERK pathways was assessed as phosphorylation of IκB, JNK, p38, and ERK by Western blot analysis. BMMs expressing hFas-WT were used as a positive control.
In examining this notion, we next sought to investigate whether mutations of these two TRAF sites affect the ability of the IVVY motif to initiate its mode of action in osteoclastogenesis; namely, IVVY motif-mediated NFATc1 expression (15, 16) and the subsequent induction of osteoclast genes (17) (Fig. 1A). In line with our recent report on the role of the IVVY motif in inducing NFATc1 expression, we revealed that double inactivation of motif 2 and motif 3 drastically impaired the ability of hFas-M23 to induce NFATc1 expression despite the presence of a normal IVVY motif (Fig. 8A). On the basis of this finding, we investigated the roles of motif 2 and motif 3 in the expression of the aforementioned osteoclast genes using BMMs expressing hFas-WT or hFas-M23 (Fig. 8B). Treatment of BMMs expressing hFas-WT or hFas-M23 with M-CSF and 10 ng/ml of α-Fas failed to stimulate the expression of MMP9, Cstk, Car2, and TRAP, replicating our previous finding that permissive RANK activation is insufficient to induce the expression of osteoclast genes (17). Furthermore, although M-CSF and 10 ng/ml of α-Fas plus TNF or IL-1 up-regulated the expression of these osteoclast genes in BMMs expressing hFas-WT, the same treatment failed to stimulate gene expression in hFas-M23 expressers (Fig. 8B). These results demonstrate that motif 2 and motif 3 are required for inducing osteoclast genes in TNF/IL-1-mediated osteoclastogenesis.
FIGURE 8.
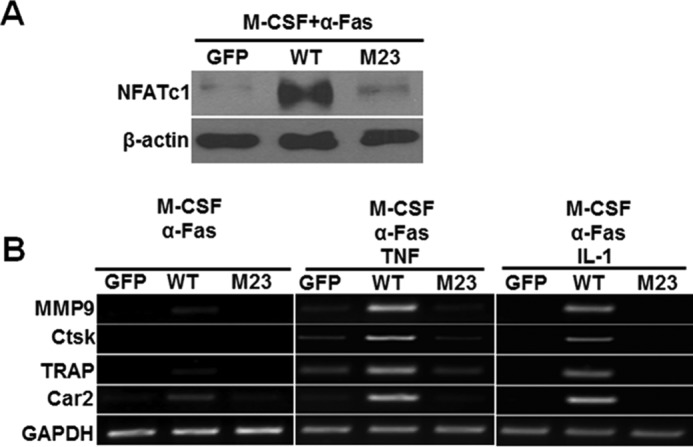
Inactivation of motif 2 and motif 3 drastically impairs the ability of RANK to up-regulate NFATc1 expression and abrogates the capacity of TNF/IL-1 to activate the expression of osteoclast genes. A, effects of inactivation of motif 2 and motif 3 on NFATc1 expression. BMMs expressing GFP, hFas-WT (WT) or hFas-M23 (M23) were treated with M-CSF (44 ng/ml) and α-Fas (100 ng/ml) for 3 days. The expression of NFATc1 was assessed by Western blot analysis using β-actin as a loading control. BMMs expressing GFP or WT were used as negative and positive controls, respectively. B, effects of inactivation of motif 2 and motif 3 on the expression of osteoclast genes. BMMs expressing GFP, hFas-WT, or hFas-M23 were treated with M-CSF (44 ng/ml) and α-Fas (10 ng/ml), M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus TNF (5 ng/ml), or M-CSF (44 ng/ml) and α-Fas (10 ng/ml) plus IL-1 (5 ng/ml) for 3 days. BMMs expressing GFP or WT were used as negative and positive controls, respectively. Gene expression was determined by semiquantitative RT-PCR using GAPDH as a loading control.
