Background: The activity of Itch and related E3 ligases is restricted by autoinhibition.
Results: Itch autoinhibition is maintained by an intramolecular interaction between its WW and HECT domains, and Ndfip1 relieves this interaction to allow trans-thiolation.
Conclusion: The primary role of Ndfip is to relieve autoinhibition of Itch and related ligases.
Significance: We describe a novel mechanism that regulates the activity of several catalytic E3 ligases.
Keywords: immunology, protein degradation, ubiquitin, ubiquitin ligase, ubiquitylation (ubiquitination)
Abstract
Nedd4-family E3 ubiquitin ligases regulate an array of biologic processes. Autoinhibition maintains these catalytic ligases in an inactive state through several mechanisms. However, although some Nedd4 family members are activated by binding to Nedd4 family-interacting proteins (Ndfips), how binding activates E3 function remains unclear. Our data reveal how these two regulatory processes are linked functionally. In the absence of Ndfip1, the Nedd4 family member Itch can bind an E2 but cannot accept ubiquitin onto its catalytic cysteine. This is because Itch is autoinhibited by an intramolecular interaction between its HECT (homologous to the E6-AP carboxy terminus domain) and two central WW domains. Ndfip1 binds these WW domains to release the HECT, allowing trans-thiolation and Itch catalytic activity. This molecular switch also regulates the closely related family member WWP2. Importantly, multiple PY motifs are required for Ndfip1 to activate Itch, functionally distinguishing Ndfips from single PY-containing substrates. These data establish a novel mechanism for control of the function of a subfamily of Nedd4 E3 ligases at the level of E2-E3 trans-thiolation.
Introduction
Catalytic homology to E6AP carboxy terminus (HECT)3 E3 ubiquitin ligases provide substrate specificity to the ubiquitylation cascade and mediate the transfer of ubiquitin to the substrate. The largest family of HECT ligases, the Nedd4 family E3 ubiquitin ligases, regulates signaling in intracellular pathways that control cancer, blood pressure, iron metabolism, and inflammation (1–9). Nedd4 family E3 ubiquitin ligases are catalytic E3s that share a conserved modular architecture (10). This includes an N-terminal C2 domain, two to four WW domains, and a HECT domain that is the defining feature of HECT E3 ligases. The catalytic HECT domain is comprised of an E2 binding “N-lobe” and a catalytic cysteine residue in the “C-lobe.” The catalytic cysteine accepts ubiquitin from an E2 in a trans-thiolation reaction to form a HECT∼Ub intermediate, rendering the ligase catalytically active (11). WW domains of Nedd4 family E3s bind several proline-rich motifs, including L/PPXY motifs and, therefore, facilitate binding of Itch to substrates (12, 13).
The catalytic function of Nedd4 E3 ligases is restrained through autoinhibition. Most Nedd4 family members appear to be autoinhibited by an intramolecular interaction between the N-terminal C2 domain and the HECT domain (14). Supporting this, removing the C2 domain from Nedd4 and Rsp5, a Nedd4 family orthologue found in yeast, releases autoinhibition. Additionally, phosphorylation can promote the activation of Itch, Nedd4, and Nedd4-2 (15–17). Among Nedd4 family members, Smurf2 is of particular interest because it not only shows C2 domain autoinhibition but is further regulated by a reduced-affinity binding site for its complimentary E2s, necessitating its interaction with the adaptor Smad7 for full function (18, 19). Recent work has provided insights into how the C2 domain mediates autoinhibition of both Nedd4 and Smurf2. Specifically, C2 domain interaction with the HECT domain maintains the ligase in a low-activity state in which the HECT domain has a reduced capacity for noncovalent binding of ubiquitin (14).
Adaptor-ligase interactions can also promote Nedd4 family catalytic activity. Such interactions are known to promote the function of the sole Nedd4-family member in yeast, Rsp5 (20–23), suggesting that this means of controlling Nedd4-family function is evolutionarily conserved. One adaptor that promotes Rsp5 function is Bsd2, the orthologue of mammalian Nedd4 family-interacting proteins (Ndfips). The polyubiquitylation activity of several Nedd4-family E3 ligases is promoted by Ndfips (4, 24). These proteins have three transmembrane-spanning regions and three L/PPXY (PY) motifs, the motifs recognized by WW domains. Ndfip1 has been shown to promote the ubiquitylation activity of several Nedd4 family members in vivo, including Itch, Nedd4-2, and WWP2 (3, 4, 24). As with many proteins that increase the function of Nedd4 family members, these proteins are commonly referred to as adaptors, but how they promote E3 function is often not known.
The Nedd4 family member Itch plays a critical role in the immune system, with known roles in hematopoietic cells, innate immune cells, and the differentiation and function of T helper cells (1, 4, 25). In CD4+ T cells, loss of Itch leads to increased TH2 polarization in vivo and in vitro, a phenomenon attributed to the increased stability of an Itch substrate, the IL-4 transcription factor JunB (25). Humans with Itch loss-of-function mutations also exhibit pleiotropic immune dysfunction (26). Strikingly, loss of Ndfip1 also leads to increased TH2 skewing controlled, in part, by accumulation of the Itch substrate JunB (4, 8). In vitro, Itch is autoinhibited (16, 24). It has been shown that phosphorylation of Itch by JNK1 can promote Itch activity (16). Additionally, Itch can be activated by its interaction with Ndfip1 (24). The dependence of Itch on Ndfip1 for function suggests that phosphorylation alone is not sufficient to promote Itch function. How intramolecular autoinhibition prevents Itch catalytic function and how Ndfip1 binding releases this has still to be determined.
Here we defined the enzymatic step at which Itch catalytic function is inhibited. Itch autoinhibition is maintained by an intramolecular interaction between its HECT and two core WW domains. This autoinhibited state prevents E2-Itch trans-thiolation from occurring, thereby rendering Itch catalytically inactive. Ndfip1 binding to Itch releases the autoinhibitory conformation that holds Itch in an “off” state. Binding of Ndfip1 to the core WW domains releases the HECT and allows the trans-thiolation reaction between the E2 and Itch to take place. Our data show that these regulatory mechanisms may be true for WWP2 and, possibly, other Nedd4 family members.
Experimental Procedures
Protein Expression and Purification
Nedd4 family member E3 ligase constructs were prepared using PCR amplification to insert restriction sites in cDNA encoding specified variants. The cDNA constructs used were as follows: mouse Itch variant 1 (catalog no. MC205232, Origene), mouse Ndfip1 (catalog no. MG202546, Origene), mouse Nedd4 (catalog no. MG222243, Origene), and mouse WWP2 (catalog no. MC202685, Origene). These inserts were ligated into an N-terminal His-MBP tag pRSF-1B plasmid. WW domain mutants were made by introducing a tryptophan-to-alanine point mutation in the hydrophobic binding pocket. Truncation mutants of Itch were constructed from mouse Itch cDNA. The ItchWW1-HECT construct contains residues 285–864. ItchWW3-HECT contains residues 397–864, and ItchHECT contains residues 486–864. Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Agilent). UbcH5B and UbcH7 fused with a small ubiquitin-like modifier and a hexahistidine tag were cloned into pRSF-duet expression vectors. GST-tagged full-length Itch and the HECT domain were cloned into pGEX 4T-1 to provide GST-Itch for affinity studies. Ndfip family constructs were prepared as described previously and ligated into the pGEX-4T-2 vector (GE Life Sciences). Plasmids were transformed into BL21(DE3) Escherichia coli and purified with nickel-nitrilotriacetic acid-agarose (Qiagen) in the case of His-MBP fusion proteins or glutathione-Sepharose 4B (GE Life Sciences) in the case of GST fusion proteins, followed by size-exclusion chromatography.
