Background: IQGAP1 is a scaffold protein essential for cellular signaling in response to external cues.
Results: Perturbation of the IQGAP1-SKAP interaction results in inhibition of directional cell migration.
Conclusion: The IQGAP1-SKAP interaction links to dynamic microtubules at the leading edge to steer cell migration.
Significance: IQGAP1 serves as a signaling hub for dynamic interaction between microtubule plus-ends and the cell cortex.
Keywords: actin, cell migration, membrane, microtubule, scaffold protein, microtubule dynamics
Abstract
Cell migration is orchestrated by dynamic interaction of microtubules with the plasma membrane cortex. However, the regulatory mechanisms underlying the cortical actin cytoskeleton and microtubule dynamics are less characterized. Our earlier study showed that small GTPase-activating proteins, IQGAPs, regulate polarized secretion in epithelial cells (1). Here, we show that IQGAP1 links dynamic microtubules to steer cell migration via interacting with the plus-end tracking protein, SKAP. Biochemical characterizations revealed that IQGAP1 and SKAP form a cognate complex and that their binding interfaces map to the WWIQ motif and the C-terminal of SKAP, respectively. The WWIQ peptide disrupts the biochemical interaction between IQGAP1 and SKAP in vitro, and perturbation of the IQGAP1-SKAP interaction in vivo using a membrane-permeable TAT-WWIQ peptide results in inhibition of directional cell migration elicited by EGF. Mechanistically, the N-terminal of SKAP binds to EB1, and its C terminus binds to IQGAP1 in migrating cells. Thus, we reason that a novel IQGAP1 complex orchestrates directional cell migration via coupling dynamic microtubule plus-ends to the cell cortex.
Introduction
Cell migration is a fundamental process for development, morphogenesis, tissue repair, and tumor metastasis. Physically, targeting and capturing of microtubules (MTs)4 plus-ends with actin cytoskeleton at special cortical regions are necessary for cell migration. Therefore, both MTs and cortical actin are necessary for efficient cell migration (2, 3).
The IQ domain-containing GTPase-activating protein (IQGAP) family proteins are multidomain containing scaffolding proteins that consist of a CH domain, a WW domain, an IQ domain, and a RasGAP-related domain (GRD). In mammals, the IQGAP family contains three proteins, IQGAP1, -2, and -3, participating in multiple biological cellular events such as cell-cell adhesion, Salmonella invasion, cytokinesis, and cell migration (1, 4–6). IQGAP1, the best characterized member of the IQGAP family, distributes at leading edges and associates with actin filaments. Functionally, IQGAP1 is necessary for cytoskeletal organization via activating Rac1 and Cdc42 to regulate actin filaments and MTs, which are essential for cell migration (7).
MT plus-end tracking proteins, referred to as +TIPs, localize to and track along the growing plus-ends of microtubules. These proteins comprise an important subgroup of the microtubule-associated proteins (MAPs) (8, 9). +TIPs regulate the dynamic behavior of microtubules as well as the interaction between microtubules and other cellular components (8, 10). Plus-end tracking proteins have emerged as important MT regulators and consequently as key factors in a wide range of cellular processes, such as MT nucleation and dynamics, transport of signaling factors, and cell migration (3, 8). Because the selective stabilization of MTs is essential for cell migration (3), +TIPs modulating MT plasticity and dynamics in cells are proposed to be regulatory factors involved in cell migration. In recent years, many +TIPs have been identified as IQGAP1-interacting proteins (7, 11). Despite decades of research, the mechanism remains partially elusive. More IQGAP1-interacting +TIPs remain to be identified and characterized. It would be of interest to identify these potential interacting proteins and elucidate their physiological role in cooperatively regulating cell migration.
+TIPs exist in a variety of forms (12–14). The crystal structure of the EB1 COOH-terminal domain reveals a novel homo-dimeric-fold comprised of a coiled coil and a four-helix bundle motif (12). A recent study reported a GTP-dependent mechanism of dimer-to-monomer transition in EB1 (15). Moreover, +TIPs functioning is controlled by conformational modifications. For example, monomeric mitotic centromere-associated kinesin (MCAK) exhibits different properties compared with dimeric MCAK. MCAK dimerization is important for its catalytic cycle by promoting MCAK binding to microtubule ends, thus enhancing the ability of MCAK to recycle for multiple rounds of microtubule depolymerization and to prevent sequestration by tubulin heterodimers (16, 17).
SKAP was originally identified as a spindle- and kinetochore-associated protein essential for faithful mitotic progression (18–20). Recently, we found that SKAP links kinetochore structural components to the spindle MTs through the Mis13-SKAP-CENP-E interaction pathway (19, 21). Knocking down SKAP by siRNA is essential for accurate kinetochore-MT attachment. Consistent with our observation, Cheeseman and colleagues (20) reported that both SKAP and its binding partner, astrin, were required for the kinetochore localization of CLASP. During the course of our study, the Gruneberg group reported that SKAP and astrin are novel +TIPs (22). However, the functionality and mechanistic role of SKAP in facilitating cell migration remain elusive.
In this report, we establish that the EB1-binding protein, SKAP, directly associates with the cell cortex-distributed scaffold protein, IQGAP1, via its C terminus. Typically, SKAP forms a dimer in vivo, and eliciting enrichment of dimeric SKAP N terminus to MT plus-ends results in MT retraction and bending. Furthermore, SKAP maintains relatively straight morphology of MTs. Moreover, both IQGAP1 and SKAP function in cell migration via the same pathway. Perturbation of the IQGAP1-SKAP interaction inhibits cell migration. Thus, the IQGAP1-SKAP-EB1 axis links dynamic microtubule plus ends to steer cell migration.
Materials and Methods
Cell Culture
HEK293T cells, from the American Type Culture Collection (ATCC, Manassas, VA) were maintained as subconfluent monolayers in advanced DMEM (Invitrogen) with 10% FBS (Hyclone) and 100 units/ml of penicillin plus 100 μg/ml of streptomycin (Invitrogen). MDA-MB-231 cells, from ATCC, were maintained in L-15 medium containing 10% FBS at 37 °C. Cells were transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol.
Plasmids
GFP- and GST-tagged SKAP, full-length and truncations, were described previously (19, 21). Bacterial expression constructs of SKAP full-length and truncations were ligated into the pMal-C2 vector (New England Biolabs, Beverly, MA). His-tagged SKAP was cloned into pETDuet-1 (Novagen). SKAP and SKAP-NT were cloned into pEGFP-C1-FKBP (ARIAD) by standard methods. Bacterial expression constructs of EB1 were cloned into pGEX-6P-1 (GE Healthcare). GFP-tagged IQGAP1 full-length and truncations were inserted into pEGFP-C2 (Clontech). A bacterial expressing construct of IQGAP1 N terminus was ligated into pMal-C2 vector (New England Biolabs). All plasmid constructs were sequenced for verification.
To generate TAT-GFP proteins to perturb the IQGAP1-SKAP interaction, we recombined the pET-22b vector with an 11-amino acid TAT sequence followed by a GFP gene and amino acid sequence competing with the IQGAP1-SKAP interaction (14). TAT-GFP-His fusion proteins were expressed and purified as described previously (14).
Affinity Purification
The SKAP-binding proteins were isolated from MDA-MB-231 cells using an anti-SKAP antibody affinity matrix as previously described (23). After extensive washing, the SAKP immunoprecipitates were fractionated on SDS-PAGE. Commassie Blue-stained bands were removed for in-gel digestion as described previously followed by mass spectrometric identification (24). Positive hits were validated using specific antibodies. The IQGAP1-binding proteins were isolated from MDA-MB-231 cells using an anti-IQGAP1 antibody affinity matrix as described above.
To detect the influence of EGF on the IQGAP1-SKAP interaction, MDA-MB-231 cells were starved over 6–8 h, followed by treatment with EGF (100 ng/ml) for 15 min. To probe the effect of TAT-GFP fusion proteins on the IQGAP1-SKAP interaction, MDA-MB-231 cells were cultured to 70–80% confluence before experimentation. Before introduction, the cells were washed with serum-free media and incubated with TAT-GFP fusion peptides at 2.5 μm using an identical concentration of TAT-GFP as a parallel control.
Expression and Purification of Recombinant Proteins
Purification of recombinant proteins was carried out as described previously (19). Briefly, the GST fusion proteins from the soluble fraction of bacteria were purified using glutathione-agarose chromatography, whereas MBP-tagged proteins were purified using amylose beads. Histidine-tagged proteins were purified using nickel-nitrilotriacetic acid-agarose beads.
For introducing TAT-GFP fusion proteins to probe the functional relevance of the IQGAP1-SKAP interaction, MDA-MB-231 cells were cultured to 20–30% confluence before experimentation. Before introduction, the cells were washed with serum-free media and incubated with TAT-GFP fusion peptides. After incubation, the cells were washed with PBS and then examined directly under fluorescence microscopy as described previously (14).
In Vitro Pulldown Assay
Pulldown assays were carried out as described previously (21). Briefly, the GST fusion proteins in the soluble fraction were purified from bacteria by glutathione-agarose chromatography, whereas MBP-tagged proteins were purified using amylose beads and then eluted by MBP elution buffer (10 mm maltose in PBS). Then, GST fusion protein-bound Sepharose beads were incubated with purified MBP-tagged fusion proteins for 1 h at 4 °C. After incubation, the beads were washed three times with PBS containing 0.25% Triton X-100 and once with PBS and then boiled in 1× SDS-PAGE sample buffer. The bound proteins were separated on an 8% SDS-polyacrylamide gel for Coomassie Blue staining and transferred onto nitrocellulose membrane for Western blotting analysis using MBP antibody.
Immunoprecipitation
pEGFP-C2 vector or GFP-tagged protein-expressing 293T cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.1% Triton X-100) on ice. Lysates were clarified by centrifugation (12,000 rpm for 20 min at 4 °C) and then incubated with GST fusion protein-bound Sepharose beads at 4 °C for 4 h. After an extensive wash, the beads were boiled in 1× SDS-PAGE sample buffer for 5 min, and the bound proteins were separated on a 10% SDS-polyacrylamide gel for transferring onto a nitrocellulose membrane for Western blotting analysis using GFP antibody.
