Abstract
The binding determinants of the human acyl-CoA binding domain-containing protein (ACBD) 6 and its function in lipid renewal of membranes were investigated. ACBD6 binds acyl-CoAs of a chain length of 6 to 20 carbons. The stoichiometry of the association could not be fitted to a 1-to-1 model. Saturation of ACBD6 by C16:0-CoA required higher concentration than less abundant acyl-CoAs. In contrast to ACBD1 and ACBD3, ligand binding did not result in the dimerization of ACBD6. The presence of fatty acids affected the binding of C18:1-CoA to ACBD6, dependent on the length, the degree of unsaturation, and the stereoisomeric conformation of their aliphatic chain. ACBD1 and ACBD6 negatively affected the formation of phosphatidylcholine (PC) and phosphatidylethanolamine in the red blood cell membrane. The acylation rate of lysophosphatidylcholine into PC catalyzed by the red cell lysophosphatidylcholine-acyltransferase 1 protein was limited by the transfer of the acyl-CoA substrate from ACBD6 to the acyltransferase enzyme. These findings provide evidence that the binding properties of ACBD6 are adapted to prevent its constant saturation by the very abundant C16:0-CoA and protect membrane systems from the detergent nature of free acyl-CoAs by controlling their release to acyl-CoA-utilizing enzymes.
Keywords: lipids, fatty acids, phospholipids, binding protein, erythrocyte, acyl-coenzyme A, acyl-coenzyme A binding domain-containing protein 6
The members of acyl-CoA binding domain-containing protein (ACBD) family are involved in the maintenance of diverse cellular functions (1–3). They are essential to a variety of cellular functions presumably via their interaction with a multitude of proteins involved in neural stem cell self-renewal, neurodegeneration, stress resistance, lipid homeostasis, intracellular vesicles trafficking, organelle formation, viral replication, and apoptotic response (2–5). Several ACBD members have been shown to interact with proteins such as GABAA (6), Numb (7), DMT1 (8), TSPO, PPM1L (9), the mHtt/Rhes complex (10), giantin/golgin-160, HNF4α (11), sterol regulatory element-binding protein (SREBP) 1 (12), plant AtEBP (13), and several viral proteins (3, 14–18). Plant ACBP members are also implicated in a multitude of function such as embryogenesis and resistance to various stresses (4, 5, 11). However, of the seven members of the mammalian family, the binding characteristics of these proteins to acyl-CoAs have only been studied in detail for member 1, DBI (aka ACBD1) (6, 19–24).
Human ACBD6 is a modular protein with an N-terminal domain carrying the acyl-CoA binding domain and a carboxy-terminal domain with two ankyrin repeats, predicted to function as anchor motif to other proteins (25). This nonconserved C-terminus domain, present in members ACBD2 to ACBD6, is lacking in ACBD1 and ACBD7. The latter appears to be a gene duplication of ACBD1. Expression of the hACBD6 protein was found to be elevated in hemangioblasts of placental tissue, in cord blood, bone marrow hematopoietic progenitor cells, and circulating CD34+ cells, as well as erythrocyte precursors (25). ACBD6 mRNA levels were higher in hematopoietic progenitors as compared with lineage-committed cells (26, 27).
To explore the binding properties of this protein to acyl-CoAs and fatty acids, a novel fluorescent binding assay was developed. This assay allowed real-time measurements of the binding activity of purified ACBD6 protein and provided the ability to assess rapid association of the protein with the ligand. We show that ACBD6-bound acyl-CoAs were the substrates for the lysophospholipid acyltransferase enzymes and that ACBD6 controls the rate of acylation in membranes by the Lands’ pathway enzymes. Comparisons were made with other ACBD proteins (ACBD1 and ACBD3) as well as mutated forms of ACBD6. We propose that the controlled release of acyl-CoA from ACBD proteins to the acyltransferase enzymes is an important determinant protecting membrane systems from the detergent nature of these compounds while maintaining substrate availability for the acylation reactions. Additional evidence that the ACBD6-mediated transfer was modulated by the complex lipids environment in the cell was provided by the intriguing finding that fatty acids that do not bind to ACBD6 could still compete for binding.
MATERIALS AND METHODS
Materials
[1-14C]C18:1-CoA (55.0 mCi/mmol) was purchased from Amersham Corp. (Arlington Heights, IL), and 1 acyl-lysophosphatidylcholine (lysoPC) and 16-NBD-16:0-CoA (N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl]amino palmitoyl CoA) from Avanti Polar Lipids Inc. (Alabaster, AL). TLC silica plates were obtained from Analtech Inc. (Newark, DE). No-weight format of disuccinimidyl suberate (DSS) was from Thermo Fisher Scientific. The acyl-CoAs and fatty acids were from Sigma-Aldrich, and all other compounds used were reagent grade.
Cloning and site-directed mutagenesis
Cloning of human ACBD3 and ACBD6 has been previously reported (25, 28). Human ACBD1 isoform1 was cloned by PCR from the cDNA clone NM_020548.5 (American Type Culture Collection; MGC:70414; IMAGE:5168729) into pET28a vector (Novagen) to yield plasmid pFK847. Site-directed mutagenesis experiments were performed with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions. Primers were designed with the QuikChange® Primer Design Program. The presence of the intended nucleotide change and the absence of unwarranted mutations were verified by full-length sequencing of the constructs.
Protein expression and purification
All proteins and mutant forms were produced as hexahistidine recombinant forms in the Escherichia coli host BL21(DE3) cells (Novagen) and purified by affinity metal chromatography as previously described (25, 28). The purified proteins were stored at −80°C in Tris-HCl 50 mM pH 8.0, NaCl 0.1 M, EDTA 5 mM, β-mercaptoethanol 5 mM, and glycerol 10% (v/v). Prior to isothermal titration calorimetry (ITC) measurements, proteins were dialyzed in the ITC buffer (see below).
[14C]acyl-CoA binding assays
Binding of radiolabeled [14C]acyl-CoA (C18:1-CoA and C20:4-CoA) to the purified proteins was determined according to the method of Augoff et al. (19). We found that the Lipidex-1000 method was unreliable when acyl-CoAs concentrations were higher than 10 µM. Lipidex-1000 resin could not remove all of the unbound ligand. Consequently, the radioactive [14C]acyl-CoA in the protein-bound fraction could not be determined accurately. This required the use of a different resin, NTA (Promega). Increasing concentrations of ligand (1 to 150 µM) were incubated with 1 µM of purified protein at 37°C for 20 min in 100 µl of 10 mM potassium phosphate pH 7.4. Tubes were chilled on ice for 5 min before the addition of 50 µl of ice-cold 50% washed NTA slurry (Promega). Tubes were rotated for 1 h at 4°C. Protein absorbed to the resin was collected by low-speed centrifugation at 2,000 g for 2 min at 4°C. The supernatant containing unbound [14C]acyl-CoA was transferred to a scintillation vial. Protein pellet was washed three times with 200 µl buffer. All three washes were pooled with the unbound fraction and counted. The pellets containing protein and acyl-CoA bound protein were transferred to a scintillation vial and counted. For competition experiments, unlabeled ligands were added during the incubation with the protein.
ITC assays
ITC measurements of the binding of acyl-CoAs and fatty acids were performed on a VP-ITC instrument (MicroCal, LLC). All experiments were performed in ammonium acetate 25 mM pH 7.4, supplemented with 0.1% Triton X-100 when fatty acid was the ligand, at 30°C. The proteins were dialyzed in the same buffer, and fresh 10 mM stocks of the ligands were prepared from powder with the dialyzing buffer. Measurements were performed with 28 injections of 10 µl of 100 µM ligand every 150 s. The cell contained the protein at an initial concentration of 10 µM. Control experiments were performed by injecting buffer into the cell containing the protein and by injecting the ligand into the cell containing buffer. Heat generated from control runs was subtracted from the data of the experimental set performed under the same conditions.