Octet BioLayer Interferometry and Binding Kinetics
Anti-GST antibody-conjugated biosensors were used to trap GST-ITCH as ligand. 1–50 μm UbcH5B and 3–250 μm UbcH7 as analytes were prepared in assay buffer (25 mm Tris (pH 7.6), 150 mm NaCl, 0.1 mg/ml bovine serum albumin, and 0.01% Tween 20) to titrate the GST-ITCH-bound biosensor to measure the binding kinetics. All experiments were carried out on an Octet Red96 (Pall ForteBio Corp., Menlo Park, CA) with four repeats at 26 °C. Both association and dissociation steps in the BioLayer interferometry assays lasted 60 s. The response shifts were extracted and fitted to an equivalent binding model to obtain the Kd values.
Ubiquitin Charging Assays
The E1 ubiquitin-activating enzyme Ube1 (Boston Biochem), the E2 ubiquitin-conjugating enzyme UbcH7 (Boston Biochem), and biotinylated ubiquitin (Boston Biochem) were incubated at 25 °C in ubiquitylation buffer (40 mm Tris (pH 7.6), 250 mm NaCl, 1.7 mm ATP, 8.3 mm MgCl2, and 3.3 μg/ml ovalbumin). After 30 min, the reaction was quenched on ice by 5-fold dilution with quenching solution (25 mm EDTA, 25 mm HEPES (pH 7.5), and 100 mm NaCl). The resultant ubiquitin-loaded E2 mixture was then introduced to the E3 mixture on ice, resulting in a final mixture concentration of 6 μm E3, 200 nm E2, 10 nm E1, and 400 nm Ub. Ndfip1 constructs introduced to the mixture were at a final concentration of 16 μm. WW domain trans-inhibition assays used a final concentration of 30 μm WW domain and 60 μm Ndfip1. Loading of ubiquitin onto E3 via thioester bond formation was stopped by quenching with an equal volume of Laemmli buffer at the indicated time points. Ubiquitylated products were resolved on SDS gels and detected using Li-Cor IRDye 800CW streptavidin (Li-Cor). Confirmation of thioester loading was obtained by introducing samples to 50 μm DTT to reduce the thioester bond.
Homogeneous E3 Ubiquitylation Assay
E3 ubiquitylation activity was monitored using a homogeneous Time-resolved-FRET assay as described previously with slight modifications (27). The reaction components were diluted in assay buffer (50 mm Tris (pH 8.0), 5 mm MgCl2, 1 mm β-mercaptoethanol, and 0.05% CHAPS) to a final volume of 20 μl and assembled in a 384-well polypropylene plate. Briefly, ubiquitylation mixture (0.2 mm ATP, 5 nm E1, and 100 nm UbcH5c) containing varying concentrations of E3 was combined with varying doses of Ndfip1/2 or peptides and homogeneous time-resolved fluorescence detection mixture (Biotin-K63TUBE (LifeSensors), Streptavidin-Terbium (CisBio), and wild-type and fluorescein-labeled ubiquitin). Time-resolved-FRET was monitored in real time using a PerkinElmer Life Sciences Envision plate reader (excitation, 340 nm; emission 1, 520 nm; emission 2, 480 nm). The TR-FRET ratio was calculated by emission520/emission480.
Jurkat Transfection
ItchWT and the ItchWW2,3A mutant were cloned into the MSCV-IRES-NGFR vector (provided by Dr. Andrew Wells, Children's Hospital of Philadelphia). Cells were transfected using the Invitrogen neon electroporation system (Invitrogen) according to the protocols of the manufacturer. Prior to transfection, Jurkat E6.1 cells (Progenra) were grown in RPMI 1640 medium (Gibco) supplemented with 10% FCS, 20 mm GlutaMAX (Gibco), and 25 mm HEPES (pH 7.0) with no antibiotics. Cells were washed with PBS and resuspended at 2.0 × 107 cells/ml in buffer R (Invitrogen) prior to electroporation. 24 h later, transfected cells were purified based on NGFR expression using phycoerythrin-NGFR antibody (BD Biosciences), anti-phycoerythrin magnetic beads, and LS columns (Miltenyi). To assess protein stability, NGFR+ cells were stimulated for 2 h with phorbol 12-myristate 13-acetate (30 ng/ml) and ionomycin (1 μg/ml), and cycloheximide (Sigma) was added to a final concentration of 10 μg/ml for an additional 2 h of stimulation. JunB was quantified using rabbit anti-JunB (Santa Cruz Biotechnology, catalog no. SC-44). The degradation of JunB in ItchWW2,3A was then compared with the degradation of JunB in ItchWT to normalize for variations in JunB expression.
HEK293T Transfection
ItchWT, ItchC832A, ItchWW2,3A, and ItchWW2,3A-C832A in the MSCV-IRES-NGFR vector were incubated with Lipofectamine 2000 (Invitrogen). 2 μg of plasmid was incubated with 20 μl of Lipofectamine 2000 for 20 min in 1 ml of Opti-MEM (Gibco) and then introduced to 70% confluent HEK293T cells on 60-mm cell culture dishes (Corning). Cells were then incubated with Lipofectamine and plasmid mixture at 37 °C in 5% CO2 for 4 h. Transfection medium was replaced with growth media, and cells were incubated at 37 °C overnight. Cycloheximide was added to a final concentration of 10 μg/ml, and time points were harvested every 2 h. Cells were washed in PBS and lysed in 50 mm Tris-HCl (pH 7.6), 1% Nonidet P-40, and 150 mm NaCl with complete protease inhibitors (Roche). Lysates were run on Bio-Rad Criterion 4–20% Tris-HCl gradient gels at 20 μg of lysate/lane.
Overlapping Peptide Array
PepSpot peptide arrays (JPT peptides) containing 81 overlapping peptides covering the mouse HECT domain (amino acids 530–864) were synthesized. Each peptide was 16 residues in length and overlapped adjacent sequences by 12 residues. The PepSpot array was activated in methanol, washed in TBS, and then blocked for 3 h in Western blocking reagent solution (Roche). The MBP-tagged WW1-WW4 cassette was then added at 10 μg/ml and incubated at 4 °C overnight. MBP was detected using a mouse monoclonal anti-MBP antibody (New England Biolabs) and IRDye 800 CW anti-mouse IgG (Li-Cor). Binding was quantified using laser densitometry on the Li-Cor Odyssey system.
Peptide Mimic Synthesis
Ndfip1 peptide mimics were synthesized by the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT). Peptides were purified using reverse-phase HPLC, and mass was confirmed by MALDI-TOF and LC-MS. Lyophilized peptides were resuspended in water, incubated at 37 °C for 1 h, and vortexed. Peptide amino acid sequences were as follows: peptide 1, N-PEQTAGDAPPPYSSITAESAAYFDY-C; peptide 2, N-GDAPPPYSSITAESAAYFDYKDESGFPKPPSYNVA-C; peptide 3, N-GDAPPPYSSITAESAAYFDYKDESGFPKPPSYNVATTLPSYDEAE-C.
Results
Itch Catalytic Function Is Restrained by a Block in E2-E3 Trans-thiolation That Is Independent of E2 Binding
Itch has been shown to be autoinhibited (24) in a way that can be relieved by interaction with Ndfip1, but the stage of the catalytic cycle at which Itch is autoinhibited is unknown. Therefore, we first asked whether Ndfip1 is required for E2-E3 trans-thiolation, a requisite step in Itch E3 activation. We incubated Itch in the presence or absence of Ndfip1 with ubiquitin-loaded E2 (UbcH7). To track ubiquitin transfer to Itch, we used biotinylated ubiquitin and visualized it using streptavidin. No ubiquitin was transferred to Itch in the absence of Ndfip1 (Fig. 1A). However, when the cytoplasmic region of Ndfip1 was added to the reaction, the transfer of ubiquitin to Itch became evident. This was confirmed to be a transfer of ubiquitin to the catalytic cysteine of Itch and not autoubiquitylation by demonstrating that the Itch∼Ub species was lost after DTT treatment (Fig. 1B). This confirmed that Itch is autoinhibited at a catalytic step prior to ubiquitin ligation or interaction with substrate.
FIGURE 1.