Antibodies
A rabbit antibody against SKAP was generated using full-length recombinant proteins from bacteria according to the standard protocol as previously described (25). The following antibodies were obtained from commercial sources: anti-EB1 mouse antibody (BD Biosciences), anti-IQGAP1 mouse antibody (BD Biosciences), anti-MCAK mouse antibody (described previously in Ref. 26), anti-GFP antibody (BD Biosciences), anti-MBP antibody (Sigma), anti-Cdc42 mouse antibody (BD Biosciences), and anti-tubulin antibody DM1A (Sigma). FITC-conjugated secondary antibodies (Pierce) and rhodamine-phalloidin (Invitrogen) were obtained commercially.
siRNAs Treatment
The siRNA sequence used for silencing SKAP was 5′-AGGCTACAAACCACTGAGTAA-3′ or a SMARTpool (L-022219-00; ThermoFisher Scientific); EB1 siRNA (5′-AAGUGAAAUUCCAAGCUAAGC-3′) (27) and IQGAP1 siRNA (5′-UGCCAUGGAUGAGAUUGGA-3′) were synthesized by Qiagen (USA). As a control, either a duplex targeting cyclophilin or a scramble sequence was used (25, 28). The 21-mer oligonucleotide RNA duplexes were synthesized by Dharmacon Research, Inc. (Boulder, CO). The small-hairpin RNAs (shRNAs) against SKAP were constructed using the same targeting sequences as their siRNAs. The pLKO.1-GFP vector was used as a control. All the siRNAs and shRNAs were transfected into cells using Lipofectamine 2000 for 72 h, and the knockdown efficiency was confirmed by Western blotting analysis and/or immunofluorescence.
Immunofluorescence Microscopy
Cells were seeded onto sterile, acid-treated 12-mm coverslips in 24-well plates (Corning Glass Works) for transfection or drug treatment (23). Cells were rinsed for 1 min with PHEM buffer (100 mm PIPES, 20 mm HEPES, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol, pH 6.9) and permeabilized for 1 min with PHEM plus 0.1% Triton X-100. Extracted cells were then fixed in freshly made 4% paraformaldehyde plus 0.05% glutaraldehyde in PHEM for 5 min and rinsed three times in PBS. After washing three times with PBST (0.05% Tween 20 in PBS), cells were blocked with 1% bovine serum albumin (Sigma) for 45 min at room temperature. Cells were subsequently incubated with the indicated primary antibodies in a humidified chamber for 1 h at room temperature. To visualize IQGAP1 and SKAP simultaneously, cells were incubated with rabbit polyclonal anti-SKAP antibody in a humidified chamber for 1 h and then washed three times in PBST. Mouse antibodies bound to IQGAP1 were visualized with rhodamine-conjugated goat anti-mouse immunoglobulin G (IgG), and binding of anti-SKAP antibody was visualized using fluorescein-conjugated goat anti-rabbit IgG. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma). To measure the EGF influence on SKAP distribution, MDA-MB-231 cells were starved over 6–8 h, followed by treatment with EGF (100 ng/ml) for 15 min. Images were acquired with a DeltaVision wide-field deconvolution microscope (Applied Precision Inc., WA), as previously described (21).
To validate the functional importance of SKAP-IQGAP1 interactions in cell migration elicited by EGF stimulation, aliquots of MDA-MB-231 cells were starved as described above followed by a pre-treatment with TAT-GFP-WWIQ peptide (0.5, 1.0, and 2.5 μm) for 30 min before addition of EGF (100 ng/ml) for 15 min. Identical concentrations of TAT-GFP recombinant protein were used as parallel controls. The treated cells were then fixed, permeabilized, and immunocytochemically stained followed by microscopic examination under a DeltaVision wide-field deconvolution microscope as above.
Deconvolution Microscopy
Deconvolution images were collected using a DeltaVision wide-field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a 100 × 1.35 NA lens was used, and optical sections were taken at intervals of 0.2 μm. Images for display were generated by projecting single optical sections as previously described (29).
Live Cell Images
For live cell and time-lapse imaging, MDA-MB-231 cells were cultured in glass-bottomed culture dishes (MatTek, MA). During imaging, cells were maintained in CO2-independent media (Invitrogen) containing 10% FBS and 2 mm glutamine in a sealed chamber at 37 °C. Images of living cells were taken with a DeltaVision microscopy system at 1 frame per 10 s. Images were prepared for publication using Photoshop (Adobe). Measurements and statistical analyses were calculated using ImageJ software (NIH) and GraphPad Prism (GraphPad software Inc.). Statistical significance was determined by Student's t test.
Gel Filtration and Molecular Mass Determination
To determine the molecular mass of purified SKAP protein in solution, we carried out a gel filtration assay using purified His-tagged SKAP as described by Ward et al. (14). Briefly, size exclusion chromatography was carried out using fast protein liquid chromatography with a Hiload 16/60 Superdex 200 PG column (GE Healthcare) previously equilibrated with PBS. Elution was performed at a flow rate of 1 ml/min. The column was calibrated with ferritin (440 kDa; RS = 6.10 nm), conalbumin (75 kDa; RS = 4.04 nm), ovalbumin (43 kDa; RS = 3.05 nm), carbonic anhydrase (29 kDa; RS = 2.55 nm), and ribonuclease (13.7 kDa; RS = 1.64 nm), which were used as standard proteins according to our recent study (14).
Scratch Assay and Single Cell Migration Analyses
For the wound healing assay, confluent MDA-MB-231 cells transfected with the indicated siRNAs and placed on coverslips were scratched with a 20-μl pipette tip, then stimulated by EGF (100 ng/ml) or 20% serum at 37 °C for the indicated time points. Images were taken with a ×10 objective under an inverted microscope (Axiovert 200) coupled with an Axiocam-HS digital camera (Carl Zeiss, Germany). The relative healing velocities were measured using ImageJ software (NIH). For assays of single cell migration, MDA-MB-231 cells were cultured in a glass-bottomed culture dish (MatTek, MA), covered with human fibronectin. During imaging, cells were kept in CO2-independent media (Invitrogen) containing 10% FBS and 2 mm glutamine in a sealed chamber at 37 °C. Images of living cells were taken with a DeltaVision microscopy system at 1 frame per 10 min. Images were prepared for publication using Photoshop (Adobe). Measurements and statistical analyses were implemented using Image-pro plus software (MEDIA) and GraphPad Prism. Statistical significance was determined by Student's t test.
Data Analyses
All fluorescence intensity measurements were carried out using MetaMorph software. Membrane ruffle fluorescence intensities were determined by measuring the integrated fluorescence intensity within a 7 × 7 pixel square positioned over a single membrane ruffle and subtracting the background intensity of a 7 × 7 pixel square positioned in a region of cytoplasm lacking membrane ruffles. Maximal projected images were used for these measurements. The relative fluorescence intensity was calculated and expressed as the ratio of SKAP over IQGAP1. To determine the significant differences between means, unpaired t tests assuming unequal variance were performed; differences were considered significant when p < 0.05.
Results
Identification of a Novel SKAP-IQGAP1 Complex in Interphase Cells
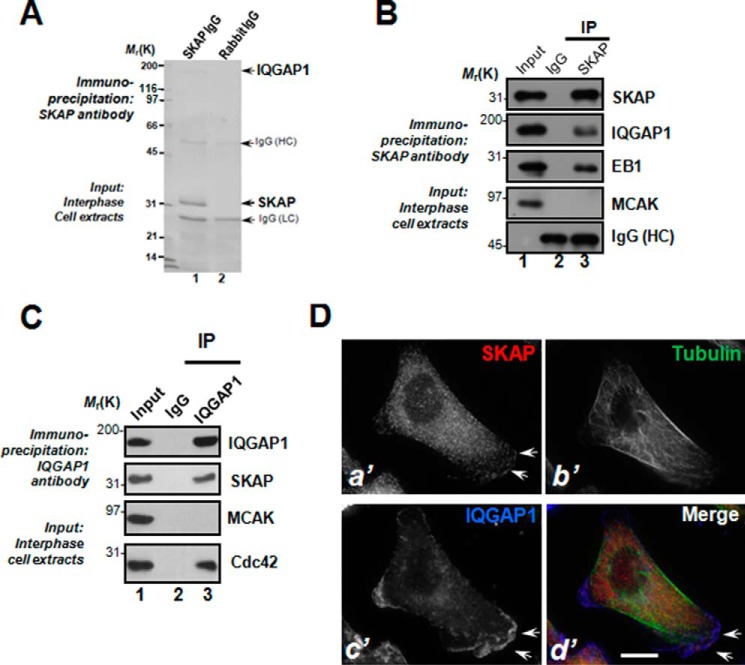
Our previous studies revealed the functional importance of SKAP in kinetochore and microtubule plus-end dynamics during mitosis (19, 21). Because SKAP protein levels remain unchanged during the cell cycle, we sought to examine the role of SKAP in interphase cells. To study the molecular association of SKAP with other microtubule end-binding proteins, we carried out an affinity isolation of a SKAP-containing protein complex followed by mass spectrometric identification of tryptic peptides derived from the complex as described previously (19, 24). As shown in Fig. 1A, anti-SKAP affinity beads coupled to an anti-SKAP antibody isolated a major polypeptide of 35 kDa in addition to three visible bands of polypeptides with approximate masses of 180, 55, and 25 kDa (lane 1). The 35-kDa polypeptide was absent from control IgG affinity matrix isolation (lane 2), and was confirmed as SKAP based on mass spectrometry and Western blotting analyses. Our mass spectrometric analyses identified the 180-kDa polypeptide as IQGAP1, a small GTPase-activating protein interacting with Cdc42 in cell migration and cellular polarity establishment (1, 30). Western blotting analyses validated that the microtubule plus-end protein EB1 also exists in the immunoprecipitates of SKAP, whereas MCAK is absent (Fig. 1B). Reciprocal immunoprecipitation using an anti-IQGAP1 antibody confirmed that SKAP forms a cognate complex with IQGAP1 (Fig. 1C, lane 3, second to the top panel). MCAK was not present in the IQGAP1 immunoprecipitates (lane 3 in the third panel), suggesting that the IQGAP1-SKAP interaction is selective. As a positive control, Cdc42 was retained by IQGAP1 immunoprecipitation (lane 3, bottom panel). Thus, we conclude that IQGAP1 forms a cognate complex with SKAP in interphase cells.