Fluorescent binding assays
Real-time measurements of 16-NBD-16:0-CoA (NBD-C16:0-CoA) binding to purified protein were performed in a fluorimeter (LS50B; Perkin Elmer) at 30°C. Stock solutions of NBD-C16:0-CoA were made in Tris-HCl 20 mM pH 7.4 at 800 µM. Reactions were performed in 200 µl of 20 mM Tris-HCl pH 7.4 with 2 µM NBD-C16:0-CoA and 5 µM ACBD6 protein and with Tween-20 at 0.8% to unquench the signal.
LysoPC acyltransferase activity and 16-NBD-16:0 sn-2-PC detection
Real-time measurements of the incorporation of NBD-C16:0-CoA into lysoPC in presence of human lysophosphatidylcholine-acyltransferase 1 (LPCAT1) produced in E. coli microsomes (29) were performed as above with 20 µM lysoPC and 30 µg of protein. After 5 min of recording, the reactions were transferred into 750 µl of CHCl3/methanol (1:2), and lipids were extracted, dried, and separated by TLC as previously described (29). Spots were detected under UV light, scraped, and extracted in CHCl3/methanol. Silica debris was removed by centrifugation, and fluorescence of NBD-labeled lipids was measured in the fluorimeter. Alternatively, a FluoChem camera equipped with Cy2 filters can be used for detection and quantification when each TLC plate is calibrated with an internal fluorescence standard.
Measurement of lysoPC and lysoPE acyltransferase activity
Incorporation of [14C]C18:1-CoA into egg lysoPC by recombinant LPCAT1 protein in E. coli membranes and by erythrocyte membranes or into lysophosphatidylethanolamine (lysoPE) by erythrocyte membrane were determined as previously described (30). Reactions were performed in glass tubes at 37°C in a shaking water bath, in 200 µl of (Tris-HCl 20 mM pH 7.4, Tween-20 0.8 mg/ml) containing 20 µM lysoPC or lysoPE and 1 µM [14C]C18:1-CoA. Reactions were initiated by the addition of 4 µg of membrane protein fractions or 50 µg ghost membrane protein, and incubated from 0 to 6 min. Three to four time points, in triplicate, were used to determine the rates of incorporation. Reactions were stopped by the addition of 200 µl of CHCl3/methanol/12 N HCl (40:40:0.26, v/v) and vigorous vortexing. Phases were separated by centrifugation at 1,000 g for 5 min, and the lipid-containing chloroform phase was dried down under N2, subsequently dissolved by vortexing in 20 µl of CHCl3/methanol (2:1, v/v). Samples were applied to TLC silica plates and developed with chloroform/methanol/acetic acid/0.9% NaCl (100:50:16:5, v/v). TLC plates were air-dried for 20 min and exposed to a PhosphoImager screen (Storm 840; Molecular Dynamics). Quantification of [14C]phosphatidylcholine (PC) and [14C]phosphatidylethanolamine (PE) formation was performed with ImageQuant software subtracting the plate background.
Acyl-CoA synthetase assay
Measurements were performed with human acyl-CoA synthetase (ACSL) 6 enzyme produced in E. coli membrane as previously described (31). Reactions were performed at 30°C in 0.1 M Tris.HCl pH 8.0, 1 mM DTT, 20 mM MgCl2, 20 mM ATP, and 0.5 mM CoA, with 4 µM [14C]C18:1-OH and 1 μg of protein. The reaction was stopped by addition of 2.25 ml of isopropanol/ heptane/2 M H2SO4 (40:10:1, v/v) and vigorous vortexing. Newly synthesized [14C]acyl-CoA was separated from unesterified [14C]fatty acid by successive addition of 1 ml of heptane and 1 ml of water and vigorous vortexing. The two phases were separated by centrifugation at 500 g for 3 min. The upper organic phase was carefully removed and collected in a scintillation vial. The lower aqueous phase was then extracted twice with 2 ml heptane saturated with 4 mg/ml palmitic acid and once with 2 ml heptane. Each of the upper phases were collected and pooled with the first one. The amounts of newly synthesized [14C]acyl-CoA and of [14C]fatty acid were measured by scintillation counting in the lower and combined upper phases, respectively. For each reaction, the total amount of [14C] present was calculated by addition of the dpm values of the upper and lower phases. Control reactions were performed with membranes obtained from cells carrying the vector pET28a, and the dpm values obtained were subtracted from those obtained with the enzyme preparations.
DSS cross-linking
The homobifunctional N-hydroxysuccinimide ester reagent DSS reacts with primary amines and was used to determine conformational changes of purified ACBD3 and ACBD6 upon binding with acyl-CoAs. DSS was bought in individual sealed microtubes of 2 mg each (Thermo Fisher Scientific), and stock solutions of 100 mM in DMSO (further diluted in DMSO as needed) were made fresh prior to each experiments. DSS was added in a 30-fold excess relative to the protein (mol/mol). Proteins were dialyzed in 10 mM potassium phosphate pH 7.4 at 4°C. Reactions were performed in 29 µl of 50 mM potassium phosphate pH 7.4 with 30 µM protein and acyl-CoAs at 40 to 400 µM at 37°C for 20 min. One microliter of DSS was added to a final concentration of 1 mM and incubated for 30 min at room temperature. Untreated control reactions were performed with addition of 1 µl of DMSO and incubated as the DSS-treated samples. Reactions were stopped by quenching of DSS with 2 µl Tris-HCl 1 M pH 7.4 for at least 15 min at room temperature. Mixtures were then boiled in SDS-PAGE loading buffer for 4 min and separated on denaturing SDS-polyacrylamide Tris-glycine gels. Proteins were detected by staining with GelCode Blue reagent (Thermo Fisher Scientific) or were transferred on nitrocellulose membrane and detected with an HRP-conjugated anti-histidine antibody (INDIA-HisProbe-HRP antibody; Thermo Fisher Scientific).