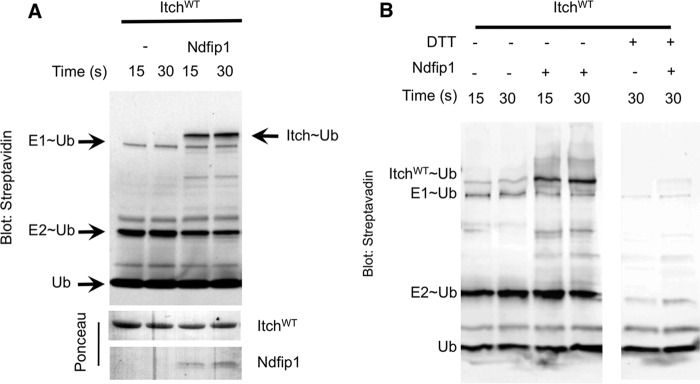
Ndfip1 promotes ubiquitin charging of Itch. A, Itch was incubated for the indicated time points with (biotinylated) ubiquitin-loaded E2 and in the presence or absence of Ndfip1. Streptavidin blotting revealed ubiquitin-charged Itch. Ponceau staining was used as a loading control. B, samples from the Itch charging experiment shown in A were divided into two tubes, and one tube was treated with 50 mm DTT. DTT treatment resulted in loss of ubiquitin charging on Itch, indicating that the interaction was mediated by the characteristic thioester bond between the C-terminal glycine tail of ubiquitin and the catalytic cysteine in the HECT domain of Itch. In all panels, similar results were obtained in at least three independent experiments.
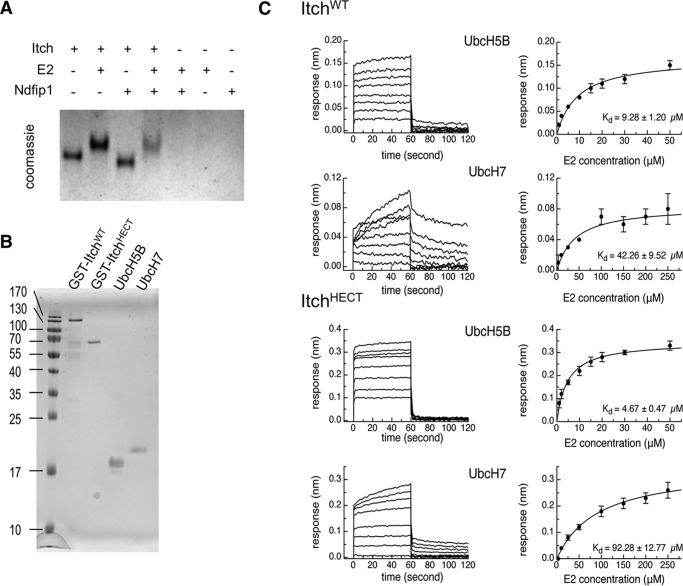
This block in Itch ubiquitin charging suggested that Itch might be unable to bind to its conjugate E2 in the absence of an adaptor, as has been shown with Smurf2. To test this, we examined Itch-E2 binding in the presence or absence of Ndfip1. Native gel analysis of Itch-E2 complexes showed that Itch does not require Ndfip1 to associate with an E2 (Fig. 2A). To further test whether autoinhibited Itch could interact with its E2, we compared the E2 binding of full-length Itch to that of ItchHECT alone, which is robustly active in the absence of Ndfip1. We tested the binding of ItchWT and ItchHECT to UbcH7 as well as UbcH5b (Fig. 2, B and C). Interestingly, ItchWT shows a marked preference for binding UbcH5B in this assay. However, it remains to be seen whether these data imply any functional preference of Itch for an E2 encountered in vivo. Nevertheless, these data show that full-length Itch can bind an E2 with similar affinity as that of the HECT domain alone, indicating that Itch can interact with an E2 even in its autoinhibited state. Therefore, unlike Smurf2, Itch autoinhibition does not appear to be controlled at the level of E2 interaction. This fits well with structural data showing that, unlike Smurf2, Itch does not have a suboptimal E2 binding pocket (18).
FIGURE 2.
Itch interacts with an E2 in the absence of Ndfip1. A, Itch-E2 complexes were analyzed by Coomassie staining under native conditions in the presence or absence of Ndfip1. B, Itch, ItchHECT, UbcH5B, and UbcH7 were recombinantly expressed and purified. C, Itch/UbcH7 and Itch/UbcH5b binding kinetics were analyzed by Octet BioLayer interferometry.
Itch Autoinhibition Is Mediated by HECT and WW Domain Interactions
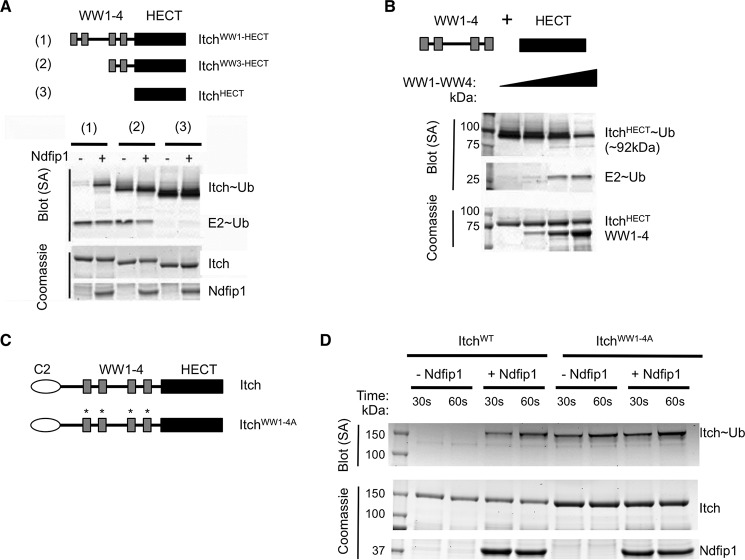
One motif proposed to maintain Itch autoinhibition lies in a proline-rich region unique to Itch, between the C2 and its most N-terminal WW domain (16, 24). To test the contribution of this region to the block in E2-E3 trans-thiolation, we generated a truncated version of Itch (ItchWW1-HECT, illustrated in Fig. 3A). Surprisingly, as with ItchWT, ubiquitin transfer to ItchWW1-HECT was promoted by, and therefore dependent on, Ndfip1 (Fig. 3, A and B). Therefore, even in the absence of the C2 domain and the proline-rich region, E2-mediated transfer of ubiquitin to the catalytic cysteine was inhibited robustly.
FIGURE 3.
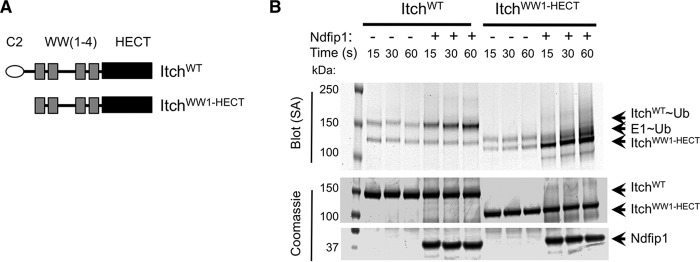
Ndfip1 is required for ubiquitin charging of ItchWW1-HECT. A, constructs generated for testing. In addition to the Itch sequence shown, these constructs contain an N-terminal His-MBP tag. B, the constructs shown in A were used in the ubiquitin charging assay with or without added Ndfip1. Ubiquitylated proteins were visualized using streptavidin. Similar results were obtained in at least three independent experiments. SA, streptavidin.
Because ItchWW1-HECT contains the WW and HECT domains, we hypothesized that autoinhibition might require the WW domains. We tested this by reducing the number of WW domains in Itch. Constructs of Itch were made that contained four, two or no WW domains (ItchWW1-HECT, ItchWW3-HECT, and ItchHECT, respectively; Fig. 4A). Consistent with our previous results, the construct containing four WW domains was inactive and relied on the addition of Ndfip1 to become efficiently charged by the E2. As shown previously, loss of all four WW domains in the ItchHECT construct leads to robust, Ndfip1-independent catalytic function. Supporting a role for multiple WW domains in blocking E2-E3 trans-thiolation, loss of two of the four WW domains led to partial relief of autoinhibition and less reliance on Ndfip1.
FIGURE 4.