FIGURE 1.
IQGAP1 is a novel SKAP interacting partner at leading edge of migrating cells. A, SKAP forms a novel complex with IQGAP in interphase cells. Aliquots of MDA-MB-231 cells were collected by centrifugation and extracted with Triton X-100-containing buffer as described under “Materials and Methods.” Clarified cell lysates were incubated with Sepharose beads affinity matrix covalently coupled with SKAP antibody and control rabbit IgG, respectively, as described under ”Materials and Methods.“ The beads were washed successively with PBS before elution with 0.2 m glycine, pH 2.3. All samples were separated by SDS-gel electrophoresis. B, SKAP immunoprecipitates from MDA-MB-231 cells in interphase were immunoblotted with antibodies against SKAP, IQGAP, MCAK, and EB1. Note that SKAP immunoprecipitation brought down IQGAP1 and EB1 but not MCAK. C, IQGAP immunoprecipitates from MDA-MB-231 cells in interphase were immunoblotted with antibodies against SKAP, IQGAP, MCAK, and Cdc42. Note that IQGAP1 immunoprecipitation brought down IQGAP1 and Cdc42 but not MCAK. D, MDA-MB-231 cells were labeled with antibodies against SKAP (red), tubulin (green), and IQGAP1 (blue). Scale bar, 10 μm. Note that the membrane ruffle-like localization of SKAP is superimposed onto that of the IQGAP1 signal and the three channels are merged, although the SKAP signal at membrane ruffles is less intense compared with IQGAP1 (d′, arrows). Bar, 10 μm.
The cortical cytoskeleton constitutes an important subcellular structure that determines cell shape and orchestrates cellular dynamics. IQGAP1 is a scaffold protein located to the cortical cytoskeleton for integration of diverse signaling pathways. The identification of IQGAP1 in the SKAP protein complex prompted us to examine SKAP localization in interphase cells relative to IQGAP1. To this end, we carried out immunofluoresence microscopic analyses of HeLa cells triply stained for microtubules, IQGAP1, and SKAP. As shown in Fig. 1D, SKAP distributes in the cytoplasm of interphase MDA-MB-231 cells and appears as comet-like structures, reminiscent of microtubule plus-ends (a′). Careful examination revealed a brief localization of SKAP around the plasma membrane region, reminiscent of the membrane ruffle structures (Fig. 1D, a′, arrows). Surprisingly, this membrane ruffle-like localization of SKAP is largely superimposed onto that of the IQGAP1 signal when signals from IQGAP1 are merged with SKAP and microtubules (d′, arrows). Thus, we conclude that SKAP interacts and co-distributes with IQGAP1 near the plasma membrane in interphase cells.
Characterization of Molecular Interactions between SKAP and IQGAP1
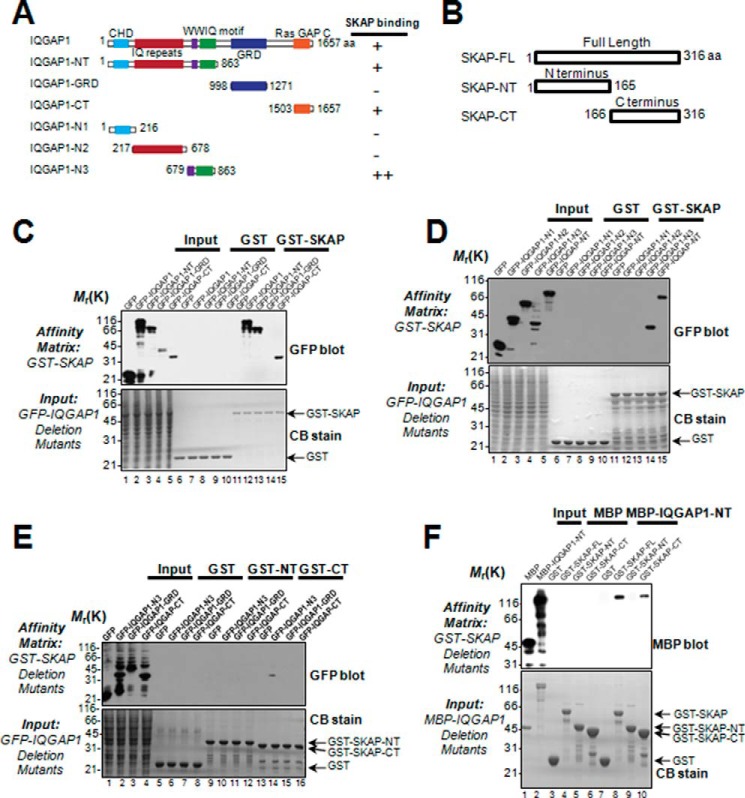
IQGAP1 is a signaling scaffold molecule containing several structure modules for orchestration of protein-protein interactions underlying the signaling cascade during establishment of cell polarity and cell migration. SKAP also contains structural determinants essential for interacting with EB1, CENP-E, and Mis13 (19, 21). If SKAP physically interacts with IQGAP1, the binding interface and perhaps structural determinants could be uncovered by biochemical characterization. To this end, we designed a series of deletion mutants of IQGAP1 and SKAP according to their structural features (Fig. 2, A and B) and expressed those proteins tagged with GFP in 293T cells.
FIGURE 2.
Biochemical characterization reveals a physical link between the SKAP C terminus and IQGAP1-NT. A, schematic drawing of IQGAP1 truncation mutants. Residue numbers at domain boundaries are indicated. B, schematic drawing of SKAP truncation mutants. Residue numbers at domain boundaries are indicated. C, SKAP binds to the IQGAP1 N (amino) terminus as well as C (carboxyl) terminus. Purified GST-SKAP proteins were used to isolate GFP-IQGAP1 deletion mutant proteins from HEK293T cell lysates and fractionated by SDS-PAGE (lower) followed by anti-GFP blotting analysis (upper). D, purified GST-SKAP proteins were used to isolate GFP-IQGAP1 N-terminal truncations from HEK293T cell lysates. Western blotting analysis with anti-GFP antibody indicated the specific associations. E, purified GST-SKAP truncations were used to absorb GFP-IQGAP1 truncations from HEK293T cell lysates. Western blotting analysis with anti-GFP antibody indicated the specific association. F, SKAP directly binds to scaffolding protein IQGAP1 via its C terminus. Using purified GST-SKAP truncations as matrices, a GST pulldown assay was performed to bind purified MBP-tagged IQGAP1-NT. Western blotting analysis with anti-MBP antibody confirmed a specific interaction.
To test whether SKAP physically binds with IQGAP1 and to define the regions of IQGAP1 involved in the interaction, we carried out a pulldown assay using GST-SKAP as an affinity matrix to absorb the GFP-IQGAP1 and its deletion mutants from 293T cell lysates. As shown in Fig. 2C (upper panel), Western blotting analyses with a monoclonal antibody of GFP demonstrated that GFP fusion proteins containing the N-terminal and C-terminal IQGAP1, in addition to full-length GFP-IQGAP1, were retained on the GST-SKAP matrix (Fig. 2C, lanes 12, 13, and 15). The GRD domain exhibited no detectable binding activity toward SKAP (lane 14), serving as a negative control for the specificity of this pulldown assay. The Coomassie Blue-stained SDS-PAGE gel exhibited equal loading of the affinity matrix (Fig. 2C, lower panel).
Because the N-terminal IQGAP1 (amino acids 1–863) contains structural modules such as the calponin homologue domain, WW domain, and IQ domains, we generated a series of deletion mutants and carried out an additional round of pulldown assays using the deletion mutants of IQGAP1 proteins as inputs. As shown in Fig. 2D (upper panel), Western blotting analysis with an anti-GFP antibody indicated that the WWIQ domain of IQGAP1, a region containing amino acids 678–863, exhibits SKAP-binding activity (lane 14).
To further pinpoint the region of SKAP involved in its binding to the WWIQ domain of IQGAP1, we expressed recombinant GST-SKAP and its deletion mutants in bacteria. Purified GST-SKAP proteins (full-length, N terminus, and C terminus) were used as affinity matrices to absorb the deletion mutants of GFP-IQGAP1. As shown in Fig. 2E, Western blotting analysis with an anti-GFP antibody indicated that the C-terminal SKAP binds to the WWIQ domain (lane 14).
To define the domain(s) required for their physical interaction, we carried out an additional GST pulldown assay using full-length SKAP and its N- and C-terminal deletion mutants as affinity matrices to absorb MBP-IQGAP1-expressing bacteria cell lysates. Immunoblotting with an anti-MBP antibody confirmed that MBP-IQGAP-NT was retained on GST-SKAP-CT, indicating a direct interaction between the SKAP C-terminal region and N-terminal IQGAP1 (Fig. 2F, lane 10). The N-terminal SKAP exhibits undetectable binding activity toward the IQGAP1 protein (lane 9). Thus, we conclude that SKAP directly interacts with the IQGAP1 WWIQ domain through its C-terminal domain.