Discussion
The objective of this work is to investigate the functional relationship of the two types of motifs in the RANK cytoplasmic domain in osteoclastogenesis. We have shown previously that inactivation of the IVVY motif exerts no impact on the activation of the NF-κB, ERK, JNK, and p38 signaling pathways in BMMs stimulated by RANKL (11). Furthermore, the IVVY motif is not involved in the activation of the NF-κB, ERK, JNK, and p38 signaling pathways because mutant RANK bearing inactivating mutations in all three TRAF sites, but with a functional IVVY motif, is unable to activate any of these signaling pathways (9). Our recent work has revealed that the IVVY motif is also essential for TNF/IL-1-induced expression of osteoclast genes and, thereby, allows these cytokines to promote osteoclastogenesis (15, 17). On the basis of these findings, we initially hypothesized that the TRAF sites and IVVY motif in the RANK cytoplasmic domain act independently to initiate distinct modes of action to promote osteoclastogenesis. However, when these two types of motifs were activated in the same cells using different chimeras in which either the TRAF sites or the IVVY motif were mutated, we found, unexpectedly, that the expressing cells could barely mediate osteoclastogenesis (Fig. 2H). This intriguing finding indicates that the TRAF-functional sites and the IVVY motif have to be in the same RANK molecule to be functional, suggesting that the TRAF sites cooperate with the IVVY motif to promote osteoclastogenesis.
Given the complex nature of the experimental manipulation involving the chimeric receptor system, we felt that it was important to address the functional cooperation between the TRAF and IVVY motifs using different assays involving simpler experimental manipulation. Therefore, we expanded upon our previous findings on the role of the IVVY motif in TNF/IL-1-mediated osteoclastogenesis to investigate whether the TRAF motifs are also required for these osteoclastogenesis assays. Specifically, these assays aimed to address whether the three RANK TRAF-functional sites function with the IVVY motif to mediate the commitment of BMMs into the osteoclast lineage, a prerequisite for TNF/IL-1-mediated osteoclastogenesis (15, 17). We revealed that these TRAF sites are critical for TNF/IL-1-mediated osteoclastogenesis (Fig. 3), which further supports the functional cooperation between the TRAF sites and the IVVY motif. However, we noted that the TRAF sites contribute to TNF/IL-1-mediated osteoclastogenesis to different extents because mutation of motif 1, motif 2, or motif 3 leads to roughly 13%, 75%, or 58% reduction, respectively, in osteoclast numbers (Fig. 4). In other words, motif 2 and motif 3 are much more potent than motif 1 in promoting TNF/IL-1-mediated osteoclastogenesis. Consistently, double mutation of motif 2 and motif 3 blocked TNF/IL-1-mediated osteoclastogenesis (Fig. 6), whereas double mutations of motif 1 with either motif 2 or motif 3 only led to a partial reduction in these osteoclastogenesis assays (Fig. 5). Because of the ability of double inactivation of motif 2 and motif 3 in blocking TNF/IL-1-mediated osteoclastogenesis, we focused on these two motifs to delineate the molecular mechanism underlying the functional cooperation between the TRAF sites and IVVY motifs in mediating TNF/IL-1-mediated osteoclastogenesis. We showed that inactivation of motif 2 and motif 3 drastically impaired the ability of RANK to induce NFATc1 expression and, consequently, abrogated the capacity of TNF/IL-1 to up-regulate osteoclast genes, which affects their ability to promote osteoclastogenesis with RANK activation (Fig. 8). These findings support the notion that motif 2 and motif 3 cooperate with the IVVY motif to up-regulate NFATc1 expression, which then regulates the osteoclast gene for osteoclastogenesis.
On the basis of the findings from this work, we propose that the functional cooperation between motif 2/motif 3 and the IVVY motif is mediated by the direct or indirect effect(s) of TRAF proteins binding to these two TRAF sites (protein Y and protein Z, Fig. 9) on the protein recruited by the IVVY motif (protein X, Fig. 9). It is likely that proteins X, Y, and Z form a signaling complex via direct or indirect interactions to mediate gene expression and, thereby, induce osteoclast lineage commitment. This notion is consistent with our finding that these two types of RANK motifs have to be on the same molecule to be functional. This is further supported by the fact that motif 2 (559–564) and motif 3 (604–609) are about 21 and 66 amino acid residues apart, respectively, from the IVVY motif. Given that the inactivation of the IVVY motif does not affect the activation of the TRAF-mediated signaling pathways in RANKL-stimulated BMMs (11), the functional cooperation between motif 2/motif 3 and the IVVY motif is likely to be unidirectional in that the function of the IVVY motif/protein X to up-regulate NFATc1 expression for the activation of osteoclast gene expression requires motifs 2/3 and protein Y/Z, but the capacity of motifs 2/3 and proteins Y/Z to activate the NF-κB and MAPK signaling pathways does not depend on the IVVY motif/protein X (Fig. 9).