RESULTS
Ligand saturation of ACBD6 in absence of oligomerization
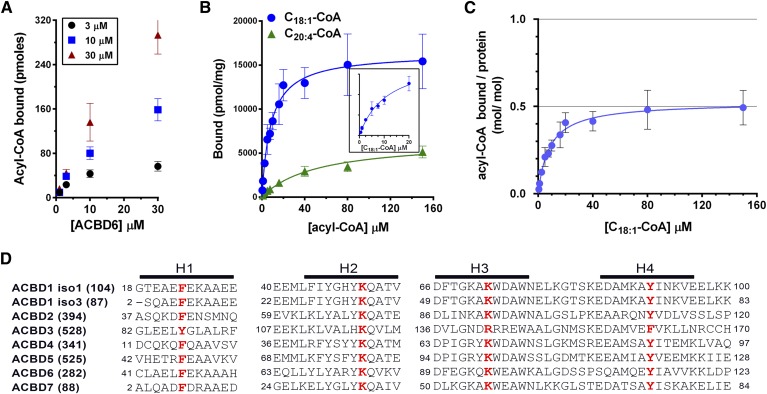
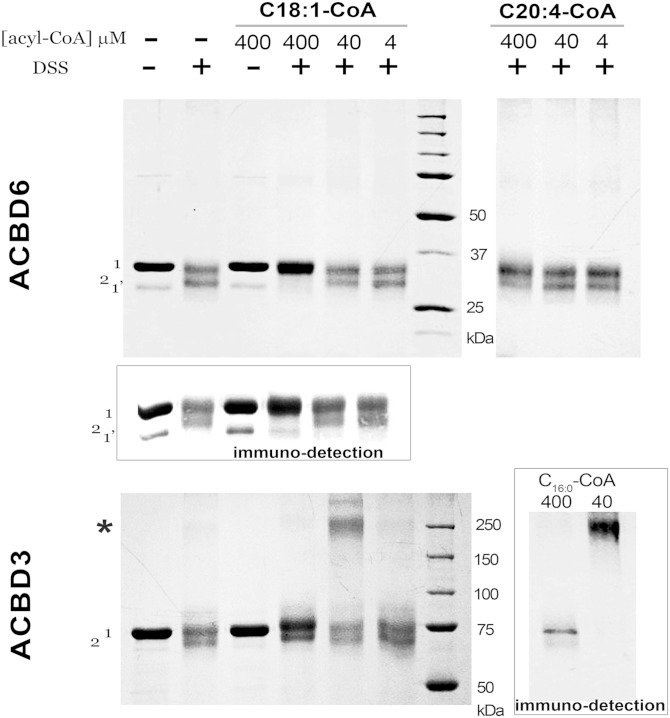
Acyl-CoA binding to ACBD6 was dependent on the ligand and protein concentration (Fig. 1A). The purified protein has a greater preference for C18:1-CoA than C20:4-CoA (Fig. 1B). The three different techniques used to measure binding (see Materials and Methods) showed that even in the presence of 10 µM acyl-CoA, 0.2 µM protein was not saturated (Fig. 1B). Saturation with an apparent stoichiometry of 0.5 (mol/mol) was attained at a much higher concentration of ligand (Fig. 1B, C). As was reported for erythrocyte ACBD1, a ligand/protein ratio of 0.5 indicated that one molecule of ligand may be bound to a dimeric form of the protein (19). Using the cross-linker DSS, high molecular mass species of hACBD3 were observed in presence of either C18:1-CoA or C16:0-CoA (Fig. 2, bottom panels). These ACBD3 oligomers were disrupted in the presence of very high concentration of acyl-CoA (400 µM). However, in contrast to ACBD1 (19) and ACBD3 (Fig. 2), ligand-bound ACBD6 did not show oligomerization under these conditions with either C18:1-CoA or C20:4-CoA (Fig. 2, top panels). An additional band (band 2) was generated by DSS in absence of ligand in both ACBD6 and ACBD3. ACBD6 band 2 was protected from DSS attack at a high concentration of C18:1-CoA but not by the weaker ligand C20:4-CoA (Fig. 2), suggesting that conformation of ACBD6-bound C18:1-CoA prevented attack of residues exposed in lipid-free ACBD6. In contrast, ACBD3 was not protected by high concentrations of C18:1-CoA (Fig. 2, left bottom panel). Together, these data indicate that binding of ligand may protect DSS modification of ACBD protein, and that this protection is both ligand and protein dependent.
Fig. 1.
Acyl-CoA binding activity of human ACBD6. A: Three different concentrations of [14C]C18:1-CoA were incubated with increasing concentrations of purified ACBD6 at 37°C for 20 min in 100 µl of 10 mM potassium phosphate pH 7.4. Unbound ligand was separated from ACBD6-bound ligand by pulling down the hexahistidine-tagged protein with NTA resin. B: Increasing concentrations of [14C]C18:1-CoA (blue circles) and [14C]C20:4-CoA(green triangle) were incubated with 1 µM ACBD6. Inset: Data of the binding of [14C]C18:1-CoA were plotted for the concentration range of 0 to 20 µM. C: Molar ratio of [14C]C18:1-CoA bound per mole of ACBD6 was plotted as function of the concentration of ligand. Errors represent the standard deviations of three measurements. D: Amino acid alignment of the predicted four helices (H1 to H4) of the ACB motif of the seven members of the ACBD family. For clarity, DBI was annotated as ACBD1 and PECI as ACBD2 in the panel. Two of the several spliced variants of ACBD1 are shown (isoform 1 and isoform 3). The number of residues of each form is indicated in parentheses. The four conserved residues predicted to be essential for acyl-CoA binding that were substituted with alanine are shown in bold red.
Fig. 2.
DSS cross-linking of ACBD3 and ACBD6 in the presence of acyl-CoAs. Increasing concentrations of the indicated acyl-CoAs were incubated in 50 mM potassium phosphate pH 7.4 with 30 µM protein at 37°C for 20 min. DSS was added to a final concentration of 1 mM for 30 min at room temperature. Following quenching of DSS, proteins were loaded on denaturing SDS-PAGE 10% gel. After electrophoresis, protein bands were detected with Gelcode Blue. The positions of the full-length protein and of an unfinished translated product present in the preparation are indicated as band 1 and 1’. Presence of cross-linked lipid-free protein is indicated as band 2. The position of high molecular mass forms observed with ACBD3 is indicated with an asterisk. Inset: Immunodetection of ACBD6 and ACBD3.
Role of conserved residues of the ACB motif in ligand binding
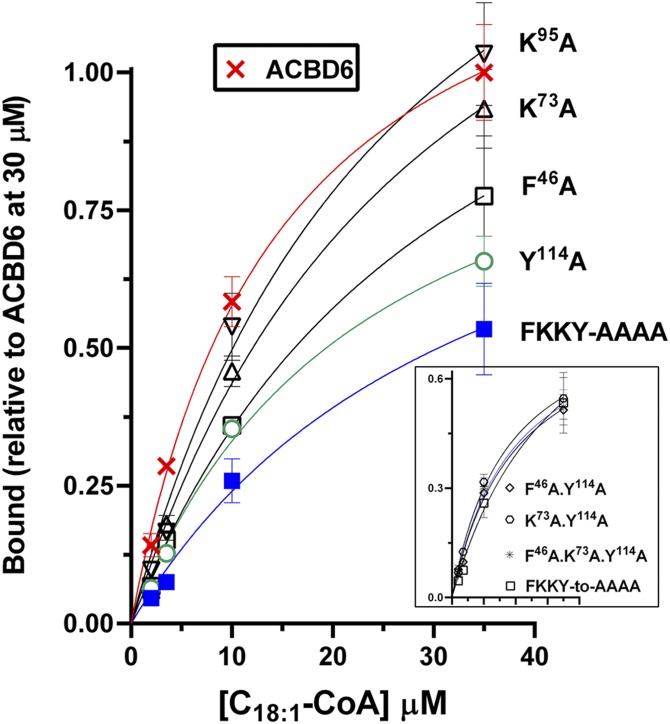
Sequence alignment and structural information provided by biochemical analysis supported evidence of conserved residues involved in binding to the acyl-CoA ligand in the four-helix bundle defining the ACB motif (Fig. 1D) (1, 5, 20, 21). One predicted essential residue in each of the four helices (21) of ACBD6 was substituted with an alanine, and binding activity of the mutant forms was determined with [14C]C18:1-CoA. As shown in Fig. 3, the conserved lysine residues K73 and K95 in helix 2 and helix 3, respectively, had no significant contribution to binding. Substitution of phenylalanine 46 (F46) in helix 1 and tyrosine 114 (Y114) in helix 4 resulted in a 30% and 40% decrease in binding, respectively. The tyrosine 114 residue seemed most critical because all mutant forms lacking it had the same low binding activity, even when combined with a modification of a nonessential residue (e.g., K73; Fig. 3, inset). The removal of all four residues still rendered a form (FKKY-AAAA) with a binding activity of 50% compared with the nonmutated protein in the presence of saturating concentration of C18:1-CoA (≥35 µM). However, at physiological concentrations (≤3.5 µM) (32), the differences between the mutant and native forms of ACBD6 will be much more pronounced.
Fig. 3.
Role of conserved residues of the ACB motif in acyl-CoA binding. Binding assays were performed as described in legend of Fig. 1 with ACBD6 (crosses) and the indicated mutant forms. Inset shows the activity of the combination of two-, three-, and four-residue substitutions. Error bars represent the standard deviations of at least three measurements.