Dimerization of the N Terminus of SKAP Promotes Its Localization to Microtubule Plus Ends
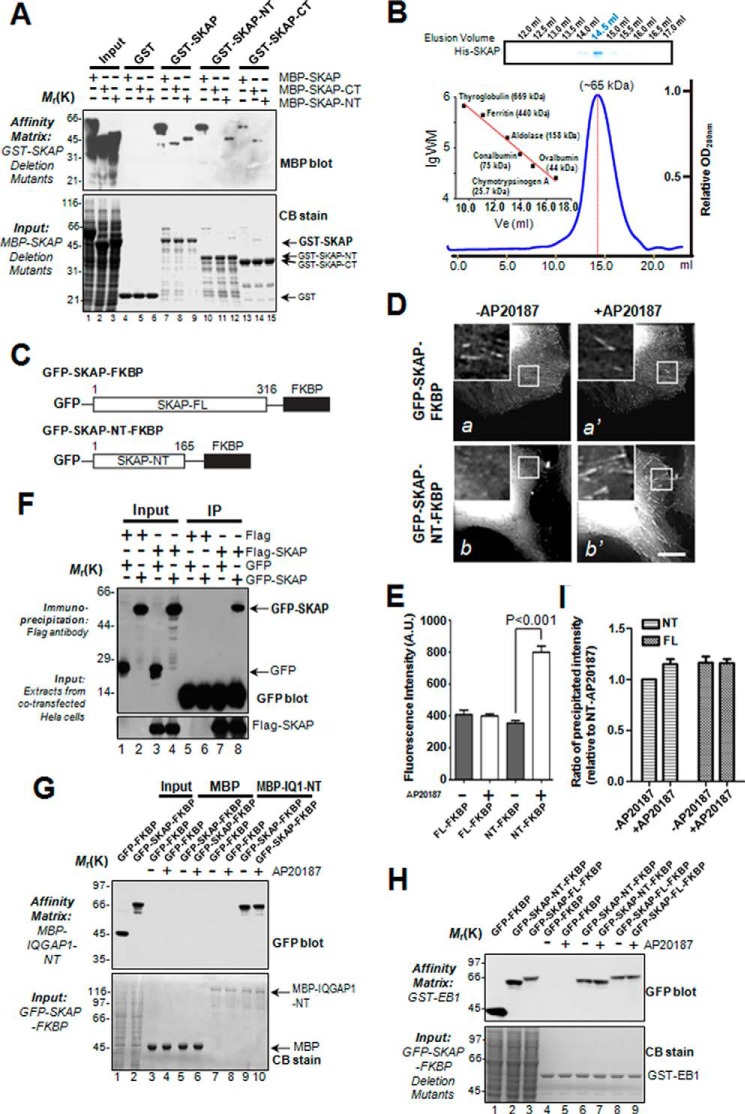
Our previous study revealed that SKAP is a MT plus-end protein (21). Many +TIPs can self-associate to form homo-dimers (12, 17). To further characterize the SKAP-IQGAP1 interaction, we tested if SKAP exhibits an intermolecular interaction and whether the intermolecular interaction of SKAP modulates its binding to IQGAP1. We generated a series of SKAP deletion mutants to perform pulldown assays. As shown in Fig. 3A, interactions were observed between FL (full-length)-FL, NT (N terminus)-NT, CT (C terminus)-CT, but not NT-CT, suggesting that SKAP proteins exhibit an intermolecular association in cis configuration. To examine whether SKAP forms a dimer in solution, we carried out a gel filtration chromatography assay as previously described (14). As shown in Fig. 3B (inset), Coomassie Blue-stained SDS-PAGE showed that His-SKAP eluted at 14.5 ml with an estimated Stokes radius of 3.85 nm, which corresponds to the dimeric form of SKAP (∼68 kDa; the calculated molecular mass of SKAP is 35.4 kDa; Fig. 3B).
FIGURE 3.
Functional dimerization of SKAP via its N terminus enhances its ability to bind microtubule plus-ends. A, purified GST-tagged SKAP full-length and its truncations were used to isolate different MBP-tagged SKAP (FL, NT, or CT) proteins. The isolated complexes were separated by gel fractionation followed by Coomassie Blue staining (lower) and anti-MBP blots (upper). B, analysis of the oligomeric state of SKAP by gel filtration chromatography. Purified His-SKAP proteins were applied to gels and then analyzed by SDS-PAGE. A gel stained with Coomassie Blue illustrates recombinant SKAP peaks at 14.5 ml with an estimated Stokes radius of 3.85 nm. The blue line trace represents the elution profile of recombinant His-SKAP protein measured by A280. C, schematic illustration of SKAP and its N terminus constructs fused to FKBP. D, representative images of GFP-SKAP-FKBP and GFP-SKAP-NT-FKBP-expressing cells before and 10 min after AP20187 treatment. E, different SKAP fusion protein signals shown in D were measured, and the intensity ratios were shown with a bar graph with reference to EB1. Statistics significance was determined by an unpaired Student's t test. Bars, mean ± S.E. from three independent experiments. In each experiment, 20 cells were measured. F, co-immunoprecipitation of exogenous SKAP from mitotic cells. Extracts from mitotic HeLa cells, transiently transfected to co-express FLAG-SKAP and GFP (lane 3) or FLAG-SKAP and GFP-SKAP (lane 4), were incubated with an anti-FLAG mouse antibody, and immunoprecipitates were resolved by SDS-PAGE followed by Western blotting analyses. Upper panel, GFP blot; lower panel, FLAG blot. G, purified MBP-IQGAP1-NT proteins were used as affinity matrices to absorb GFP-SKAP-FKBP fusion proteins from HEK293T cells treated with or without AP20187. The affinity matrices were extensively washed, and associated proteins were fractionated by SDS-PAGE (lower) followed by anti-GFP blotting analysis (upper). H, purified GST-EB1 proteins were used as affinity matrices to absorb GFP-SKAP-FKBP fusion proteins from HEK293T cells treated with or without AP20187. The affinity matrices were extensively washed, and associated proteins were fractionated by SDS-PAGE (lower) followed by anti-GFP blotting analysis (upper). I, quantitative analysis of binding efficiency of GFP-SKAP-FKBP fusion proteins retained on EB1 shown in H. Bars, mean ± S.E. from three independent experiments. Note that chemically induced dimerization promotes binding of N-terminal SKAP but not full-length SKAP to EB1.
To examine the functional relevance of dimerization, we constructed a chemically inducible dimerization system as recently described (31), in which the C-terminal of SKAP was replaced by the homo-dimerization domain of the FK506-binding protein (FKBP; strategy illustrated in Fig. 3C). The FKBP-driven dimerization is initiated by a rapamycin derivative, AP20187 (31). As shown in Fig. 3D, the fluorescence signal of GFP-SKAP-NT-FKBP was detected after the addition of AP20187 in live cells (b and b′), which was significantly different from the control group (GFP-SKAP-FKBP), in which only a minimal increase in intensity could be measured after addition of AP20187 (a and a′). The localization of SKAP-NT to the microtubule plus-ends significantly increased after drug treatment (Fig. 3D).
To quantify these results, the microtubule plus-end fluorescence signal of each protein was analyzed corresponding to treatments. The fluorescence signal ratio of GFP-SKAP-NT-FKBP, but not full-length, was significantly increased after the chemical induction (Fig. 3E; p < 0.001), suggesting that SKAP-NT is not sufficient for spontaneous dimerization in vivo, even though it exhibits a homo-dimeric interaction in vitro. The chemically induced dimerization of SKAP-NT promotes a stable localization and tracking onto microtubule plus-ends. Based on these results, we conclude that dimerization of SKAP-NT is essential for a stable localization to microtubule plus-ends in vivo.
Interestingly, the fluorescence signal ratio of GFP-SKAP-FKBP has no obvious change after the chemical induction, which led us to presume that SKAP itself is inherently a dimer in vivo. To verify our hypothesis, FLAG-SKAP and GFP-SKAP were co-transfected into cells to perform a co-immunoprecipitation assay. As shown in Fig. 3F (upper panel), Western blotting analyses with a monoclonal antibody of GFP demonstrated that GFP-SKAP, but not GFP, associated with FLAG-SKAP (lane 8), indicating that SKAP is indeed a dimer in vivo.
To further understand the relationship between SKAP and IQGAP1, we carried out a pulldown assay using MBP-IQGAP1-NT as an affinity matrix to absorb the GFP-SKAP-FKBP from 293T cell lysates treated with or without AP20187. The result shows that the FKBP-driven dimerization of SKAP has no discernible effect on its interaction with IQGAP1 (Fig. 3G). Considering that SKAP is inherently a dimer in vivo, we conclude that SKAP associates with IQGAP1 as a dimer.
We next sought to understand the mechanism of action underlying this dimerization-elicited stable plus-end tracking and localization. To test if dimerization of SKAP promotes its binding to EB1, we performed a GST pulldown assay in which GST-EB1 was used as an affinity matrix to absorb GFP-SKAP-FKBP and GFP-SKAP-NT-FKBP from 293T cells treated with or without the chemical inducer AP20187 (Fig. 3H). Densitometric analyses of SKAP protein bound to GST-EB1 indicated that the chemically induced SKAP dimer binds to EB1 better, with a 20% higher efficiency (Fig. 3, H and I), supporting the notion that dimerization of SKAP-NT promotes a stable and efficient tracking on microtubule plus-ends.
SKAP Is Important for Keeping MT Relatively Straight Morphology
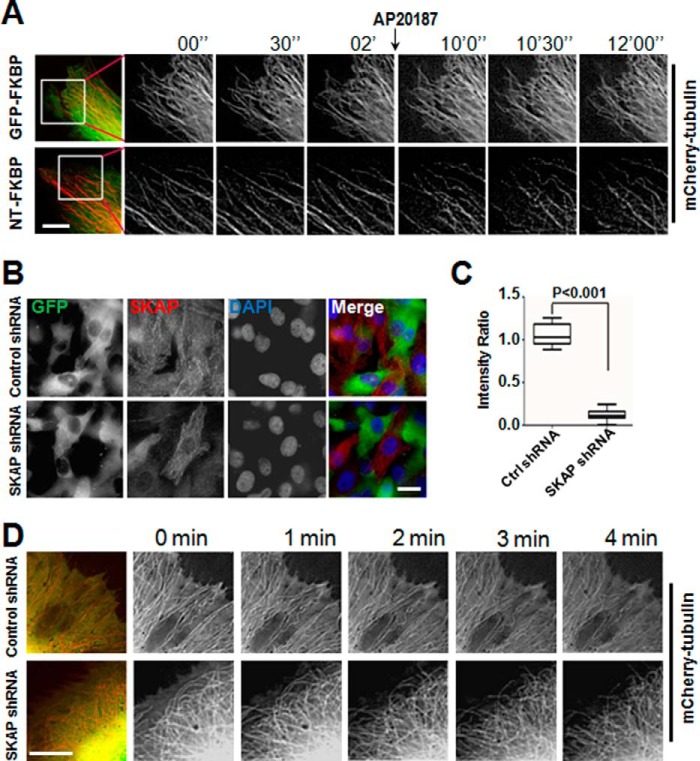
SKAP-NT is responsible for binding with microtubules (18, 20). The enhancement of SKAP-NT localization to microtubule plus-ends led us to examine the influence of dimerization of SKAP-NT on MT morphology. To this end, real-time imaging was used to photograph mCherry-tubulin and GFP-SKAP-NT-FKBP co-transfected into MDA-MB-231 cells to examine the shape of MTs. The results showed that most of the relatively straight morphology of MTs in GFP-SKAP-NT-FKBP-expressing cells, but not in GFP-FKBP-expressing cells, became retracted and bent upon the addition of AP20187 (Fig. 4A).