FIGURE 9.
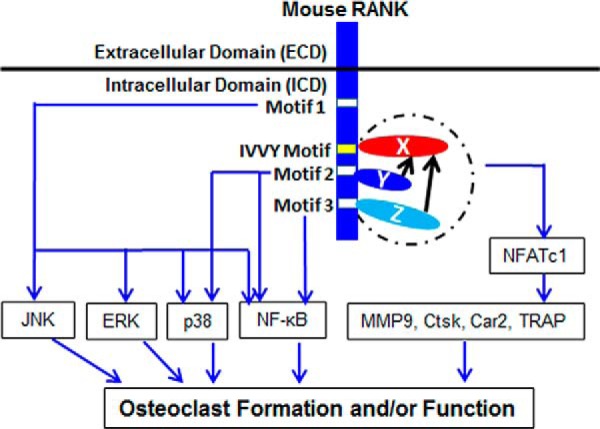
Proposed model for the molecular basis of the functional cooperation between motif 2/motif 3 and the IVVY motif. X, the protein binding to the IVVY motif to mediate osteoclast lineage commitment; Y, the TRAF protein binding to motif 2; Z, the TRAF protein binding to motif 3; dashed circle, the proposed formation of the signaling complex by proteins X, Y, and Z. Black arrows highlight the unidirectional nature of the functional cooperation between motif 2/motif 3 and the IVVY motif.
The IVYY motif has been shown to regulate the late stage of osteoclastogenesis by recruiting early estrogen-induced gene 1 (EEIG 1), which forms a complex with GRB2-associated binding protein 2 (Gab2), resulting in the activation of NFATc1 via the phospholipase Cγ2 (PLCγ2) signaling pathway (14, 16). Also, the IVVY motif can promote the function of mature osteoclasts through indirect interaction with Vav3 to regulate the osteoclast cytoskeleton (13). However, the protein that binds to the IVVY motif to regulate osteoclast lineage commitment has not yet been identified. Moreover, the TRAF proteins binding to motif 2 and motif 3 have not been identified convincingly. An early in vitro study showed that motif 2 interacts with TRAF3 and that motif 3 is capable of binding TRAF1, TRAF2, and TRAF5 (35). Another in vitro study showed that neither TRAF1 nor TRAF3 interacts with RANK (36). Therefore, a better understanding of the molecular mechanism underlying the functional cooperation between motif 2/motif 3 and the IVVY motif depends on the identification of the protein that binds to the IVVY motif to regulate osteoclast lineage commitment as well as the unambiguous elucidation of proteins binding to motif 2 and motif 3.
In conclusion, our data reveal, for the first time, that the IVVY and TRAF motifs of RANK do not function independently in mediating osteoclastogenesis. The function of the IVVY motif requires the presence of intact TRAF sites in the same RANK molecule. Moreover, this functional cooperation is critical for osteoclastogenesis and the induction of osteoclast genes. These findings provide novel insights into the mechanism by which the RANKL/RANK system regulates osteoclastogenesis and suggests that RANK signaling in osteoclasts is more complex than previously thought. Nevertheless, the precise molecular mechanism underlying the functional cooperation between the TRAF and IVVY motifs remains to be determined. As an important next step in addressing this issue, future research should be directed toward identifying the protein(s) with which the IVVY motif interacts and TRAF proteins binding to motif 2 and motif 3 to promote osteoclastogenesis.
Author Contributions
J. J., S. Wang, and X. F. designed the study. J. J., S. Wang, Z. S., and J. L. carried out experiments and collected data. J. J., S. Wang, S. Wei, and X. F. analyzed and interpreted data. J. J., S. Wang, S. Wei, and X. F. prepared the manuscript. All authors approved the final version of the manuscript.
This work was supported, in whole or in part, by NIAMS/National Institutes of Health Grant AR47830 (to X. F.) and a NIAMS graduate research supplement to Grant AR47830 (to J. J.). This work was also supported by a Within Our Reach innovative basic research grant from the Research and Education Foundation of the American College of Rheumatology (to X. F.), Grants 3087110 and 80171939 from the National Natural Science Foundation of China (to S. Wang), Grant 11C33150710 from the Guangzhou Science and Technology Program (to S. Wang), and Grant 201102A212024 from the Guangzhou Medical Science and Technology Project (to S. Wang). The authors declare that they have no conflicts of interest with the contents of this article.