Itch autoinhibition is due to an intramolecular interaction between its WW and HECT domains. A, constructs were generated that lack two or all four WW domains, as illustrated. Constructs were assessed for autoinhibition using the ubiquitin charging assay. Streptavidin shows ubiquitylated proteins, and Coomassie shows loading controls. B, cassettes containing all four WW domains or the HECT domain were generated as depicted. Ubiquitin charging of the HECT domain was assessed in the presence of increasing concentrations of the WW domain cassette (His-MBP-HECT is ∼84.2 kDa). Again, streptavidin shows ubiquitylated proteins, and Coomassie shows loading controls. C, full-length Itch and a construct in which the following point mutations were made in the binding pocket of each WW domain: W315A, W347A, W427A, and Y467A (indicated by asterisks and, together, termed ItchWW1–4A). D, these constructs were analyzed for their ability to accept ubiquitin in the presence or absence of Ndfip1 using the ubiquitin charging assay. In A, B, and D, similar results were obtained in at least three independent experiments. SA, streptavidin.
To test whether WW domains could prevent ubiquitin charging, we next asked whether Itch WW domains could inhibit ubiquitin charging of the HECT domain in trans. Increasing concentrations of Itch WW domains 1–4 (WW1–4, Fig. 4B) were added to the ItchHECT charging reaction. This addition led to reduced ubiquitin charging of the HECT domain by the E2-conjugating enzyme (Fig. 4B), supporting that WW domains block trans-thiolation of the catalytic cysteine in the HECT domain. To directly test the importance of the Itch WW domains in autoinhibition, we introduced point mutations in each of the four WW domains of the full-length Itch construct (ItchWW1–4A, Fig. 4C). These four point mutations were sufficient to relieve autoinhibition and allow ubiquitin charging of Itch (Fig. 4D). Addition of Ndfip1 to this reaction had no effect. Therefore, Itch autoinhibition is mediated by an intramolecular interaction between its HECT and WW domains.
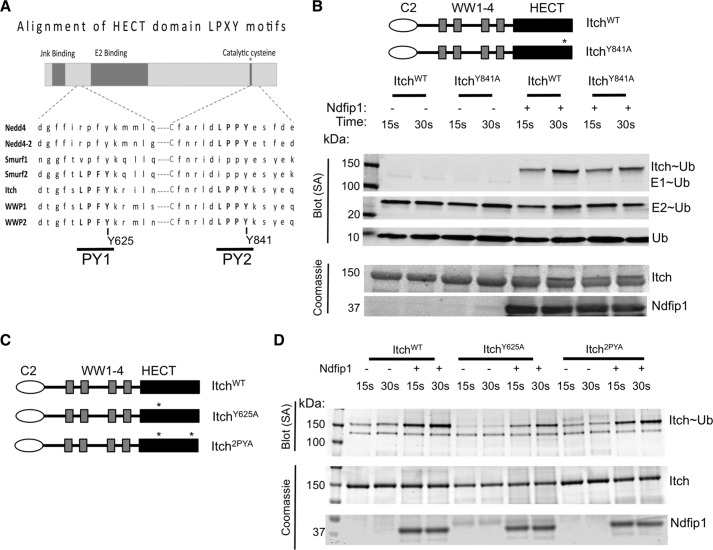
HECT Domain PY Motifs Are Not Sufficient for Itch Autoinhibition
It has been proposed that autoinhibition of Nedd4-2 is due to a PY motif in its HECT domain (17, 28, 29). Notably, this PY motif is buried in the C-lobe conformation observed in all HECT domain structures solved to date, although an isolated peptide binds the NEDD4L WW3 domain as in other LPXY motif WW domain crystals (17, 29). A comparison of the Nedd4 family HECT domains revealed that this PY motif and a second PY motif are conserved among multiple Nedd4 family members (Fig. 5A). Because PY motifs can bind Nedd4 family WW domains, we asked whether one or both of these HECT domain PY motifs mediated Itch autoinhibition. To determine this, we tested ubiquitin charging of full-length Itch with point mutations in these conserved HECT PY motifs (ItchY625A, ItchY841A, and Itch2PYA, Fig. 5, B–D). The amino acid sequence for Itch to which we refer can be accessed through the UniProtKB database under UniProtKB accession no. Q8C863.2 (30). We determined that these mutations in the Itch HECT PY motifs did not relieve autoinhibition, suggesting that either these PY motifs played no role or that these PY motifs worked together with other motifs.
FIGURE 5.
The PY motifs in the HECT domain are not sufficient for Itch autoinhibition. A, alignment of the HECT domains of Nedd4 family members illustrates the conservation of two PY motifs. B, a point mutation in Itch, changing the tyrosine in the second HECT PY to alanine. This mutant was tested using the ubiquitin charging reaction in the presence or absence of Ndfip1 and visualized by streptavidin. Loading controls for Itch and Ndfip1 are also shown. C, tyrosine-to-alanine point mutations in the HECT PY1 were generated, as was a mutant containing both PY Tyr → Ala mutations, as illustrated in C. D, Mutants shown in C were tested as described in B. In B and D, similar results were obtained in at least three independent experiments. *, site of mutation; SA, streptavidin.
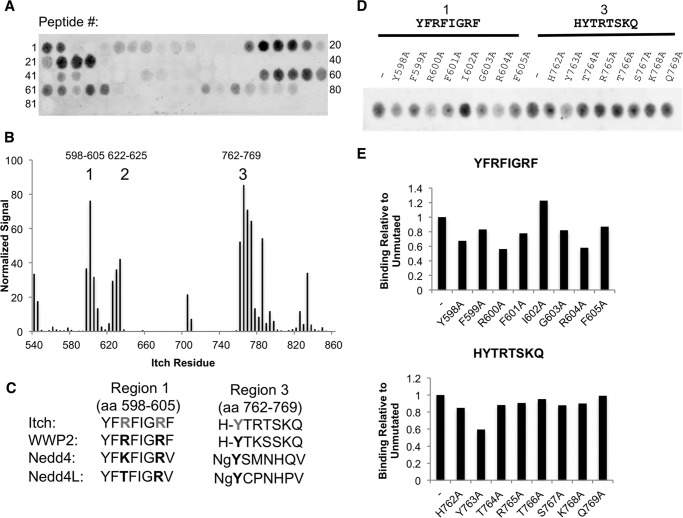
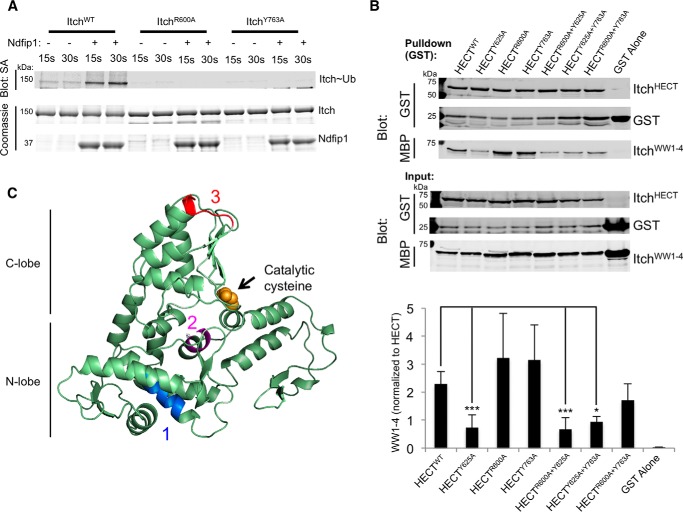
To investigate potential noncanonical or cryptic WW domain binding motifs in the HECT domain, an overlapping peptide array was incubated with the MBP tagged WW1–4 region, and binding of the WW domains to the peptide array was detected using an antibody recognizing MBP (Fig. 6A). We identified three regions with the potential to bind Itch WW domains. Region 1 was a YFRFIGRF motif (residues 598–605), region 2 was the LPFY motif we analyzed in Fig. 5 (residues 622–625), and region 3 was a HYTRTSKQ motif (residues 762–769) (Fig. 6, B and C). Because we determined previously that mutating the tyrosine in region 2 was not sufficient to relieve autoinhibition, we investigated the contributions of the individual residues in motifs of regions 1 and 3 by alanine scanning (Fig. 6D). In region 1, mutation of Arg-600 or Arg-604 resulted in diminished binding of the WW domain cassette, whereas, in region 3, mutation of the Tyr-763 residue resulted in decreased WW domain binding (Fig. 6, D and E), demonstrating a potential to mediate autoinhibition.