FIGURE 4.
Suppression of SKAP or perturbation of SKAP function alters microtubule plasticity in cells. A, induced dimerization of SKAP-NT resulted in MT retraction and bending. Real-time imaging of MT in MDA-MB-231 cells expressing GFP-FKBP or GFP-SKAP-NT-FKBP proteins treated with or without AP20187. Scale bar, 10 μm. B, the efficiency of SKAP shRNA. The protein level of SKAP was significantly decreased after 72 h in cells transfected with SKAP shRNA. Scale bar, 20 μm. C, statistical analysis of the relative intensity of SKAP in control and SKAP-suppressed cells. Approximately 30 cells were analyzed for each condition in three independent experiments. The error bar represents S.D. D, suppression of SKAP resulted in MT retraction and bending. Real-time imaging of MT in MDA-MB-231 cells co-transfected with mCherry-tubulin plasmid and the control or SKAP shRNA. Scale bar, 10 μm.
These results indicate that SKAP-NT may function in a dominant-negative manner to perturb the plasticity control of MTs. We speculated that knockdown of SKAP would result in similar morphologies of MTs. To validate our hypothesis, an shRNA targeting the SKAP sequence was used to knock down SKAP expression in cells by transient transfection. An immunofluorescence assay and quantitative analysis indicated a knockdown efficiency of at least 80% (Fig. 4, B and C). The results show that the morphology of MTs in SKAP-depleted cells became retracted and bent rather than remaining relatively straight as the phenotypic MTs induced by SKAP-NT dimerization (Fig. 4D), thus validating our hypothesis.
SKAP Participates in Directional Cell Migration
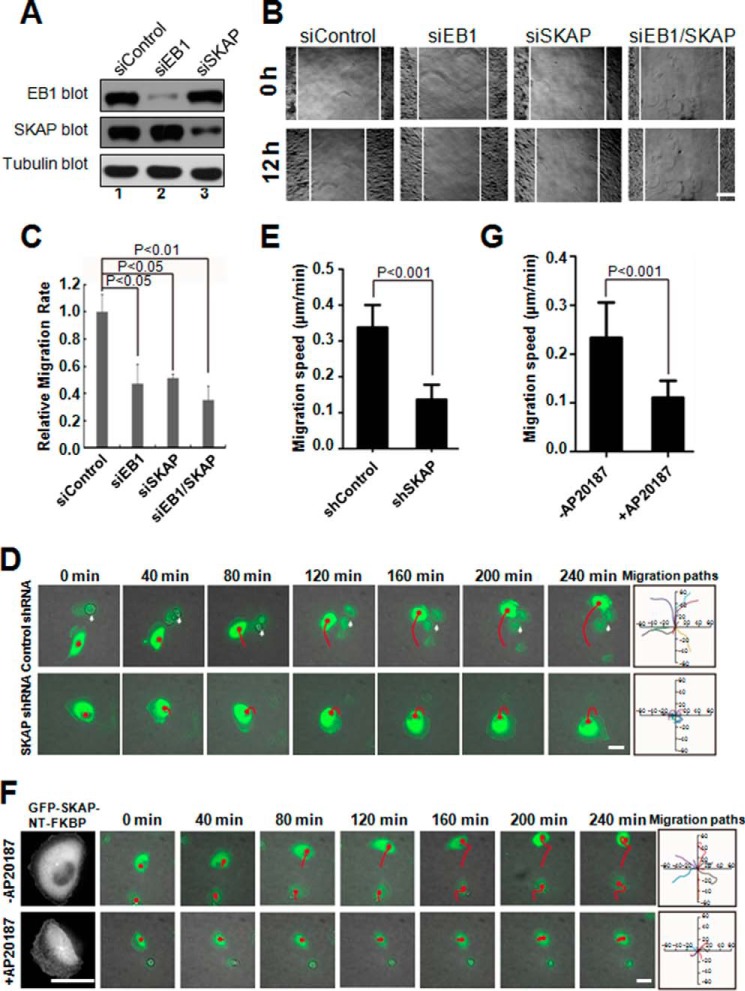
Several +TIPs modulate cell migration in different manners (3, 8). To examine the function of endogenous SKAP underlying cell migration, MDA-MB-231 cells were depleted of SKAP by transfection with siRNA duplexes. Because EB1 is involved in cell migration (32), we included EB1 as a positive control. Western blotting analysis revealed that SKAP was efficiently depleted by specific siRNAs but not by scramble sequences, whereas the protein levels of tubulin and EB1 were unaffected (Fig. 5A, lane 3). Our quantitative analyses show knockdown efficiencies of 82 and 89% for SKAP and EB1, respectively (Fig. 5A).
FIGURE 5.
SKAP is essential for EGF-elicited directional cell migration. A, efficiency and specificity of SKAP and EB1 siRNA. The protein levels of SKAP and EB1 were significantly decreased in cells transfected with corresponding siRNAs. B, suppression of SKAP inhibited cell migration during wound healing. Cells were treated with the indicated siRNAs for 72 h and then were wounded, followed by visualization using phase-contrast microscopy at the indicated time points. Scale bar, 100 μm. C, relative migration rates shown in B were calculated, and the bar graph was made. Bars indicate mean ± S.E. from 3 independent experiments. Statistics significance was determined by an unpaired Student's t test. D, MDA-MB-231 cells transfected with control or SKAP shRNAs (green) followed by real-time imaging at 10-min intervals. Migration tracks of transfected cells are shown as red lines, white arrows indicate mitotic cells. The migration paths of randomly picked transfected cells are presented for each group. Scale bar, 30 μm. E, relative migration speeds shown in D were calculated and graphed. Bars indicate mean ± S.D. from 3 independent experiments. Statistics significance was determined by an unpaired Student's t test. F, real-time imaging of cell migration in MDA-MB-231 cells expressing GFP-SKAP-NT-FKBP proteins treated with or without AP20187. Migration tracks of transfected cells are shown as red lines. The migration paths of randomly picked transfected cells are presented for each group. Scale bars, 30 μm. G, relative migration speeds shown in F were calculated and graphed. Bars indicate S.D. from three independent experiments. Statistics significance was determined by an unpaired Student's t test.
To probe whether SKAP regulates cell migration, we used a wound-healing assay to judge if suppression of SKAP alters the cell dynamics. Specifically, aliquots of MDA-MD-231 cells were transfected with SKAP siRNAs, followed by starvation and generating a linear scratch by a sterile pipette tip as previously described (24). As shown in Fig. 5B, suppression of SKAP protein inhibited cell migration, as determined by the wound healing assay. Our quantitative analyses show that suppression of SKAP alone or together with EB1 reduced the relative migration velocities of cells toward the opposite side by 53.2, 45.9, and 62.7%, respectively, compared with that of the scrambled siRNA (Fig. 5C). These results demonstrate that SKAP functions in cell migration and suggest that endogenous SKAP is a regulator responsible for the EGF-stimulated cell migration.
To further validate these results at the single cell level, MDA-MB-231 cells transfected with control shRNA (green) or SKAP shRNA (green) were starved and then stimulated by serum to migrate. Suppression of SKAP induced a measurable defect in cell migration (Fig. 5D). To quantify these results, the migration speed of cells transfected with control shRNA or SKAP shRNA was calculated (Fig. 5E). Next, we examined the influence of SKAP-NT enrichment to MT plus-ends on cell migration. Time-lapse microscopy showed that cell migration of GFP-SKAP-NT-FKBP-transfected cells treated with AP20187 was impaired (Fig. 5F). To quantify these results, the migration speed was calculated (Fig. 5G). Compared with transfected cells without treatment of AP20187, the migration speed was significantly reduced in GFP-SKAP-NT-FKBP-expressing cells treated with AP20187, indicating that persistent localization of SKAP-NT on MT plus-ends serves as a dominant-negative suppression of cell migration. Thus, we conclude that SKAP is required for directional migration of MDA-MB-231 cells.
The IQGAP1-SKAP Interaction Is Essential in Cell Migration
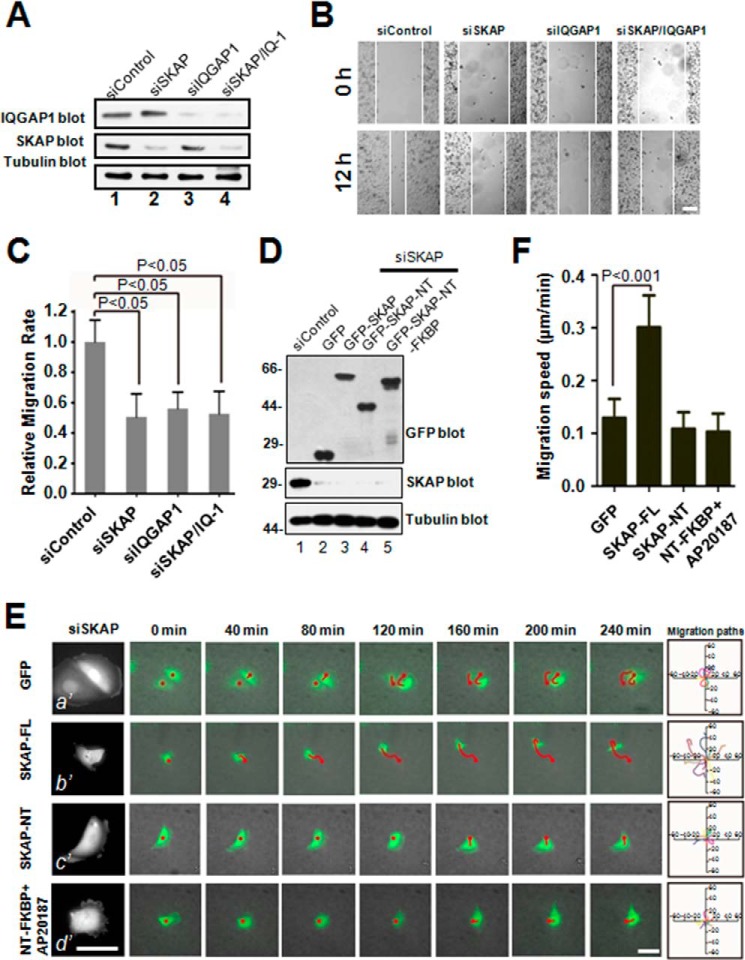
The IQGAP1-SKAP interaction led us to investigate whether the involvement of SKAP in cell migration is through the IQGAP1 process. To this end, corresponding siRNAs were used to knock down the expression of SKAP or IQGAP1 in MDA-MB-231 cells. Western blotting analysis revealed that SKAP and IQGAP1 were efficiently depleted by the specific siRNA but not by scrambled sequences (Fig. 6A). In wound-healing assays, knockdown of SKAP and IQGAP1 individually or simultaneously decreased the relative migration velocities of cells toward the opposite side by 47.8, 43.2, and 47.2%, respectively, compared with that of the scrambled siRNA (Fig. 6, B and C), indicating that SKAP and IQGAP1 functions in cell migration by the same pathway.