Acyl-chain-length determinants
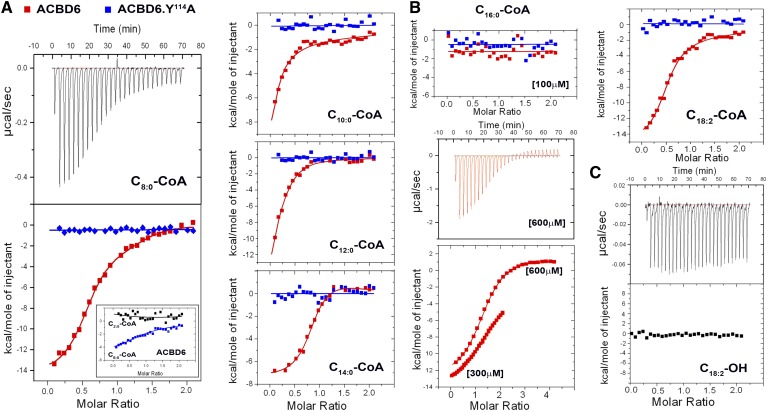
Binding of acyl-CoAs of various aliphatic chain lengths was assessed by ITC measurements. Compared with the mutant ACBD6.Y114A, enthalpy-driven binding of C8:0-CoA, C10:0-CoA, C12:0-CoA, C14:0-CoA, and C18:2-CoA was detected with ACBD6 (Fig. 4). A weak binding was detected with short C6:0-CoA species but not with acetyl-CoA (C2; Fig. 4A, inset). Binding to C20:4-CoA (Fig. 1B) could not be measured by ITC due to the high amount of heat generated by injection of the compound alone into buffer (supplementary Fig. 1). Under similar conditions, binding of C16:0-CoA was not detected (Fig. 4B). However, binding was observed when the concentration of C16:0-CoA was increased from 100 µM to 600 µM. Weak binding of C16:0-CoA to ACBD6 was also observed with the Lipidex-1000 and isoelectrofocusing methods (25). Thus, a broad range of acyl-CoA molecules of medium (≥6) to long (≤20) chain length can bind to ACBD6. No binding to fatty acids was detected (Fig. 4C) (25).
Fig. 4.
Aliphatic chain length requirement for binding. ITC measurements were performed in ammonium acetate 25 mM pH 7.4, supplemented with 0.1% Triton X-100 when fatty acid was the ligand, at 30°C. The proteins were dialyzed in the same buffer, and fresh 10 mM stocks of the acyl-CoA ligands were prepared from powder with the dialyzing buffer. Measurements were performed by 28 injections of ligand at 100 µM into the chamber containing 10 µM ACBD6 (red) and the mutant form ACBD6.Y114A (blue). A: ACBD6 bind medium-chain acyl-CoA. B: The concentration of C16:0-CoA was increased from 100 µM to 300 µM and to 600 µM with 30 µM ACBD6 in the chamber. C: ACBD6 does not bind fatty acid.
Binding competition by fatty acids
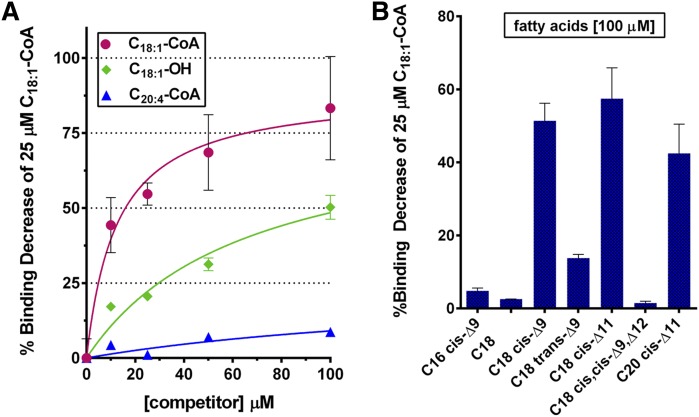
As mentioned, binding of fatty acids to ACBD6 could not be detected with the three techniques we tested (Lipidex-1000 and NTA resins and ITC). This finding raised the question of whether association to the acyl chain was highly specific with respect to the presence of CoA or whether the lack of detectable binding to an acyl-OH was due to an unstable fatty acid-ACBD6 complex. To explore this further, competition experiments were performed to assess binding properties of the protein toward fatty acids. As shown in Fig. 5, binding of [14C]C18:1-CoA to ACBD6 was measured in the presence of increasing concentrations of C18:1-CoA, C20:4-CoA, and the fatty acid C18:1-OH. As anticipated, C18:1-CoA efficiently competed with binding of [14C]C18:1-CoA. The weaker ligand C20:4-CoA had little effect on the binding of [14C]C18:1-CoA. Surprisingly, C18:1-OH strongly competed with [14C]C18:1-CoA binding. A 50% reduction was reached with only a 4-fold excess of C18:1-OH (Fig. 5A). Requirements for length, unsaturation, and stereoisomer conformation of the aliphatic chain for competition of the radiolabeled C18 cis-Δ9-CoA species ([14C]C18:1-CoA) were also assessed. Saturated (stearic acid; C18:0), monounsaturated trans isomer (elaidic acid; C18 trans-Δ9), and polyunsaturated cis isomer (linoleic acid; C18 cis,cis-Δ9,Δ12) did not affect or weakly affected binding (Fig. 5B). However, two monounsaturated cis isomer species, asclepic acid (C18 cis-Δ11) and gadoleic acid (C20 cis-Δ11), competed with binding of [14C]C18 cis-Δ9-CoA with similar efficiency as oleic acid itself (C18 cis-Δ9). Interestingly, the monounsaturated cis isomer species palmitoleic acid (C16 cis-Δ9) was not a competitor, which provided further evidence of the low affinity of ACBD6 for acyl chain length of 16 carbons. Thus, interference of binding of a monounsaturated cis stereoisomer acyl-CoA species by fatty acids was specific of monounsaturated species in the cis conformation, with less stringency for the chain length. These results provide the first evidence that stereoisomeric conformation of the aliphatic chain plays an essential role in binding to ACBD6 protein (supplementary Fig. 2A).
Fig. 5.
Competition of binding of [14C]C18:1-CoA. Binding assays were performed as described in legend of Fig. 1, and 0.1% Triton X-100 was added to the buffer when fatty acids were used. A: Binding of 25 µM [14C]C18:1-CoA to ACBD6 was measured in the presence of increasing concentrations of C18:1-CoA (red circles), C20:4-CoA (blue triangles), and C18:1-OH (green diamonds). B: Binding of 25 µM [14C]C18:1-CoA to ACBD6 was measured in the presence of 100 µM of the indicated fatty acids. Binding activity values are reported as percentage of the difference of the amount of bound [14C]C18:1-CoA to ACBD6 in presence of the unlabeled species relative to the amount obtained in their absence. Error bars represent the standard deviations of at least three measurements.