FIGURE 6.
WW domains can bind several regions in the HECT domain. A, peptide arrays were blocked and then incubated with MBP-tagged ItchWW1–4. Binding was then analyzed by detection of the MBP tag. B, quantification of A aligned to Itch HECT domain residues. C, alignment of putative binding regions 1 and 3 between Nedd4 family members. aa, amino acids. D, peptide array displaying alanine mutants of binding regions seen in the overlapping peptide array were blocked and then incubated with MBP-ItchWW1–4. Binding was then analyzed by detection of MBP. E, quantification of D.
We attempted to determine the contributions of these motifs to autoinhibition by making point mutations in the HECT domain, mutating residue Arg-600 to an alanine in region 1 or residue Tyr-763 to an alanine in region 3. We then tested the activity of these mutants in a ubiquitin charging assay (Fig. 7A). Unfortunately, these constructs were not functional because they were not charged with ubiquitin even upon inclusion of Ndfip1 (Fig. 7A). We therefore developed a panel of HECT domains in which either single or multiple binding motif residues were mutated to alanine to test how these regions could potentially impact binding. We coexpressed these HECT domain mutants with ItchWW1–4 and assayed coprecipitation of ItchWW1–4 with the HECT domain (Fig. 7B). Using this approach, we determined that mutations of region 2 impaired WW-HECT binding, whereas mutations in regions 1 or 3 revealed no significant difference in coprecipitation of ItchWW1–4.
FIGURE 7.
Binding analysis reveals a motif in the HECT domain required for WW domain interactions. A, ubiquitin charging of ItchWT, ItchR600A, and ItchY763A in the presence or absence of Ndfip1. B, top panel, MBP-tagged ItchWW1–4 was coexpressed with a panel of GST-tagged ItchHECT mutants (GST-HECT is ∼75 kDa), and coprecipitation of MBP-ItchWW1–4 with each HECT was analyzed. Bottom panel, quantification of blots. ItchWW1–4 coprecipitation was normalized to ItchHECT. Data are mean ± S.E., two-tailed Student's t test. *, p < 0.05; ***, p < 0.001. C, Itch HECT domain structure highlighting region 1 in blue, 2 in pink, and 3 in red. In A and B, similar results were obtained in at least three independent experiments. SA, streptavidin.
As with the previously identified region of NEDD4L that binds the WW3 domain, all sequences we identified are embedded in folded structures within the ITCH HECT domain. Notably, computational studies of NEDD4L revealed the potential for local unfolding of the HECT domain to reveal cryptic linear motifs that could mediate autoinhibition (29). Although it is impossible to model these regions in the context of interaction with a WW domain, we highlighted their relative locations (Fig. 7C). Overall, our data raise the possibility that region 2 could play an important role mediating HECT-WW binding. Future structural studies are needed to determine how this region or other, still unidentified regions in the HECT domain could mediate autoinhibition.
Ndfip1 Binds Itch WW Domains to Mediate Autoinhibition
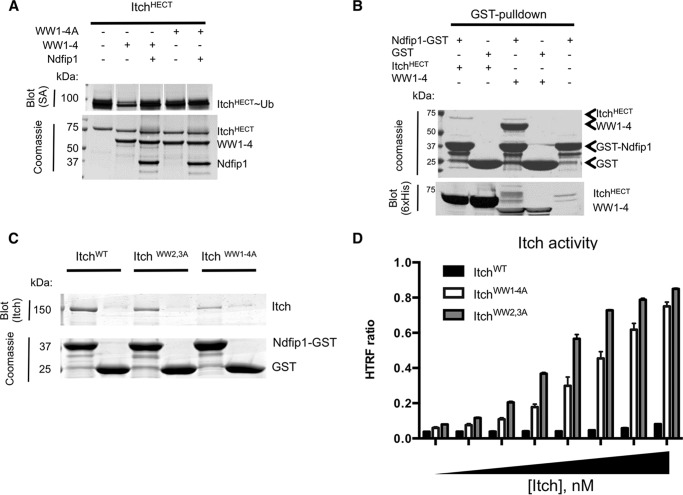
To determine how Ndfip1 relieves autoinhibition, we asked whether the addition of Ndfip1 could prevent WW domain-mediated inhibition of the ItchHECT domain in trans. We repeated the experiment in Fig. 4B but, this time, in the presence of Ndfip1 (Fig. 8A). Ndfip1 prevented WW domain-mediated inhibition of the ItchHECT domain. Furthermore, the WW domain cassette lacking functional WW domains was unable to support autoinhibition, and addition of Ndfip1 to this reaction had no effect. This fits with published data showing that Ndfip1 binds the WW domains of Itch (24). Nevertheless, to show that Ndfip1 binds exclusively to the WW domains and not the HECT domain, we coexpressed various combinations of these cassettes and used a pulldown approach to test whether Ndfip1 could also bind the HECT domain. Consistent with previous studies, we found that Ndfip1 associated with WW1–4 but not the Itch HECT domain (Fig. 8B).
FIGURE 8.
Ndfip1 relieves autoinhibition by binding Itch WW domains. A, ubiquitin charging of the HECT domain was tested alone (first lane) or in the presence of WT WW domains (WW1–4, second and third lanes) or mutant WW domains (WW1–4A, fourth and fifth lanes) and in the presence or absence of Ndfip1 (third and fifth lanes). Detection of the ubiquitin-charged HECT domain was by streptavidin. B, GST-tagged Ndfip1 or GST alone was pulled down from cells also expressing the Itch His-MBP-HECT or WW1–4 domains. Enriched proteins were visualized by Coomassie staining, and domain expression was confirmed in bacterial lysates using anti-His antibody. Cells expressing GST-Ndfip1, but no Itch domain, provided a negative control. C. GST-tagged Ndfip1 or GST alone was pulled down from bacteria expressing Itch, ItchWW2,3A, or ItchWW1–4A. Proteins were visualized by Coomassie and Itch construct expression was confirmed using anti-Itch. D, the activity of Itch, ItchWW2,3A, and ItchWW1–4A was assayed using a homogeneous FRET assay. In all panels, similar results were obtained in at least three independent experiments. HTRF, homogeneous time-resolved FRET; SA, streptavidin.
To determine which WW domains were preferred for Ndfip1 interactions, we tested several mutants of Itch that contained point mutations in one or two WW domains. We identified one mutant that showed a significant decrease in Ndfip1 binding. Specifically, mutating WW domains 2 and 3 (ItchWW2,3A) was sufficient to interrupt most of the interaction between Itch and Ndfip1 (Fig. 8C). To determine whether this minimal mutation was also sufficient to relieve autoinhibition of Itch, we used an autoubiquitylation assay that measured ubiquitin chain formation by Itch. This approach can analyze the formation of ubiquitin chains over time by measuring the FRET that occurs when ubiquitin chains are generated. This high-throughput assay also offers the advantage of monitoring the time courses of ubiquitin chain formation with many variables assayed simultaneously. Using this assay, we determined that ItchWW2,3A was active in the absence of Ndfip1 (Fig. 8D). This was in stark contrast to ItchWT, which was inactive for the duration of the assay, a finding consistent with our ubiquitin charging results showing no trans-thiolation in the absence of Ndfip1. Importantly, ItchWW2,3A was as active as ItchWW1–4A, the mutant determined previously to be relieved of autoinhibition. These data indicate that Ndfip1 activates Itch by relieving an intramolecular interaction between the Itch WW and HECT domains that restricts its catalytic function.