FIGURE 6.
The IQGAP1-SKAP interaction links the MTs to cell cortex in cell migration. A, characterization of the efficiency and specificity of various siRNA oligonuclotides in MDA-MB-231 cells. B, in MDA-MB-231 cells, suppression of SKAP, IQGAP1, or SKAP/IQGAP1 exhibits the same defect in directional migration. Cells were treated with the indicated siRNAs for 72 h and scratched, followed by visualization with phase-contrast microscopy at the indicated time points. Scale bar, 100 μm. C, relative migration rates in B were calculated and graphed. Bars indicate S.D. from three independent experiments. Statistics significance was evaluated by an unpaired Student's t test. D, characterization of SKAP siRNA-resistant plasmids. Aliquots of MDA-MB-231 cells, transfected with three GFP-SKAP constructs, express at comparable levels judged by anti-GFP blotting analysis. E, real-time imaging of cell migration in MDA-MB-231 cells co-transfected with SKAP siRNA and the indicated SKAP siRNA-resistant plasmids. Migration tracks of transfected cells are shown as red lines. The migration paths of randomly picked transfected cells are presented for each group. Scale bars, 30 μm. F, relative migration speeds in E were calculated and graphed. Bars indicate S.D. from three independent experiments. Statistics significance was determined by an unpaired Student's t test.
Because of the requirement of the N terminus of SKAP for its MT plus-end localization and its C terminus for binding to IQGAP1, we speculated that SKAP is involved in cell migration by bridging MT plus-ends with IQGAP1 at the cell cortex. To test this hypothesis without the complication of endogenous SKAP, we carried out a rescue assay in which constructs expressing siRNA-resistant GFP-tagged SKAP full-length, NT SKAP, and NT-FKBP SKAP were introduced into aliquots of MDA-MB-231 cells depleted of endogenous SKAP by an siRNA targeted to SKAP. In cells transfected with SKAP siRNA virtually no endogenous SKAP protein was detected (Fig. 6D, lanes 2–5), indicating a high efficiency of knockdown. Western blotting analyses of the GFP tag demonstrated a comparable level of GFP-SKAP protein expression from various transfected constructs (Fig. 6D, top panel). Our early studies demonstrated that exogenous GFP-SKAP and its various deletion mutants are typically expressed at a level 2-fold of that for endogenous SKAP proteins (19, 21).
Having demonstrated the ability of various GFP-SKAP proteins expressed in the absence of endogenous SKAP, we sought to test how the SKAP deletion mutants alter cell migration. As shown in Fig. 6E, time-lapse microscopy showed that the migration of SKAP-depleted and GFP tag-expressing cells was impaired (a′), and that this phenotype was rescued by full-length SKAP (b′) but not by SKAP-NT (c′) or NT-FKBP SKAP (d′) (Fig. 6E). Quantitative analyses showed that the migration speed was rescued to the comparable level of scramble siRNA-transfected cells only in the cells expressing full-length GFP-SKAP (Fig. 6F), indicating that integration of the IQGAP1-SKAP interaction is essential for EGF-elicited cell migration.
The IQGAP1-SKAP Interaction Links Dynamic MT Plus-ends to the Cell Cortex to Steer Cell Migration
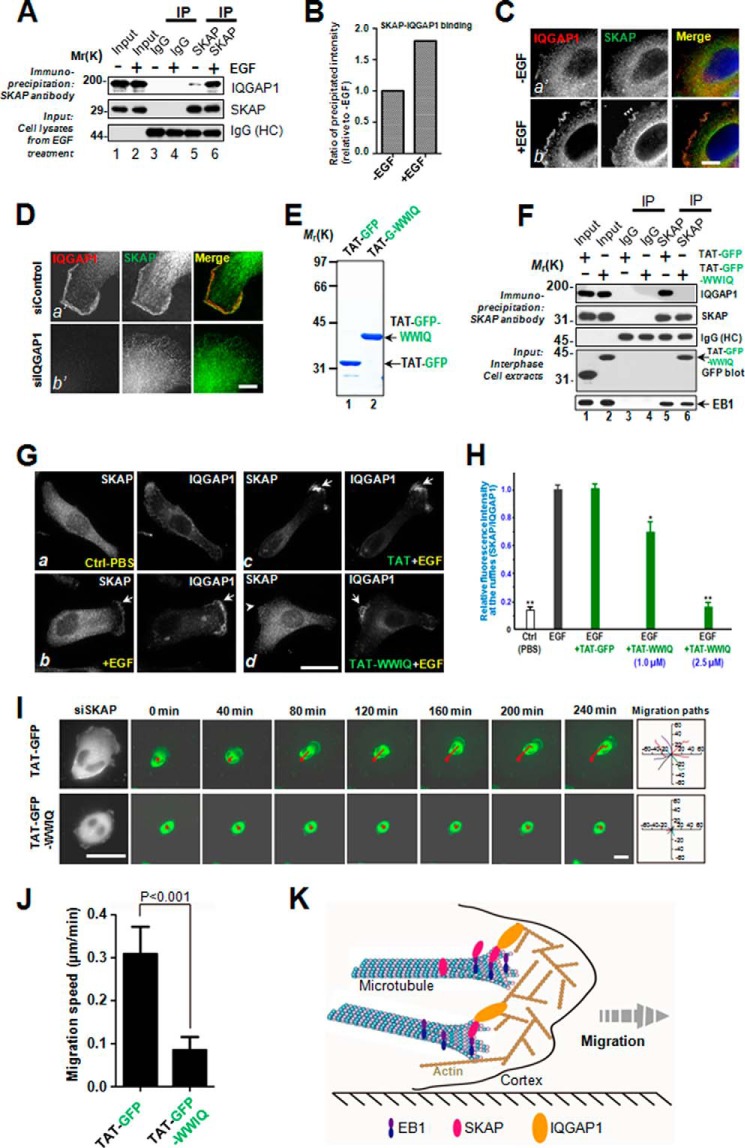
To further define the mechanism of action underlying EGF-elicited cell migration and the role of IQGAP1 in this process, we tested whether the interaction of SKAP and IQGAP1 is regulated by EGF stimulation. To this end, we carried out SKAP immunoprecipitation using MDA-MB-231 cells treated with or without EGF before preparation of cell lysates. As shown in Fig. 7A, Western blotting analyses show that EGF addition resulted in a higher proportion of IQGAP1 from EGF-stimulated preparations (lane 6). Quantitative analyses confirmed that EGF stimulation promotes the association of SKAP with IQGAP1 (Fig. 7B). Because IQGAP1 is a signaling scaffold located at the cell cortex, the EGF-promoted IQGAP1-SKAP interaction prompted us to examine the subcellular location of IQGAP1 relative to SKAP in EGF-starved and stimulated cells. As shown in Fig. 7C, EGF stimulation produces characteristic membrane ruffles that are enriched in IQGAP1 (b′, arrows). Examination of SKAP revealed a super-imposition of IQGAP1 and SKAP on the membrane ruffles (b′, arrows). Their co-distribution and the membrane ruffles are absent in the unstimulated preparation (a′).
FIGURE 7.
Perturbation of IQGAP1-SKAP interaction inhibits cell migration. A, SKAP immunoprecipitates from MDA-MB-231 cells treated without or with EGF in interphase were immunoblotted with SKAP and IQGAP1 antibodies. Nonspecific rabbit IgG-coupled beads were used as a control. B, measurements of the ratio of precipitated intensity of IQGAP1 in A. C, representative immunofluorescence images of MDA-MB-231 cells treated without or with EGF were fixed and stained for SKAP (green) and IQGAP1 (red). Scale bar, 10 μm. D, representative immunofluorescence images of MDA-MB-231 cells transfected with the indicated siRNAs. At 72 h post-transfection, MDA-MB-231 cells were fixed and stained for SKAP (green) and IQGAP1 (red). Scale bar, 10 μm. E, Coomassie Blue-stained SDS-PAGE gel was used to assess the quality of purified recombinant TAT-GFP-His6 and TAT-GAP-WWIQ proteins. Bacteria expressed TAT-GFP-peptide and TAT-GFP-H6 (control) were purified with nickel-nitrilotriacetic acid affinity chromatography and desalted into DMEM. Protein concentration was determined by Bradford assays. F, SKAP immunoprecipitates from MDA-MB-231 cells incubated with TAT-GFP or TAT-GFP-WWIQ peptide in interphase were immunoblotted with SKAP, IQGAP1, EB1, and GFP antibodies. Nonspecific rabbit IgG-coupled beads were used as a control. Note that the EB1-SKAP interaction was not altered by the addition of the TAT-GFP-WWIQ peptide but that the endogenous SKAP-IQGAP1 interaction was perturbed by the addition of the TAT-GFP-WWIQ peptide. G, TAT-GFP-peptide recombinant WWIQ peptide disrupted SKAP-IQGAP1 association and librated SKAP from IQGAP1-containing membrane ruffles. Aliquots of TAT-GFP or TAT-GFP-WWIQ (2.5 μm) were added to cultured MDA-MB-231 cells for 30 min followed by EGF stimulation for 10 min. After the stimulation, treated cells were subjected to fixation, immunocytochemistry, and examination. Note that incubation of the TAT-GFP-WWIQ peptide liberated SKAP from membrane ruffle localization (d, arrowhead). Scale bars, 10 μm. H, statistical analyses of the TAT-GFP-WWIQ peptide liberation of SKAP from IQGAP1-positive membrane ruffles. The TAT-GFP-WWIQ peptide liberated SKAP from membrane ruffles in a dose-dependent manner (**, p < 0.001 for 2.5 μm TAT-GFP-WWIQ peptide; *, p < 0.05 for 1.0 μm TAT-GFP-WWIQ peptide). I, real-time imaging of cell migration in MDA-MB-231 cells with the TAT-GFP or TAT-GFP-WWIQ peptide. Trajectories of transfected cells are shown as red lines. The migration paths of randomly picked transfected cells are presented for each group. Scale bars, 30 μm. J, relative migration speeds in I were calculated and graphed. Bars indicate S.D. from three independent experiments. Statistics significance was determined by an unpaired Student's t test. K, proposed working model accounting for the SKAP function in directional cell migration. In short, the SKAP-IQGAP1 complex serves as a novel link to orchestrate directional cell migration via coupling dynamic microtubule plus-ends to the cell cortex.