Fast equilibrium and rapid exchange of bound acyl-CoA
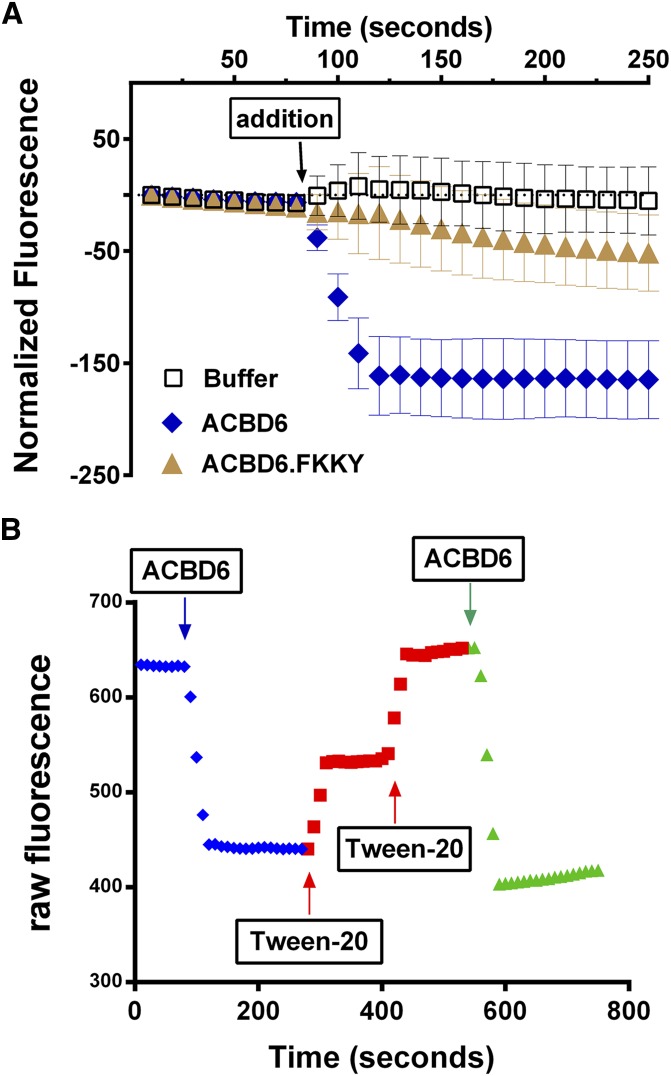
To gain insight into the association-dissociation kinetics of the ACBD6 and its ligand, we developed a novel fluorescently based method using the compound 16-NBD-C16:0-CoA (supplementary Fig. 2B). In the absence of detergent, NBD-C16:0-CoA fluorescence is low due to self-quenching. Addition of the detergent Tween-20 or albumin unquenched the fluorescence and increased the signal in a dose-dependent manner (supplementary Fig. 3). Real-time measurements of NBD-C16:0-CoA binding to ACBD6 protein were performed by monitoring the decrease of the Tween-20 unquenched fluorescence of NBD-C16:0-CoA in the presence of the protein. Addition of ACBD6 resulted in an almost immediate decrease of the fluorescence signal, which rapidly reached a new equilibrium (Fig. 6A). The decrease in the fluorescence was the result of quenching following binding of the NBD-C16:0-CoA to ACBD6. The mutant ACBD6.FKKY-AAAA, which poorly binds the ligand, decreased the fluorescence by a small amount (Fig. 6A). Addition of the detergent Tween-20 following quenching by ACBD6 led to a quick increase of the signal (Fig. 6B). This effect of Tween on fluorescence was not the result of denaturation of ACBD6, but of a release of NBD-C16:0-CoA from ACBD6 to its environment. A second injection of the protein into this mild-detergent environment successfully brought down the fluorescence signal. These findings provided evidence of a rapid and reversible exchange of the NBD-C16:0-CoA between the protein and detergent micelles.
Fig. 6.
Real-time acyl-CoA binding measurements. Real-time fluorescence measurements of 16-NBD-C16:0-CoA binding to purified protein were performed in 200 µl of Tris-HCl 20 mM pH 7.4 with Tween-20 at 0.8% at 30°C with 2 µM 16-NBD-C16:0-CoA. A: ACBD6 (blue diamonds) and the mutant form ACBD6.FKKY-AAAA (gold triangles) were injected during measurement at the indicated time at a final concentration of 5 µM. Injection of the same volume of buffer (open squares) was used as control. Error bars represent the standard deviations of at least three measurements. B: A series of injections of ACBD6 and Tween-20 were performed in the same cuvette and changes in fluorescence were monitored as a function of time.
Acyl-CoA bound to ACBD6 is slowly released to LPCAT1
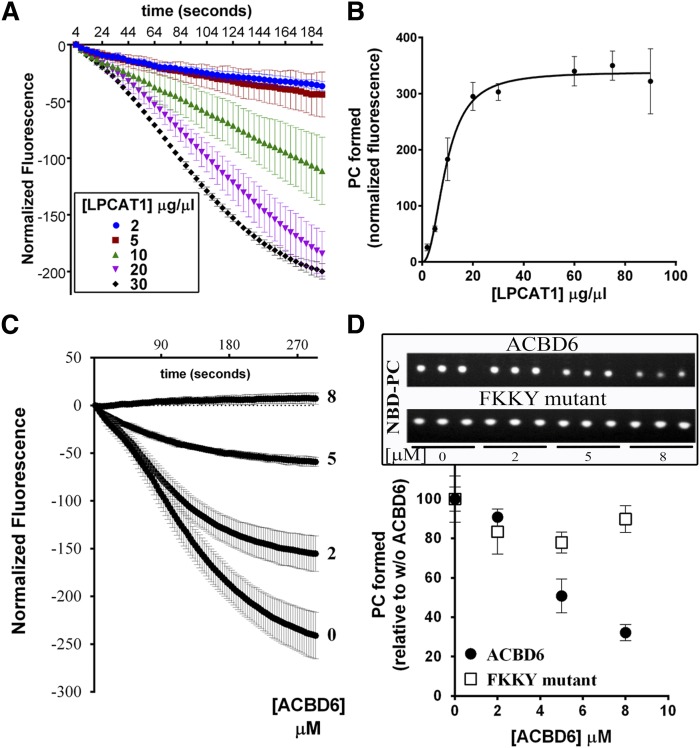
The acylation of lysoPC with NBD-C16:0-CoA by LPCAT1 was measured in absence and presence of ACBD6. Both NBD-C16:0-CoA and lysoPC are detergents. In the mixed micellar structure, the fluorescence of NBD-C16:0-CoA is high. Formation of NBD-PC leads to the formation of vesicles and quenching of the fluorescence. The change in fluorescence was monitored over time in the presence of LPCAT1 (Fig. 7A). In the absence of ACBD6, increasing amounts of NBD-C16:0-CoA were incorporated into the lysoPC acceptor with increasing concentration of enzyme (Fig. 7A, B). The presence of ACBD6 in the reaction resulted in the apparent decrease of the formation of PC (Fig. 7C and supplementary Fig. 3). To directly show the formation of PC, the reaction mixtures were collected after the 5 min measurement, the lipids were extracted, and fluorescently labeled-compounds were identified after TLC separation. Quantification of the product confirmed the ACBD6-dependent decrease of the formation of NBD-PC by LPCAT1 (Fig. 7D). The mutant form ACBD6.FKKK-AAAA, which did not bind the substrate NBD-C16:0-CoA (Fig. 6A), had very little effect on the production of PC (Fig. 7D and supplementary Fig. 2). The lack of inhibition of PC formation by this form provided further evidence that the availability of the acyl-CoA substrate for LPCAT1 was limited by binding to ACBD6.
Fig. 7.
Effect of ACBD6 on the acyl-CoA transfer to lysoPC by LPCAT1. Real-time measurements of the incorporation of 16-NBD-C16:0-CoA into lysoPC were performed with 20 µM lysoPC and with 2 µM 16-NBD-C16:0-CoA as described in legend of Fig. 6. After 5 min of recording, the reactions (200 µl) were transferred into 750 µl of CHCl3/methanol (1:2), and lipids were extracted and NBD-PC was quantified as described in the Materials and Methods. A: Reactions were performed with increasing concentration of LPCAT1 microsomes. B: The amount of PC formed was plotted as a function of the concentration of LPCAT1. C: Reactions were performed with 30 µg of LPCAT1 microsomes and the indicated concentration of ACBD6 (0 to 8 µM). D: The amount of PC formed was plotted as a function of the concentration of ACBD6 or the mutant ACBD6.FKKY-AAAA. Inset: UV shadow imaging of the separated NBD-PC molecules by TLC. Reactions were performed in triplicate. Error bars represent the standard deviations of at least three measurements.