Mutating WW Domains 2 and 3 Results in Increased Itch Function in Cells
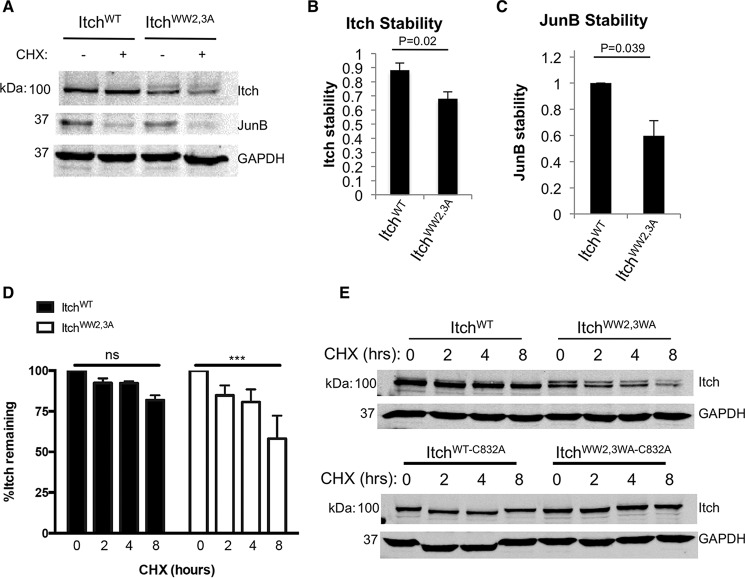
We next asked whether relief of autoinhibition was sufficient to increase Itch activity in cells. Itch has been shown to promote ubiquitin-mediated degradation of JunB, an AP-1 transcription factor component that promotes IL-2 and IL-4 production (4, 8, 16, 24). To examine the effects of ItchWW2,3A on JunB stability, we expressed ItchWT or ItchWW2,3A in Jurkat cells, an immortalized T lymphocyte line, and assessed JunB stability after stimulation (Fig. 9A). Both ItchWT and ItchWW2,3A showed high levels of expression. However, ItchWW2,3A levels were consistently lower than levels of the ItchWT counterpart. Cycloheximide treatment indicated that this was due to increased degradation of ItchWW2,3A (Fig. 9B). Despite the reduced level of ItchWW2,3A, we observed that JunB degradation was significantly increased in cells expressing ItchWW2,3A compared with those expressing ItchWT (Fig. 9C). Supporting this, expression of these mutants in HEK293T cells also showed that activated Itch was less stable, whereas the catalytically inactive version of ItchWW2,3A was stabilized (Fig. 9, D and E). This highlights that ItchWW2,3A instability is mediated by its catalytic activity because high ItchWW2,3A activity resulted in its instability. Furthermore, this indicates that, although Ndfip1 might act to recruit an E2 (31) under some circumstances, the primary function of Ndfip1 is to relieve Itch autoinhibition.
FIGURE 9.
Relieving autoinhibition of Itch is sufficient to facilitate substrate degradation. A, Jurkat cells transfected with ItchWT or ItchWW2,3A were stimulated and then treated with cycloheximide (CHX). Cells were harvested, and the expression of Itch and JunB was analyzed by Western blot. B, Itch stability over 4-h cycloheximide treatment normalized to GAPDH expression. Data are mean ± S.D., two-tailed Student's t test. C, degradation of JunB relative to expression of Itch or ItchWW2,3A mutant. JunB levels were normalized to GAPDH. Loss of JunB following cycloheximide treatment is shown as a -fold change of ItchWW2,3A relative to cells expressing ItchWT. Data are mean ± S.E., single-sample Student's t test. D and E, Itch stability was analyzed in HEK293T cells as in A but with the addition of constructs in which Itch catalytic activity was ablated (C832A mutation). The percentage of remaining Itch was quantified for ItchWT and ItchWW2,3A at the indicated time point. Data are mean ± S.E. ***, p ≤ 0.001; ns, not significant; two-way analysis of variance. In all panels, similar results were obtained in at least three independent experiments.
Ndfip1 Is Required for Some but Not All Nedd4 Family Members
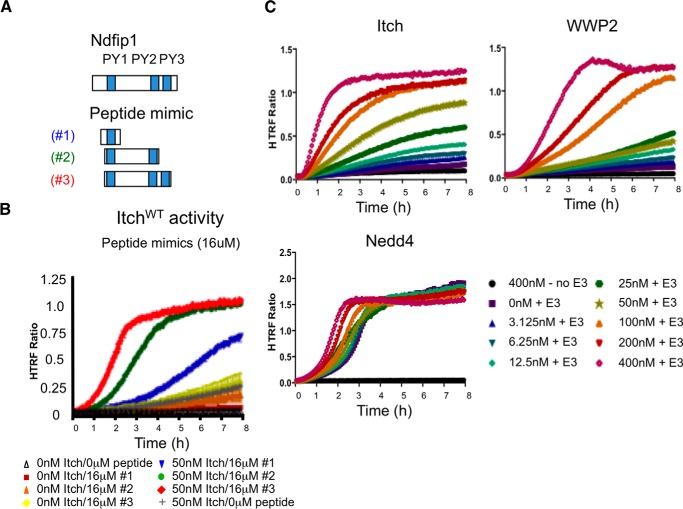
To identify the minimal motifs in Ndfip1 needed to activate Itch, peptide mimics of Ndfip1 were tested. We generated synthetic peptides that contain one, two, or all three PY motifs of Ndfip1 (Fig. 10A). There is a striking increase in Itch activity as the number of PY motifs is increased (Fig. 10B). Although these data show that Ndfip1 promotes Itch activation, all Nedd4 family members contain between two and four WW domains. Therefore, the intramolecular interaction mediated by WW domains could be a common means of Nedd4 family autoinhibition (10). To test whether Ndfip1 could promote the activity of other Nedd4 family members, we again employed the FRET-based assay to monitor ubiquitin chain formation. Addition of Ndfip1 substantially activated both ITCH and WWP2 in a dose-dependent manner. Additionally, we tested the prototypic family member Nedd4. Interestingly, Nedd4 was active in these assays without the addition of Ndfip1. The addition of Ndfip1 modestly but consistently potentiated the formation of ubiquitin chains (Fig. 10C), but these differences were only observed at early time points. These differences could be explained by the increased activity of E3s when associated with potential substrates as described in Kamadurai et al. (32). On the basis of these data, we propose a model of how Itch and a subset of Nedd4 family members are autoinhibited and can be activated by Ndfip1 (Fig. 11). This model advances our understanding of how Itch and related Nedd4 family members are held in an off position and how they are activated by defining the structural features of autoinhibition and Ndfip1 activation.
FIGURE 10.
Ndfip1 and its peptide mimics are required to activate a distinct subset of Nedd4 family members. A, peptides were generated that spanned one, two, or all three PY motifs of Ndfip1. B, the peptides shown in A were tested by analyzing Itch activity. Multiple plate reads were analyzed, with each plate read occurring at 4-min intervals. Data are representative of a minimum of three individual experiments. C, Ndfip1 was analyzed for its ability to activate Itch, WWP2, and Nedd4 using a homogeneous FRET assay. In B and C, similar results were obtained in at least three independent experiments. HTRF, homogeneous Time-Resolved FRET.
FIGURE 11.