If IQGAP1 were a signaling scaffold at the cortex responsible for SKAP localization, suppression of IQGAP1 would diminish the localization of SKAP to the cortex. Consistent with our rationale, suppression of IQGAP1 indeed reduced the localization of SKAP at the membrane ruffles (Fig. 7D, b′) but not in scramble siRNA-transfected cells (a′), demonstrating that IQGAP1 provides a linkage for SKAP with the cell cortex.
To assess the functional effect of the SKAP-IQGAP1 interaction in cell migration, a membrane-permeable peptide containing the WWIQ domain of IQGAP1 (679–863 amino acids) was constructed to modulate the endogenous SKAP-IQGAP1 interaction. This was achieved by introducing an 11-amino acid peptide derived from the TAT protein transduction domain into a fusion protein containing amino acids involved in the binding interface between SKAP and IQGAP1, as described previously (1, 14, 33). The recombinant protein was histidine-tagged and purified to homogeneity using nickel-affinity beads (Fig. 7E, TAT-GFP-WWIQ, lane 2). As predicted, the recombinant TAT-GFP-WWIQ protein (2.5 μm) disrupted the SKAP-IQGAP1 association in vivo, as judged by an anti-SKAP immunoprecipitation (Fig. 7F, lane 6). As a negative control, TAT-GFP did not interfere with the interaction of SKAP-IQGAP1 (Fig. 7F, lane 5). In addition, the TAT-GFP-WWIQ protein did not alter the EB1-SKAP interaction (Fig. 7F, bottom panel), demonstrating a selective alteration of the SKAP-IQGAP1 interaction by the TAT-WWIQ peptide. The results show the effect of the TAT-GFP-WWIQ recombinant protein in competing with full-length IQGAP1 for association with SKAP.
Our trial experiments showed that the internalization of TAT-GFP and TAT-GFP-WWIQ proteins was seen in virtually all cells in culture at 30 min after the addition of TAT-GFP fusion proteins. Having demonstrated the ability of the TAT-GFP-WWIQ peptide in perturbing the endogenous SKAP-IQGAP1 interaction, we sought to test whether the TAT-GFP-WWIQ peptide perturbs the co-distribution of SKAP with IQGAP1 to the membrane ruffles elicited by EGF stimulation. To this end, aliquots of MDA-MB-231 cells were starved for 7 h, followed by pretreatment of TAT-GFP or TAT-GFP-WWIQ peptides for 30 min. The TAT-treated cells were then exposed to EGF (100 ng/ml) for 15 min. These cells were then fixed and stained for SKAP and IQGAP followed by microscopic analyses. As shown in Fig. 7G, IQGAP1 and SKAP exhibit a minimal co-distribution profile at the plasma membrane in cells starved from EGF (a). Addition of EGF into the starved cells resulted in apparent membrane ruffles (Fig. 7G, b, arrows). The localization of SKAP is superimposed on that of IQGAP1 in EGF-stimulated cells. However, this co-distribution was perturbed by TAT-GFP-WWIQ peptide (Fig. 7G, d, arrowhead) but not by the TAT-GFP peptide (Fig. 7G, c, arrows). Our quantitative analyses show that addition of the TAT-GFP-WWIQ peptide induced a dose-dependent liberation of SKAP into the cytoplasmic “soluble” fraction as judged by the disappearance of the SKAP signal from the EGF-elicited membrane ruffles (Fig. 7H).
To determine whether the IQGAP1-SKAP interaction is required for cell migration, aliquots of MDA-MB-231 cells were exposed to TAT-GFP-WWIQ or TAT-GFP for 30 min before imaging cell migration elicited by EGF; real-time imaging of the cells began 5 min after EGF addition (Fig. 7I). The results showed that treatment of cells with TAT-GFP-WWIQ perturbed EGF-elicited cell migration relative to cells treated with the TAT-GFP control. This inhibitory action is relatively specific, as a low concentration of TAT-GFP peptide did not interfere with mitotic progression.5 In addition, the TAT-GFP peptide did not induce cell death or alter cell fate. Our quantitative analyses show that TAT-GFP-WWIQ significantly reduced the velocity of cell migration (Fig. 7J; p < 0.001). Thus, the SKAP-IQGAP1 interaction is required for cell migration. Therefore, we conclude that the IQGAP1-SKAP interaction provides a regulatory linkage between the MT plus-ends and cell cortex to steer cell migration.
Discussion
IQGAP1 is a signaling scaffold protein for integration of protein-protein interactions essential for molecular dynamics and cellular plasticity (34, 35). Our results show that IQGAP1 links dynamic microtubules via interacting with SKAP to steer cell migration. The functional relevance of the biochemical interaction between IQGAP1 and SKAP was validated using a membrane-permeable TAT-WWIQ peptide, which results in an inhibition of directional cell migration. Our findings identify a molecular mechanism orchestrating the dynamics of cortical microtubule attachment during directional cell migration and support a role of the IQGAP-SKAP-EB1 signaling hub in dynamic membrane-microtubule interactions in response to extracellular cues (36).
The accurate regulation of cell migration is critical to developmental morphogenesis, tissue homeostasis, and tumor metastasis. The MT plus-end tracking proteins establish a complex structure platform that serves as a molecular engine to regulate MT dynamics and thereby orchestrate cellular events such as cell migration (8, 10). However, it has remained elusive how unique +TIPs are specifically regulated in response to extracellular environmental changes to ensure an accurate direction of cell migration. In this study, we explored the IQGAP1-SKAP physiological function in the regulation of cell migration.
Previous studies have found that many +TIPs accumulate at the cell cortex and function in cell migration through different pathways (8). It is reasonable that SKAP is also involved in cell migration. Our data suggest, as expected, that SKAP depletion induces a defect in cell migration. Importantly, endogenous SKAP accumulates at the cell leading edge in MDA-MB-231 cells, suggesting that SKAP interacts with proteins distributed at the cell cortex. Many +TIPs are involved in cell migration by interaction with the cortical cytoskeleton protein, IQGAP1 (7, 11, 37). Our results also reveal that SKAP binds to IQGAP1 and serves as a linkage between MT plus-ends and the cell cortex. It is noteworthy that the +TIPs usually bind to the C terminus of IQGAP1. However, we found that the middle region (WWIQ domain) of IQGAP1 is responsible for the interaction with SKAP. These results demonstrate that IQGAP1 interacts with +TIPs through different regions. Perhaps these regions of IQGAP1 are involved in cell migration by binding to different +TIPs. It would be worthwhile to investigate whether they have a synergistic interaction with IQGAP1 to regulate cell migration.
Our previous research revealed that the localization of SKAP to MT plus-ends is in an EB1-dependent manner by direct interaction via its N terminus (21). Interestingly, SKAP-NT itself exhibits a weak localization to MT plus ends. This phenotype suggests that SKAP localization to MT plus-ends is regulated by its C terminus, even though SKAP-CT has no binding affinity to EB1. The C terminus of SKAP is likely responsible for SKAP dimerization, for there are two coiled-coil regions within the C terminus. We found that SKAP is a dimer and that artificially induced dimerization enhances localization of SKAP-NT to MT plus-ends. EB1 has also been reported to be a dimer (12). Thus, it would be of interest in the future to elucidate the structural basis of the SKAP interaction with EB1. Elucidation of the structural basis of this interaction will enable us to consolidate the EB1-SKAP interaction and regulation into a mechanistic model by which the dimerization regulates the localization of SKAP to MT plus-ends.
Most +TIPs affect MT dynamics. For example, end-binding proteins (EBs) promote MT dynamics and growth, and suppress catastrophes (38, 39). Particularly, EB1, a well studied member of the EB family, promotes MT polymerization and prevents MTs from pausing (40). Other +TIPs have different functions, such as the cytoplasmic linker proteins (CLIPs) and CLIP-associated proteins (CLASPs), which increase rescue frequency (41, 42), whereas the MT depolymerase MCAK promotes the disassembly of MTs. A recent report argued that SKAP is a novel MT plus-end tracking protein and promotes MT growth in vitro (22). Knockdown of SKAP had significant influence on MT growth and shortening rate as well as MT stability in vivo.
Recently, we developed a new method for optically imaging intracellular protein interactions at the nanometer scale of spatial resolution in live cells using photoactivatable complementary fluorescent (PACF) proteins (43), based on photoactivation of complementary fluorescent proteins. Upon maturation, sparse subsets of PACF molecules were activated, localized, and then bleached (36). This feature provides benefits for super-resolution microscopic analyses of photoactivation localization and spatially matched molecular interaction. The aggregate position information from all PACF subsets was then assembled into a nano-scale map, which allows imaging of a single molecule copy of specific target protein-protein interactions in space and time in fixed and live cells. Using PACF, we obtained precise localization of the dynamic microtubule plus-end hub protein EB1 dimers and their distinct distributions at the leading edges and cell bodies of migrating cells (43). Thus, in the future, it would be of interest to employ PACF-based analyses to probe how dimeric SKAP is precisely located to IQGAP1 in leading and trailing edges of migrating cells and how different microtubule plus-end tracking proteins function in cell migration at the single molecule level. This will enable us to conduct single molecule dynamics mapping at the single microtubule scale during cell migration.