Acyl-CoA transfer but not acyl-CoA synthesis is affected by ACBD6
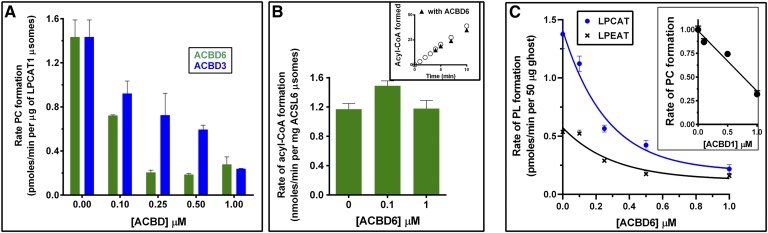
Decrease of PC formation by LPCAT1 from NBD-C16:0-CoA in the presence of ACBD6 was also observed with [14C]C18:1-CoA (Fig. 8A). This finding indicated that the slow transfer of the fluorescent ligand from ACBD6 to LPCAT1 was not the consequence of the presence of the bulky NBD group at the beginning of the aliphatic chain (supplementary Fig. 2). Slowing of the acylation rate of lysoPC was also not unique to ACBD6. Purified ACBD3 decreased formation of PC by LPCAT1 similarly in a concentration-dependent fashion (Fig. 8A). The consequence of the fast binding of the ligand to ACBD6 (Fig. 6) on the synthesis of acyl-CoA by the red blood cell (RBC) long-chain ACSL6 enzyme was investigated (33). Although, it was speculated that binding of the product of the reaction to an acyl-CoA carrier should stimulate esterification of fatty acids, presumably by preventing feedback inhibition (2, 20, 21, 23), we did not detect an effect of ACBD6 on the rate of synthesis of oleoyl-CoA (Fig. 8B). Presence of ACBD6 at the beginning of the reaction or its addition during the formation of C18:1-CoA did not result in a stimulation of the esterification reaction.
Fig. 8.
Release of acyl-CoA from ACBD proteins regulates the acyltransferase reaction. A: Acyltransferase reactions were performed at 37°C with 20 µM lysoPC and 1 µM [14C]C18:1-CoA. Prior to addition of 4 µg of LPCAT1 microsomes, ACBD3 or ACBD6 was added at the indicated concentration for 20 min. B: ACSL activity of human ACSL6 enzyme were measured at 30°C with 2 µg of microsomes and 4 µM [14C]C18:1-OH. Inset: Reaction was initiated in the absence of ACBD6 for 3.5 min. ACBD6 was then added at a final concentration of 5 µM (black triangles). C: Acylation of lysoPC and lysoPE by erythrocyte membranes was performed as in A in the presence of increasing concentration of ACBD6. Inset: Rate of acylation of lysoPC in the presence of increasing concentration of ACBD1.
Reacylation of the two most abundant phospholipids, PC and PE, in the RBC membrane is carried out by LPCAT1 and an unidentified lysophosphatidylethanolamine-acyltransferase (LPEAT) enzyme. The rates of transfer of [14C]C18:1-CoA on lysoPC and on lysoPE by the acylating enzymes of the RBC membrane were reduced in the presence of ACBD6 (Fig. 8C). In particular, the lysoPC-acyl-CoA transferase reaction of the RBC was very sensitive to ACBD6 (Fig. 8A). Of the seven members of the ACBD family, ACBD1 and ACBD6 appear to be the only acyl-CoA carriers present in human erythrocytes (25, 34). Reduction of the rate of PC formation was also observed when purified ACBD1 protein was incubated with RBC membranes (Fig. 8C, inset).
DISCUSSION
Organization and composition of phospholipids in membrane bilayers is maintained by a set of metabolic enzymes, binding proteins, and transbilayer transport proteins. Lipid binding proteins facilitate the transfer of the hydrophobic lipid molecules through the water phase between membranes and modulate their availability to lipid metabolic enzymes. Moreover, molecules like lysophospholipids, fatty acids, and their activated forms (acyl-CoA) are powerful detergents that need to be controlled to avoid breaches in membrane lipid bilayer integrity. One of the functions of the acyl-CoA binding motif of the ACBD proteins is to bind medium- to long-chain acyl-CoA molecules generated to provide substrates in the Lands’ pathway (35) (Fig. 9). However, the diversity of cellular functions affected by these proteins implicates their interaction with other lipid species, enzymes, and regulators. The role of the ligand and of its binding to the N-terminal conserved domain on their association to other proteins is still largely unknown (2, 3). The addition of ACBD6 to membranes resulted in the reduction of the acyltransferase reaction catalyzed by LPCAT1 and LPEAT. ACBD6 also controlled activity of the LPCAT enzyme of the pathogen Chlamydia trachomatis in infected human cells (28). In addition, mouse ACBD1 was shown to reduce activity of acyl-CoA:cholesterol acyltransferase (ACAT) (11), and bovine ACBD1 decreased long-chain fatty acid synthesis (36). The inhibitory effect of ACBD3 on SREBP1 was reduced by truncation of the ACB motif (12). The detailed evaluation of the role of the ligands in the interactions of these binding proteins with other proteins might elucidate the precise function of the ACBD family.
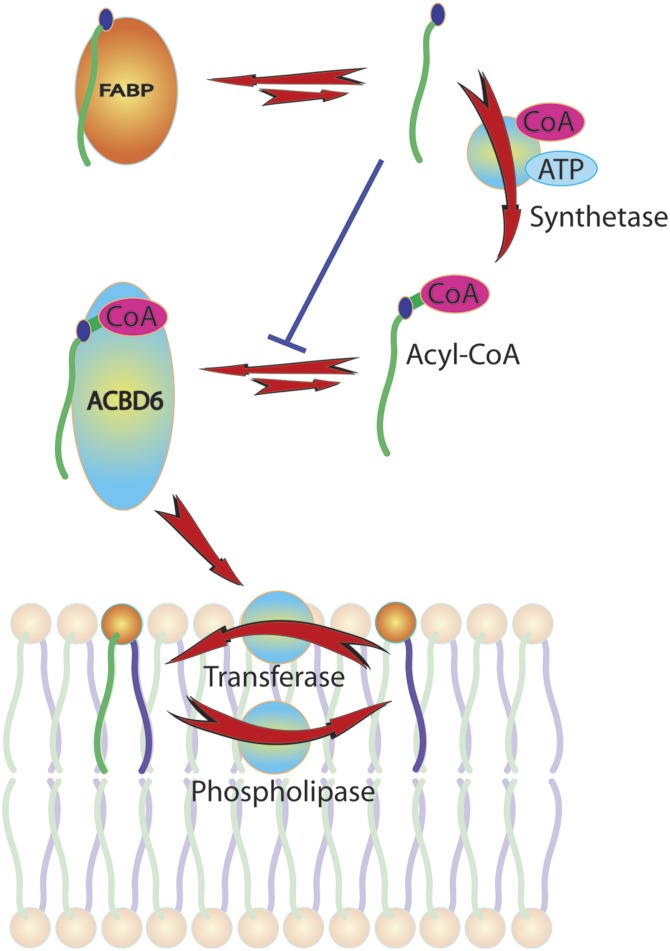
Fig. 9.
Model of the role of ACBD6 in the Lands’ pathway. Glycerophospholipids are deacylated by the action of phospholipase A2 and generate lysophospholipid molecules. Both dietary and de novo synthesized fatty acids are activated to acyl-CoA molecules by members of the ACSL family. Long-chain acyl-CoAs in the cytosol bind to ACBD6 and are released to the lysophospholipid:acyl-CoA acyltransferase enzymes to generate phospholipid from lysophosphlipid. The transfer of free acyl-CoA to ACBD6 is extremely rapid, whereas the transfer from ACBD6 to the acyltransferase is slow. This property of ACBD6 limits the amount of free acyl-CoA and protects membranes from its detergent-like activity. The pool of free fatty acids in the cytosol, which impairs loading of acyl-CoA on the ACBD6 protein, is kept low by the action of fatty acid binding proteins (FABPs), their conversion into acyl-CoAs, and further utilization of acyl-CoA such as the formation of acyl-carnitine molecules (not shown).