- M-CSF
- macrophage/monocyte-colony stimulating factor
- TRAF
- TNF receptor-associated factor
- RANK
- receptor activator of nuclear factor κB
- RANKL
- receptor activator of nuclear factor κB ligand
- BMM
- bone marrow macrophage
- α-MEM
- α-minimal essential medium.
References
- 1. Teitelbaum S. L. (2000) Bone resorption by osteoclasts. Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 2. Feng X., McDonald J. M. (2011) Disorders of bone remodeling. Annu. Rev. Pathol. 6, 121–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teitelbaum S. L. (2006) Osteoclasts: culprits in inflammatory osteolysis. Arthritis Res. Ther. 8, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 5. Ross F. P., Teitelbaum S. L. (2005) αvβ3 and macrophage colony-stimulating factor: partners in osteoclast biology. Immunol. Rev. 208, 88–105 [DOI] [PubMed] [Google Scholar]
- 6. Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 7. Anderson D. M., Maraskovsky E., Billingsley W. L., Dougall W. C., Tometsko M. E., Roux E. R., Teepe M. C., DuBose R. F., Cosman D., Galibert L. (1997) A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390, 175–179 [DOI] [PubMed] [Google Scholar]
- 8. Armstrong A. P., Tometsko M. E., Glaccum M., Sutherland C. L., Cosman D., Dougall W. C. (2002) A RANK/TRAF6-dependent signal transduction pathway is essential for osteoclast cytoskeletal organization and resorptive function. J. Biol. Chem. 277, 44347–44356 [DOI] [PubMed] [Google Scholar]
- 9. Liu W., Xu D., Yang H., Xu H., Shi Z., Cao X., Takeshita S., Liu J., Teale M., Feng X. (2004) Functional identification of three receptor activator of NF-κB cytoplasmic motifs mediating osteoclast differentiation and function. J. Biol. Chem. 279, 54759–54769 [DOI] [PubMed] [Google Scholar]
- 10. Feng X. (2005) Regulatory roles and molecular signaling of TNF family members in osteoclasts. Gene 350, 1–13 [DOI] [PubMed] [Google Scholar]
- 11. Xu D., Wang S., Liu W., Liu J., Feng X. (2006) A novel receptor activator of NF-κB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J. Biol. Chem. 281, 4678–4690 [DOI] [PubMed] [Google Scholar]
- 12. Guerrini M. M., Sobacchi C., Cassani B., Abinun M., Kilic S. S., Pangrazio A., Moratto D., Mazzolari E., Clayton-Smith J., Orchard P., Coxon F. P., Helfrich M. H., Crockett J. C., Mellis D., Vellodi A., Tezcan I., Notarangelo L. D., Rogers M. J., Vezzoni P., Villa A., Frattini A. (2008) Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am. J. Hum. Genet. 83, 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim H., Choi H. K., Shin J. H., Kim K. H., Huh J. Y., Lee S. A., Ko C. Y., Kim H. S., Shin H. I., Lee H. J., Jeong D., Kim N., Choi Y., Lee S. Y. (2009) Selective inhibition of RANK blocks osteoclast maturation and function and prevents bone loss in mice. J. Clin. Invest. 119, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taguchi Y., Gohda J., Koga T., Takayanagi H., Inoue J. (2009) A unique domain in RANK is required for Gab2 and PLCγ2 binding to establish osteoclastogenic signals. Genes Cells 14, 1331–1345 [DOI] [PubMed] [Google Scholar]
- 15. Jules J., Zhang P., Ashley J. W., Wei S., Shi Z., Liu J., Michalek S. M., Feng X. (2012) Molecular basis of requirement of receptor activator of nuclear factor κB signaling for interleukin 1-mediated osteoclastogenesis. J. Biol. Chem. 287, 15728–15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi H. K., Kang H. R., Jung E., Kim T. E., Lin J. J., Lee S. Y. (2013) Early estrogen-induced gene 1, a novel RANK signaling component, is essential for osteoclastogenesis. Cell Res. 23, 524–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jules J., Shi Z., Liu J., Xu D., Wang S., Feng X. (2010) Receptor activator of NF-{kappa}B (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-α (TNF)-mediated osteoclastogenesis. J. Biol. Chem. 285, 37427–37435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dinarello C. A. (2009) Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550 [DOI] [PubMed] [Google Scholar]