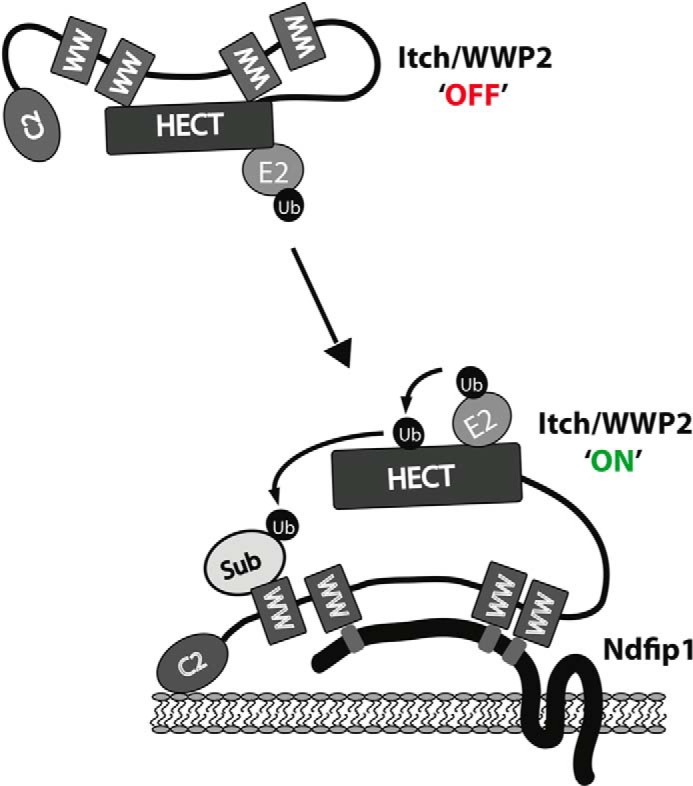
Model showing how Itch is autoinhibited and how Ndfip1 activates Itch. WW domains 2 and 3 of Itch bind to the HECT domain, mediating autoinhibition. Ndfip1 binds multiple WW domains through its PY motifs and relieves this autoinhibition, leaving other WW domains available to recruit substrate. This relief of autoinhibition allows the Itch HECT domain to accept ubiquitin on its catalytic cysteine and, subsequently, ubiquitylate substrate.
Discussion
Proper orchestration of the ubiquitylation cascade is critical for cellular functions. Catalytic E3 ubiquitin ligases, which provide substrate specificity and can also catalyze the addition of ubiquitin to target proteins, are highly regulated through various autoinhibitory mechanisms. RING E3s can be inhibited through intramolecular associations that obstruct functions required for the ligase activity, such as substrate binding in UBR1 (33), dimerization in cIAP1 (34), and E2 binding in Cbl-b (35). Even the multisubunit RING E3 Skp-Cullin-F-box (SCF) can be inhibited by the interaction of the extreme C terminus of CUL1 with the Rbx1 RING domain (36). Thioester-forming E3 ligases such as RING-between-RINGs and bacterial HECT-like E3s are subject to autoinhibition through multiple means, including E2 recruitment/binding, and impairment of E3∼Ub thioester intermediate formation. In RING-between-RINGs, distinct regions mask the active site cysteine requiring structural rearrangements for activity (37–40), whereas, in bacterial SspH1, the inhibitory leucine-rich repeat domain inhibits ubiquitin transfer to targets after E3 trans-thiolation (41). Of the thioester-forming Nedd4 family members, Smurf2 has the most defined mechanisms of autoinhibition. Smurf2 function is tightly orchestrated by its adaptor Smad7 (42) because Smurf2 activity is restrained by both an intramolecular interaction and by a suboptimal E2-binding site within the HECT domain. Smad7 activates Smurf2 in two ways: by bringing it together with its E2 (18) and by directing it to the plasma membrane, where the C2 releases the HECT domain. It has been shown recently that the C2 domain limits Smurf2 function by maintaining the HECT domain in a low-activity state in which it does not form a thioester bond with ubiquitin, preventing downstream conjugation of ubiquitin to the substrate (14). Although crystal structures have provided insights into potential intramolecular interactions, the intrinsically unstructured nature of regions within Nedd4 family members has prevented a complete picture of autoinhibited structures because only individual, stable domains can be crystallized.
Activation of Nedd4 family members can be accomplished through changes in the intracellular milieu, phosphorylation, and adaptor protein interactions. Intramolecular inhibitory interactions between the C2 and HECT domains of NEDD4 are relieved by phosphorylation of two tyrosine residues, one found in the C2 domain and another in the HECT domain, activating NEDD4 catalytic activity (17). Nedd4 autoinhibition is relieved by C2 domain-mediated calcium binding, which may also promote membrane association (43), whereas Nedd4-2 phosphorylation by serum and glucocorticoid-regulated kinase promotes SGK ubiquitylation and subsequent degradation (15). Smurf2, as described previously, is regulated both by Smad7 binding (18, 19) and an intramolecular interaction between the C2 and HECT domains.
Itch can be activated by phosphorylation and by association with adaptor proteins (4, 16, 24). Itch is activated by JNK1-mediated phosphorylation, an event that has been mapped to a unique region in Itch between its C2 and first WW domain (16). Itch is also regulated by interactions with adaptor proteins, such as Nedd4 family interacting proteins 1 and 2. Therefore, we took a reductive approach to isolate the mechanism by which Ndfip1 binding alleviates Itch autoinhibition.
Here we provide new information on how autoinhibition prevents the activation of several related Nedd4 family E3 ubiquitin ligases and show how Ndfip1 activates Itch, two processes that are linked functionally. Our data reveal the regions in Itch that mediate autoinhibition, namely WW domains 2 and 3, and that autoinhibition is achieved via blocking E2-E3 trans-thiolation. These two WW domains bind the Itch HECT domain, preventing the transfer of ubiquitin to the catalytic cysteine of Itch. This autoinhibition holds Itch in an off position and maintains Itch stability. Supporting this, we showed that Itch activation is tightly linked to its stability (Fig. 9). Additionally, we determined that loss of both Ndfip1 and Ndfip2 in T cells leads to a significant stabilization of Itch protein and concomitant loss of Itch function (data not shown). Therefore, regulation of cellular Itch levels depends on adapter proteins such as Ndfip1 and Ndfip2.
Although it is clear that two Itch WW domains bind the HECT domain, the precise regions in the HECT domain that are used for binding remain unclear. Related challenges have been encountered in attempts to understand mechanisms underlying autoinhibition of other Nedd4 family HECT E3s, although recent breakthrough studies revealed the potential for local flexibility to expose otherwise folded linear motifs within HECT domains (17, 28, 29). We determined one region, a PY motif (amino acids 622–625), that supports binding between the HECT and WW domains. However, mutation of this region was not sufficient to relieve autoinhibition. Considering that two WW domains are needed to maintain autoinhibition, it is likely that the WW domains bind the PY motif as well as another region. Using a peptide array, we identified two other motifs in the HECT domain that were able bind the Itch WW domains. However, subsequent mutations of these regions did not reduce HECT-WW binding. Therefore, in the future, a more detailed structural analysis will be needed to precisely define HECT-WW binding.
We show that Ndfip1 binding to Itch relieves autoinhibition, allowing the trans-thiolation reaction between E2 and E3 to occur. Additionally, we show that multiple PY motifs are required for Ndfip1 to activate Itch. We propose that the number of PY binding motifs that a protein contains could functionally distinguish an activator of Itch from a substrate. Interestingly, there is evidence that viral proteins may coopt this mechanism because the Epstein-Barr virus LMP2A requires two PY motifs to activate the human Itch homologue AIP4 (44). Supporting this, we tested Ndfip2 and determined that it, like Ndfip1, promotes ubiquitin charging of Itch. Therefore, our data clarify that, in the autoinhibited state, Itch can bind to an E2, and it is not until interaction with Ndfip1 that it is capable of ubiquitylation. Our model also illustrates that Ndfip1 binds to the same WW domains that are necessary for autoinhibition. This may suggest a paradigm for other E3-adapter interactions. Additionally, these mechanistic insights also apply to another Nedd4 family member, WWP2.
Although, at this point, it is not possible to make a detailed structural model, this work builds on emerging principles on the basis of structural and other studies of HECT E3s. Our work reveals new intramolecular interactions that target the HECT domain to mediate autoinhibition. Previous work has shown that Nedd4, Nedd4L (Nedd4-2), and Smurf2 mediate autoinhibition through a C2 domain-HECT domain interaction (14, 17, 19, 29). We show that Itch autoinhibition is mediated by WW-HECT domain interactions. Identification of such distinct mechanisms reveals how cells have evolved unique ways to activate different Nedd-4 family members. Furthermore, these distinct inhibitory mechanisms could be used to develop therapeutics that activate some, but not all, Nedd4 family E3 ubiquitin ligases, allowing specificity and, therefore, resulting in fewer off-target effects.