As reported by others, definition of the binding parameters of these proteins is complicated by the detergent-like nature of acyl-CoA molecules, which prevented their addition to a saturating level to determine their binding constant (37). Our data indicate that the binding properties of human ACBD6 protein are different from ACBD1, which was reported to display a molar ligand/protein stoichiometry of 1 and a binding constant of ∼10 nM (24). The high ligand concentrations required to reach saturation of ACBD6 were not compatible with the removal binding capacity of the Lipidex-1000 resin, but both the NTA and ITC methods provided a similar binding constant of ∼10 µM. The ITC profiles obtained with ACBD6 were different from those obtained with purified ACBD1. ITC measurements monitor heat changes of ligand binding to the protein in a succession of small-volume injections of concentrated ligand. Typically, several successive injections are required to progressively saturate all the sites of the protein in the chamber. For ACBD1, the protein was saturated in a single injection event (20, 21, 38). ACBD6 titration required several injections to progressively saturate the binding site(s) of the protein and strongly supports the difference in binding affinity of these two proteins. At those high concentration of acyl-CoA, formation of micelles and other lipid structures will complicate the modeling required to define the binding affinities of the site or sites on the protein (20, 25). In an effort to correct for the signal generated by the formation of these structures, control injections with only ligand were performed to generate lipid structures in the absence of the binding protein. However, accurate signal correction cannot be obtained because the formation of these protein-free structures will be affected by the presence of the protein. As can be seen on the thermogram plots of ACBD6 at high concentration of C16:0-CoA (Fig. 4B, upper graph), ligand injections prevented the formation of an equilibrium and disturbed the system even in absence of binding. A similar finding was observed for ACBD1 when saturation was reached with C12:0-CoA (20, 21). As shown previously with C18:1-CoA (25), ITC profiles obtained with several acyl-CoAs tested in this study (Fig. 4) were best fitted in a model including two sets of sites with different binding constants (supplementary Fig. 1). Binding of C18:1-CoA to regulator of antimicrobial-assisted survival (RaaS) was also resolved as a bimolecular interaction (39). It cannot be excluded that a fraction of purified protein with no or a different binding capacity accounts for a second category of sites. This possibility could explain the results obtained with the NTA resin method (Fig. 1) and the calculated stoichiometry of 0.5. Displacement of binding and mathematical modeling was used to assess the binding parameters of ACBD1 (20, 24, 38). ACBD1 binds nanomolar amounts of ligand. Also in this case, it cannot be excluded that a fraction of the purified ACBD1 was already loaded with ligand during the ITC measurements, which would have prevented its titration at low ligand concentration and would have resulted in the sudden saturation of the preparation following a single injection (20, 21).
Despite those uncertainties, the binding constant values for acyl-CoAs to ACBD6 were in the range of 1 to 10 µM. These findings are consistent with the value obtained with radiolabeled C18:1-CoA binding and the NTA slurry method (Fig. 1B) and Lipidex-1000 resin (25). Binding of C18:1-CoA to bacterial RaaS was calculated at 3.65 µM (39). As measured by displacement of the electron paramagnetic resonance (EPR) signal of the spin-labeled C12:0-CoA bound to bovine ACBD1, concentration of acyl-CoAs with chain lengths of 10 to 24 were also in the range of 1 to 30 µM (24). Unexpected stoichiometry of 0.77 was defined for the binding of C14:0-CoA to trypanosome ACBP protein (40) and of 0.5 for binding of C16:0-CoA to erythrocyte ACBD1 (19). Stoichiometry values of the plant ACBP4 and ACBP5 for C18:1-CoA were 0.5 and 0.4, respectively (41). Stoichiometry values of bacterial fatty acid degradation regulator (FadR) to C14:0-CoA, C16:0-CoA, and C18:1-CoA were 0.6, 0.8, and 0.7, respectively (42). Our findings are consistent with the fact that stoichiometry might not be well defined due to the complexity of the binding of acyl-CoA molecules to proteins (43, 44) and, in some cases, as a consequence of an inadequate methodology to measure very high binding constants (21, 22).
The RBC lacks de novo synthesis, but phospholipids are renewed constantly via the Lands’ pathway (35). Fatty acids are activated by ACSL (33), and reacylation of the two most abundant phospholipids, PC and PE, in the RBC membrane is carried out by LPCAT1 and an unidentified LPEAT enzyme (35). The molecular species of phospholipids in the RBC like most mammalian membranes carry mainly saturated fatty acids like palmitic acid in the sn-1 position and unsaturated fatty acids like oleic and linoleic acid at the more metabolic active sn-2 position (45). To maintain composition in the constantly changing substrate environment of plasma, a system is in place that involves the enzymes in the Lands’ pathway and binding proteins. These proteins have to adapt to the complex lipid-protein interactions in the membrane as well as the lipid composition in the plasma. The finding that acyl-CoA binding to ACBD6 was rapid and dynamic supports the proposed role of these proteins as intracellular acyl-CoA carriers to modulate the activity of membrane-bound acyl-CoA utilizing enzymes (38, 46). The slowing of the acylation of PC and PE in presence of the ACBD proteins provides further evidence that in a cell the function of these proteins might primarily be to protect membranes and acyl-CoA-utilizing enzymes from free acyl-CoAs (23). The controlled ligand release from ACBD6 to the acyltransferase enzymes should protect them from the detergent nature of their substrate without limiting its availability. Evidence that this ACBD6-mediated transfer is influenced by the complex lipid environment in the cell was further provided by the intriguing finding that some but not all fatty acids modulated the binding of oleoyl-CoA. In E. coli, the DNA-binding activity of the acyl-CoA binding transcriptional regulators FadR and fatty acid biosynthesis regulator (FabR) are affected by the relative ratio of unsaturated to saturated fatty acids (47, 48). Thus, selective competition of acyl-CoA binding to human ACBD6 by nonligand molecules might indicate a sensory function of these proteins in lipid homeostasis of the cell.
Supplementary Material
Footnotes
Abbreviations:
- 16-NBD-16:0-CoA
- N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl]amino palmitoyl CoA
- ACBD
- acyl-CoA binding domain-containing protein
- ACSL
- long-chain acyl-CoA synthetase
- DSS
- disuccinimidyl suberate
- ITC
- isothermal titration calorimetry
- LPCAT1
- lysophosphatidylcholine-acyltransferase 1
- LPEAT
- lysophosphatidylethanolamine-acyltransferase
- lysoPC
- lysophosphatidylcholine
- lysoPE
- lysophosphatidylethanolamine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- RBC
- red blood cell
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Burton M., Rose T. M., Faergeman N. J., Knudsen J. 2005. Evolution of the acyl-CoA binding protein (ACBP). Biochem. J. 392: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faergeman N. J., Wadum M., Feddersen S., Burton M., Kragelund B. B., Knudsen J. 2007. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol. Cell. Biochem. 299: 55–65. [DOI] [PubMed] [Google Scholar]
- 3.Fan J., Liu J., Culty M., Papadopoulos V. 2010. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog. Lipid Res. 49: 218–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao S., Chye M. L. 2009. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 47: 479–484. [DOI] [PubMed] [Google Scholar]
- 5.Xiao S., Chye M. L. 2011. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 50: 141–151. [DOI] [PubMed] [Google Scholar]
- 6.Knudsen J., Mandrup S., Rasmussen J. T., Andreasen P. H., Poulsen F., Kristiansen K. 1993. The function of acyl-CoA-binding protein (ACBP)/diazepam binding inhibitor (DBI). Mol. Cell. Biochem. 123: 129–138. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y., Atkins J. B., Rompani S. B., Bancescu D. L., Petersen P. H., Tang H., Zou K., Stewart S. B., Zhong W. 2007. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 129: 163–178. [DOI] [PubMed] [Google Scholar]