In summary, we present evidence that IQGAP1 directly interacts with SKAP. The +TIP protein SKAP self-associates to form a homo-dimer. Induced dimerization of SKAP-NT enhances its localization to microtubule plus-ends in vivo. Persistent enrichment of SKAP-NT to MT plus-ends results in MT retraction and bending, consistent with that in SKAP-suppressed cells. Our results also demonstrate that SKAP promotes microtubule dynamics and participates in directional cell migration. Subsequently, we found that IQGAP1 and SKAP exist in the same complex to regulate cell migration. Thus, we concluded that the IQGAP1-SKAP-EB1 interaction serves as a bona fide linkage between MTs and the cell cortex in cell migration (Fig. 7I). Interestingly, many +TIPs stabilize MTs at the cell cortex via connecting actin filaments or interacting with cortical cytoskeleton protein IQGAP1 (3, 7, 37, 44, 45) and are involved in cell directional migration (3, 7, 37). However, more sophisticated mechanisms need to be delineated. Furthermore, it would be exciting and challenging ahead to visualize and compare the single microtubule dynamics and corresponding cortex during cell migration.
Author Contributions
D. C., X. D., and X. L. conceived the project. D. C., Z. S., W. W., H. W., S. A., and D. W. designed and performed most biochemical experiments. J. Z., B. Q., X. L., X. Z., and X. D. designed and performed cell biological characterization. L. L., G. A., X. W., D. W., and C. J., performed in vitro reconstitution experiment and data analyses. D. C., D. L. H., X. D., and X. Y. wrote the manuscript, and all authors have read and approved the manuscript.
Acknowledgments
We thank members of our groups for stimulating discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK56292, CA164133, and G12RR03034, Chinese 973 Project Grants 2012CB917204, 2013CB911203, and 2014CB964803, Chinese Academy of Science Grant KSCX2-YW-H-10, Anhui Province Key Project Grant 08040102005, International Collaboration Grant MOST2009DFA31010, Chinese Natural Science Foundation Grants 31320103904, 91313303, 31430054, 90913016, 31171300, and 31100992, 31471275, 31301121, MOE20113402130010, Fundamental Research Funds for Central Universities Grants WK2060190018 and WK2340000021, and MOE Innovative Team IRT13038. The authors declare no competing financial interests.
D. Cao and X. Wang, unpublished observation.
- MT
- microtubule
- IQGAP
- IQ domain-containing GTPase-activating protein
- MCAK
- mitotic centromere-associated kinesin
- FKBP
- FK506-binding protein
- MBP
- maltose-binding protein
- EB
- end-binding protein
- PACF
- photoactivatable complementary fluorescent.
References
- 1. Zhou R., Guo Z., Watson C., Chen E., Kong R., Wang W., Yao X. (2003) Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell 14, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krause M., Gautreau A. (2014) Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590 [DOI] [PubMed] [Google Scholar]
- 3. Watanabe T., Noritake J., Kaibuchi K. (2005) Regulation of microtubules in cell migration. Trends Cell Biol. 15, 76–83 [DOI] [PubMed] [Google Scholar]
- 4. Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005) IQGAP1: a key regulator of adhesion and migration. J. Cell Sci. 118, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 5. Brown M. D., Bry L., Li Z., Sacks D. B. (2007) IQGAP1 regulates Salmonella invasion through interactions with actin, Rac1, and Cdc42. J. Biol. Chem. 282, 30265–30272 [DOI] [PubMed] [Google Scholar]
- 6. Adachi M., Kawasaki A., Nojima H., Nishida E., Tsukita S. (2014) Involvement of IQGAP family proteins in the regulation of mammalian cell cytokinesis. Genes Cells 19, 803–820 [DOI] [PubMed] [Google Scholar]
- 7. Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. (2002) Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109, 873–885 [DOI] [PubMed] [Google Scholar]
- 8. Akhmanova A., Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 9. Carvalho P., Tirnauer J. S., Pellman D. (2003) Surfing on microtubule ends. Trends Cell Biol. 13, 229–237 [DOI] [PubMed] [Google Scholar]
- 10. Galjart N. (2010) Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528–537 [DOI] [PubMed] [Google Scholar]
- 11. Watanabe T., Noritake J., Kakeno M., Matsui T., Harada T., Wang S., Itoh N., Sato K., Matsuzawa K., Iwamatsu A., Galjart N., Kaibuchi K. (2009) Phosphorylation of CLASP2 by GSK-3β regulates its interaction with IQGAP1, EB1 and microtubules. J. Cell Sci. 122, 2969–2979 [DOI] [PubMed] [Google Scholar]
- 12. Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., Vale R. D. (2005) Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 168, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drabek K., van Ham M., Stepanova T., Draegestein K., van Horssen R., Sayas C. L., Akhmanova A., Ten Hagen T., Smits R., Fodde R., Grosveld F., Galjart N. (2006) Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr. Biol. 16, 2259–2264 [DOI] [PubMed] [Google Scholar]
- 14. Ward T., Wang M., Liu X., Wang Z., Xia P., Chu Y., Wang X., Liu L., Jiang K., Yu H., Yan M., Wang J., Hill D. L., Huang Y., Zhu T., Yao X. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gireesh K. K., Sreeja J. S., Chakraborti S., Singh P., Thomas G. E., Gupta H., Manna T. (2014) Microtubule +TIP protein EB1 binds to GTP and undergoes dissociation from dimer to monomer on binding GTP. Biochemistry 53, 5551–5557 [DOI] [PubMed] [Google Scholar]
- 16. Moore A. T., Rankin K. E., von Dassow G., Peris L., Wagenbach M., Ovechkina Y., Andrieux A., Job D., Wordeman L. (2005) MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 169, 391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hertzer K. M., Ems-McClung S. C., Kline-Smith S. L., Lipkin T. G., Gilbert S. P., Walczak C. E. (2006) Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol. Biol. Cell 17, 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang L., Seki A., Fang G. (2009) SKAP associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle 8, 2819–2827 [DOI] [PubMed] [Google Scholar]
- 19. Huang Y., Wang W., Yao P., Wang X., Liu X., Zhuang X., Yan F., Zhou J., Du J., Ward T., Zou H., Zhang J., Fang G., Ding X., Dou Z., Yao X. (2012) CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidt J. C., Kiyomitsu T., Hori T., Backer C. B., Fukagawa T., Cheeseman I. M. (2010) Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J. Cell Biol. 191, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., Ding X., Huang Y., Wang D., Yao X. (2012) Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287, 39380–39390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunsch A. K., Linnane E., Barr F. A., Gruneberg U. (2011) The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 192, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 24. Fang Z., Miao Y., Ding X., Deng H., Liu S., Wang F., Zhou R., Watson C., Fu C., Hu Q., Lillard J. W. Jr., Powell M., Chen Y., Forte J. G., Yao X. (2006) Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4. Mol. Cell. Proteomics 5, 1437–1449 [DOI] [PubMed] [Google Scholar]
- 25. Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A. W., Teng M., Cleveland D. W., Goldberg M. L., Niu L., Yao X. (2004) Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279, 54590–54598 [DOI] [PubMed] [Google Scholar]
- 26. Jiang K., Wang J., Liu J., Ward T., Wordeman L., Davidson A., Wang F., Yao X. (2009) TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 29. Kops G. J. (2009) Dividing the goods: co-ordination of chromosome biorientation and mitotic checkpoint signalling by mitotic kinases. Biochem. Soc. Trans. 37, 971–975 [DOI] [PubMed] [Google Scholar]
- 30. Smith J. M., Hedman A. C., Sacks D. B. (2015) IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 25, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao L., Jin C., Chu Y., Varghese C., Hua S., Yan F., Miao Y., Liu J., Mann D., Ding X., Zhang J., Wang Z., Dou Z., Yao X. (2010) Dimerization of CPAP orchestrates centrosome cohesion plasticity. J. Biol. Chem. 285, 2488–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell. Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Wang Z., Jiang K., Zhang L., Zhao L., Hua S., Yan F., Yang Y., Wang D., Fu C., Ding X., Guo Z., Yao X. (2009) PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 284, 23059–23071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rokavec M., Li H., Jiang L., Hermeking H. (2014) The p53/miR-34/IQGAP1 axis in development and diseases. J. Mol. Cell. Biol. 6, 214–230 [DOI] [PubMed] [Google Scholar]
- 35. Xia P., Zhou J., Song X., Wu B., Liu X., Li D., Zhang S., Wang Z., Yu H., Ward T., Zhang J., Li Y., Wang X., Chen Y., Guo Z., Yao X. (2014) Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell. Biol. 6, 240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 [DOI] [PubMed] [Google Scholar]
- 37. Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004) Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 7, 871–883 [DOI] [PubMed] [Google Scholar]
- 38. Komarova Y., De Groot C. O., Grigoriev I., Gouveia S. M., Munteanu E. L., Schober J. M., Honnappa S., Buey R. M., Hoogenraad C. C., Dogterom M., Borisy G. G., Steinmetz M. O., Akhmanova A. (2009) Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tirnauer J. S., Bierer B. E. (2000) EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tirnauer J. S., Grego S., Salmon E. D., Mitchison T. J. (2002) EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell 13, 3614–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. (2005) CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Komarova Y. A., Akhmanova A. S., Kojima S., Galjart N., Borisy G. G. (2002) Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xia P., Liu X., Wu B., Zhang S., Song X., Yao P. Y., Lippincott-Schwartz J., Yao X. (2014) Super-resolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell 25, 4166–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moseley J. B., Bartolini F., Okada K., Wen Y., Gundersen G. G., Goode B. L. (2007) Regulated binding of adenomatous polyposis coli protein to actin. J. Biol. Chem. 282, 12661–12668 [DOI] [PubMed] [Google Scholar]
- 45. Tsvetkov A. S., Samsonov A., Akhmanova A., Galjart N., Popov S. V. (2007) Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton 64, 519–530 [DOI] [PubMed] [Google Scholar]