Author Contributions
C. R., B. A. S., and P. M. O. conceived and coordinated the study and wrote the paper. H. K. and K. P. W designed, performed, and analyzed the experiments shown in Fig. 2. S. K., and E. E. M. designed, performed, and analyzed the experiments shown in Fig. 10. C. R. designed, performed, and analyzed the experiments shown in all figures. C. E. O. provided advice, and M. Y. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Jimmy Vo and Alan Deng for technical assistance, Dr. Andrew Wells for reagents, and Dr. Steven Seeholzer and Dr. Hua Ding of the Children's Hospital of Philadelphia Protein Core for advice and assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R37GM065930 (to B. A. S.). This work was also supported by an American Asthma Foundation scholar award and an Institute for Translational Medicine and Therapeutics award (to P. M. O.), by American Lebanese Syrian Associated Charities, and by the Howard Hughes Medical Institute (to B. A. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- HECT
- homologous to the E6-AP carboxyl terminus domain
- Ub
- ubiquitin
- Ndfip
- Nedd4 family-interacting protein
- NGFR
- nerve growth factor receptor
- RING
- Really Interesting New Gene
- MBP
- maltose-binding protein.
References
- 1. Ahmed N., Zeng M., Sinha I., Polin L., Wei W.-Z., Rathinam C., Flavell R., Massoumi R., Venuprasad K. (2011) The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat. Immunol. 12, 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhalla V., Oyster N. M., Fitch A. C., Wijngaarden M. A., Neumann D., Schlattner U., Pearce D., Hallows K. R. (2006) AMP-activated kinase inhibits the epithelial Na+ channel through functional regulation of the ubiquitin ligase Nedd4-2. J. Biol. Chem. 281, 26159–26169 [DOI] [PubMed] [Google Scholar]
- 3. Foot N. J., Dalton H. E., Shearwin-Whyatt L. M., Dorstyn L., Tan S. S., Yang B., Kumar S. (2008) Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood 112, 4268–4275 [DOI] [PubMed] [Google Scholar]
- 4. Oliver P. M., Cao X., Worthen G. S., Shi P., Briones N., MacLeod M., White J., Kirby P., Kappler J., Marrack P., Yang B. (2006) Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rotin D., Staub O. (2012) Nedd4-2 and the regulation of epithelial sodium transport. Front. Physiol. 3, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang B., Kumar S. (2010) Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee I. H., Campbell C. R., Song S. H., Day M. L., Kumar S., Cook D. I., Dinudom A. (2009) The activity of the epithelial sodium channels is regulated by caveolin-1 via a Nedd4-2-dependent mechanism. J. Biol. Chem. 284, 12663–12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beal A. M., Ramos-Hernández N., Riling C. R., Nowelsky E. A., Oliver P. M. (2012) TGF-β induces the expression of the adaptor Ndfip1 to silence IL-4 production during iTreg cell differentiation. Nat. Immunol. 13, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramon H. E., Riling C. R., Bradfield J., Yang B., Hakonarson H., Oliver P. M. (2011) The ubiquitin ligase adaptor Ndfip1 regulates T cell-mediated gastrointestinal inflammation and inflammatory bowel disease susceptibility. Mucosal Immunol. 4, 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ingham R. J., Gish G., Pawson T. (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 11. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 12. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 13. Jolliffe C. N., Harvey K. F., Haines B. P., Parasivam G., Kumar S. (2000) Identification of multiple proteins expressed in murine embryos as binding partners for the WW domains of the ubiquitin-protein ligase Nedd4. Biochem. J. 351, 557–565 [PMC free article] [PubMed] [Google Scholar]
- 14. Mari S., Ruetalo N., Maspero E., Stoffregen M. C., Pasqualato S., Polo S., Wiesner S. (2014) Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 22, 1639–1649 [DOI] [PubMed] [Google Scholar]
- 15. Zhou R., Snyder P. M. (2005) Nedd4-2 phosphorylation induces serum and glucocorticoid-regulated kinase (SGK) ubiquitination and degradation. J. Biol. Chem. 280, 4518–4523 [DOI] [PubMed] [Google Scholar]
- 16. Gallagher E., Gao M., Liu Y. C., Karin M. (2006) Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. U.S.A. 103, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Persaud A., Alberts P., Mari S., Tong J., Murchie R., Maspero E., Safi F., Moran M. F., Polo S., Rotin D. (2014) Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci. Signal. 7, ra95. [DOI] [PubMed] [Google Scholar]
- 18. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 19. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 20. Léon S., Erpapazoglou Z., Haguenauer-Tsapis R. (2008) Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell 19, 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hettema E. H., Valdez-Taubas J., Pelham H. R. (2004) Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nikko E., Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 24. Mund T., Pelham H. R. (2009) Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 10, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang D., Elly C., Gao B., Fang N., Altman Y., Joazeiro C., Hunter T., Copeland N., Jenkins N., Liu Y. C. (2002) Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3, 281–287 [DOI] [PubMed] [Google Scholar]
- 26. Lohr N. J., Molleston J. P., Strauss K. A., Torres-Martinez W., Sherman E. A., Squires R. H., Rider N. L., Chikwava K. R., Cummings O. W., Morton D. H., Puffenberger E. G. (2010) Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 86, 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marblestone J. G., Larocque J. P., Mattern M. R., Leach C. A. (2012) Analysis of ubiquitin E3 ligase activity using selective polyubiquitin binding proteins. Biochim. Biophys. Acta 1823, 2094–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bruce M. C., Kanelis V., Fouladkou F., Debonneville A., Staub O., Rotin D. (2008) Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 415, 155–163 [DOI] [PubMed] [Google Scholar]
- 29. Escobedo A., Gomes T., Aragón E., Martín-Malpartida P., Ruiz L., Macias M. J. (2014) Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 22, 1446–1457 [DOI] [PubMed] [Google Scholar]
- 30. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kathania M., Zeng M., Yadav V. N., Moghaddam S. J., Yang B., Venuprasad K. (2015) Ndfip1 regulates Itch ligase activity and airway inflammation via UbcH7. J. Immunol. 194, 2160–2167 [DOI] [PubMed] [Google Scholar]
- 32. Kamadurai H. B., Qiu Y., Deng A., Harrison J. S., Macdonald C., Actis M., Rodrigues P., Miller D. J., Souphron J., Lewis S. M., Kurinov I., Fujii N., Hammel M., Piper R., Kuhlman B., Schulman B. A. (2013) Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife 2, e00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A. (2002) Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. U.S.A. 99, 14110–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez J., John S. W., Tenev T., Rautureau G. J., Hinds M. G., Francalanci F., Wilson R., Broemer M., Santoro M. M., Day C. L., Meier P. (2011) CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Mol. Cell 42, 569–583 [DOI] [PubMed] [Google Scholar]
- 35. Dou H., Buetow L., Hock A., Sibbet G. J., Vousden K. H., Huang D. T. (2012) Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z. Q. (2008) Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. U.S.A. 105, 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duda D. M., Olszewski J. L., Schuermann J. P., Kurinov I., Miller D. J., Nourse A., Alpi A. F., Schulman B. A. (2013) Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trempe J. F., Sauvé V., Grenier K., Seirafi M., Tang M. Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., Nagar B., Fon E. A., Gehring K. (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 [DOI] [PubMed] [Google Scholar]
- 39. Riley B. E., Lougheed J. C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K., Diep L., Zhang Z., Chiou S., Bova M., Artis D. R., Yao N., Baker J., Yednock T., Johnston J. A. (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wauer T., Komander D. (2013) Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chou Y. C., Keszei A. F., Rohde J. R., Tyers M., Sicheri F. (2012) Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases. J. Biol. Chem. 287, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan X., Chen Y. G. (2011) Smad7: not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem. J. 434, 1–10 [DOI] [PubMed] [Google Scholar]
- 43. Wang J., Peng Q., Lin Q., Childress C., Carey D., Yang W. (2010) Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J. Biol. Chem. 285, 12279–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeda M., Ikeda A., Longnecker R. (2001) PY motifs of Epstein-Barr virus LMP2A regulate protein stability and phosphorylation of LMP2A-associated proteins. J. Virol. 75, 5711–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]