- 19. Beyaert R. F. (1998) in Cytokines (Mires-Sluis A., Thorpe R., eds), pp. 335–360, Academic Press Limited, London [Google Scholar]
- 20. Pacifici R. (1998) Cytokines, estrogen, and postmenopausal osteoporosis: the second decade. Endocrinology 139, 2659–2661 [DOI] [PubMed] [Google Scholar]
- 21. Strand V., Kavanaugh A. F. (2004) The role of interleukin-1 in bone resorption in rheumatoid arthritis. Rheumatology 43, iii10–iii16 [DOI] [PubMed] [Google Scholar]
- 22. Graves D. T., Cochran D. (2003) The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J. Periodontol. 74, 391–401 [DOI] [PubMed] [Google Scholar]
- 23. Lam J., Takeshita S., Barker J. E., Kanagawa O., Ross F. P., Teitelbaum S. L. (2000) TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Invest. 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li P., Schwarz E. M., O'Keefe R. J., Ma L., Boyce B. F., Xing L. (2004) RANK signaling is not required for TNFα-mediated increase in CD11(hi) osteoclast precursors but is essential for mature osteoclast formation in TNFα-mediated inflammatory arthritis. J. Bone Miner. Res. 19, 207–213 [DOI] [PubMed] [Google Scholar]
- 25. Ma T., Miyanishi K., Suen A., Epstein N. J., Tomita T., Smith R. L., Goodman S. B. (2004) Human interleukin-1-induced murine osteoclastogenesis is dependent on RANKL, but independent of TNF-α. Cytokine 26, 138–144 [DOI] [PubMed] [Google Scholar]
- 26. Wei S., Kitaura H., Zhou P., Ross F. P., Teitelbaum S. L. (2005) IL-1 mediates TNF-induced osteoclastogenesis. J. Clin. Invest. 115, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeshita S., Kaji K., Kudo A. (2000) Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 15, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 28. Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu D., Shi Z., McDonald J., Pan G., Cao X., Yu X., Feng X. (2004) Development of a chimaeric receptor approach to study signalling by tumour necrosis factor receptor family members. Biochem. J. 383, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng X., Novack D. V., Faccio R., Ory D. S., Aya K., Boyer M. I., McHugh K. P., Ross F. P., Teitelbaum S. L. (2001) A Glanzmann's mutation in β 3 integrin specifically impairs osteoclast function. J. Clin. Invest. 107, 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bodmer J. L., Schneider P., Tschopp J. (2002) The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27, 19–26 [DOI] [PubMed] [Google Scholar]
- 32. Inoue J., Ishida T., Tsukamoto N., Kobayashi N., Naito A., Azuma S., Yamamoto T. (2000) Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp. Cell Res. 254, 14–24 [DOI] [PubMed] [Google Scholar]
- 33. Asagiri M., Takayanagi H. (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 34. Liu W., Wang S., Wei S., Sun L., Feng X. (2005) Receptor activator of NF-κB (RANK) cytoplasmic motif, 369PFQEP373, plays a predominant role in osteoclast survival in part by activating Akt/PKB and its downstream effector AFX/FOXO4. J. Biol. Chem. 280, 43064–43072 [DOI] [PubMed] [Google Scholar]
- 35. Galibert L., Tometsko M. E., Anderson D. M., Cosman D., Dougall W. C. (1998) The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-κB, a member of the TNFR superfamily. J. Biol. Chem. 273, 34120–34127 [DOI] [PubMed] [Google Scholar]
- 36. Hsu H., Lacey D. L., Dunstan C. R., Solovyev I., Colombero A., Timms E., Tan H. L., Elliott G., Kelley M. J., Sarosi I., Wang L., Xia X. Z., Elliott R., Chiu L., Black T., Scully S., Capparelli C., Morony S., Shimamoto G., Bass M. B., Boyle W. J. (1999) Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. U.S.A. 96, 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]