- 8.Okazaki Y., Ma Y., Yeh M., Yin H., Li Z., Yeh K. Y., Glass J. 2012. DMT1 (IRE) expression in intestinal and erythroid cells is regulated by peripheral benzodiazepine receptor-associated protein 7. Am. J. Physiol. Gastrointest. Liver Physiol. 302: G1180–G1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinoda Y., Fujita K., Saito S., Matsui H., Kanto Y., Nagaura Y., Fukunaga K., Tamura S., Kobayashi T. 2012. Acyl-CoA binding domain containing 3 (ACBD3) recruits the protein phosphatase PPM1L to ER-Golgi membrane contact sites. FEBS Lett. 586: 3024–3029. [DOI] [PubMed] [Google Scholar]
- 10.Sbodio J. I., Paul B. D., Machamer C. E., Snyder S. H. 2013. Golgi protein ACBD3 mediates neurotoxicity associated with Huntington’s disease. Cell Reports. 4: 890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrescu A. D., Payne H. R., Boedecker A., Chao H., Hertz R., Bar-Tana J., Schroeder F., Kier A. B. 2003. Physical and functional interaction of Acyl-CoA-binding protein with hepatocyte nuclear factor-4 alpha. J. Biol. Chem. 278: 51813–51824. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Patel V., Bang S., Cohen N., Millar J., Kim S. F. 2012. Maturation and activity of sterol regulatory element binding protein 1 is inhibited by acyl-CoA binding domain containing 3. PLoS One. 7: e49906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H. Y., Chye M. L. 2004. Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54: 233–243. [DOI] [PubMed] [Google Scholar]
- 14.Dorobantu C. M., Ford-Siltz L. A., Sittig S. P., Lanke K. H., Belov G. A., van Kuppeveld F. J., van der Schaar H. M. 2015. GBF1- and ACBD3-independent recruitment of PI4KIIIbeta to replication sites by rhinovirus 3A proteins. J. Virol. 89: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greninger A. L., Knudsen G. M., Betegon M., Burlingame A. L., DeRisi J. L. 2013. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. MBio. 4: e00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Z., Yang X., Yang G., Zhang L. 2014. Hepatitis C virus NS5A competes with PI4KB for binding to ACBD3 in a genotype-dependent manner. Antiviral Res. 107: 50–55. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa-Sasaki K., Sasaki J., Taniguchi K. 2014. A complex comprising phosphatidylinositol 4-kinase IIIbeta, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex. J. Virol. 88: 6586–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki J., Ishikawa K., Arita M., Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31: 754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Augoff K., Kolondra A., Chorzalska A., Lach A., Grabowski K., Sikorski A. F. 2010. Expression, purification and functional characterization of recombinant human acyl-CoA-binding protein (ACBP) from erythroid cells. Acta Biochim. Pol. 57: 533–540. [PubMed] [Google Scholar]
- 20.Faergeman N. J., Sigurskjold B. W., Kragelund B. B., Andersen K. V., Knudsen J. 1996. Thermodynamics of ligand binding to acyl-coenzyme A binding protein studied by titration calorimetry. Biochemistry. 35: 14118–14126. [DOI] [PubMed] [Google Scholar]
- 21.Kragelund B. B., Knudsen J., Poulsen F. M. 1999. Acyl-coenzyme A binding protein (ACBP). Biochim. Biophys. Acta. 1441: 150–161. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen J. T., Borchers T., Knudsen J. 1990. Comparison of the binding affinities of acyl-CoA-binding protein and fatty-acid-binding protein for long-chain acyl-CoA esters. Biochem. J. 265: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen J. T., Rosendal J., Knudsen J. 1993. Interaction of acyl-CoA binding protein (ACBP) on processes for which acyl-CoA is a substrate, product or inhibitor. Biochem. J. 292: 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosendal J., Ertbjerg P., Knudsen J. 1993. Characterization of ligand binding to acyl-CoA-binding protein. Biochem. J. 290: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soupene E., Serikov V., Kuypers F. A. 2008. Characterization of an acyl-coenzyme A binding protein predominantly expressed in human primitive progenitor cells. J. Lipid Res. 49: 1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y. C., Wu Q., Chen J., Xuan Z., Jung Y. C., Zhang M. Q., Rowley J. D., Wang S. M. 2009. The transcriptome of human CD34+ hematopoietic stem-progenitor cells. Proc. Natl. Acad. Sci. USA. 106: 8278–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto M. L., Clark T. A., Gee S. L., Kang J. A., Schweitzer A. C., Wickrema A., Conboy J. G. 2009. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 113: 3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soupene E., Wang D., Kuypers F. A. 2015. Remodeling of host phosphatidylcholine by Chlamydia acyltransferase is regulated by acyl-CoA binding protein ACBD6 associated with lipid droplets. MicrobiologyOpen. 4: 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soupene E., Kuypers F. A. 2012. Phosphatidylcholine formation by LPCAT1 is regulated by Ca(2+) and the redox status of the cell. BMC Biochem. 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soupene E., Fyrst H., Kuypers F. A. 2008. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. USA. 105: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soupene E., Dinh N. P., Siliakus M., Kuypers F. A. 2010. Activity of the acyl-CoA synthetase ACSL6 isoforms: role of the fatty acid Gate-domains. BMC Biochem. 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen J., Neergaard T. B., Gaigg B., Jensen M. V., Hansen J. K. 2000. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J. Nutr. 130: 294S–298S. [DOI] [PubMed] [Google Scholar]
- 33.Soupene E., Kuypers F. A. 2006. Multiple erythroid isoforms of human long-chain acyl-CoA synthetases are produced by switch of the fatty acid gate domains. BMC Mol. Biol. 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fyrst H., Knudsen J., Schott M. A., Lubin B. H., Kuypers F. A. 1995. Detection of acyl-CoA-binding protein in human red blood cells and investigation of its role in membrane phospholipid renewal. Biochem. J. 306: 793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lands W. E. 1960. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235: 2233–2237. [PubMed] [Google Scholar]
- 36.Mandrup S., Hojrup P., Kristiansen K., Knudsen J. 1991. Gene synthesis, expression in Escherichia coli, purification and characterization of the recombinant bovine acyl-CoA-binding protein. Biochem. J. 276: 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abo-Hashema K. A., Cake M. H., Lukas M. A., Knudsen J. 1999. Evaluation of the affinity and turnover number of both hepatic mitochondrial and microsomal carnitine acyltransferases: relevance to intracellular partitioning of acyl-CoAs. Biochemistry. 38: 15840–15847. [DOI] [PubMed] [Google Scholar]
- 38.Abo-Hashema K. A., Cake M. H., Lukas M. A., Knudsen J. 2001. The interaction of acyl-CoA with acyl-CoA binding protein and carnitine palmitoyltransferase I. Int. J. Biochem. Cell Biol. 33: 807–815. [DOI] [PubMed] [Google Scholar]
- 39.Turapov O., Waddell S. J., Burke B., Glenn S., Sarybaeva A. A., Tudo G., Labesse G., Young D. I., Young M., Andrew P. W., et al. 2014. Oleoyl coenzyme A regulates interaction of transcriptional regulator RaaS (Rv1219c) with DNA in mycobacteria. J. Biol. Chem. 289: 25241–25249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne K. G., Ferguson M. A. 2000. Cloning, expression, and characterization of the acyl-CoA-binding protein in African trypanosomes. J. Biol. Chem. 275: 12503–12508. [DOI] [PubMed] [Google Scholar]
- 41.Leung K. C., Li H. Y., Mishra G., Chye M. L. 2004. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol. Biol. 55: 297–309. [DOI] [PubMed] [Google Scholar]
- 42.DiRusso C. C., Tsvetnitsky V., Hojrup P., Knudsen J. 1998. Fatty acyl-CoA binding domain of the transcription factor FadR. Characterization by deletion, affinity labeling, and isothermal titration calorimetry. J. Biol. Chem. 273: 33652–33659. [DOI] [PubMed] [Google Scholar]
- 43.Richards E. W., Hamm M. W., Fletcher J. E., Otto D. A. 1990. The binding of palmitoyl-CoA to bovine serum albumin. Biochim. Biophys. Acta. 1044: 361–367. [DOI] [PubMed] [Google Scholar]
- 44.Richards E. W., Hamm M. W., Otto D. A. 1991. The effect of palmitoyl-CoA binding to albumin on the apparent kinetic behavior of carnitine palmitoyltransferase I. Biochim. Biophys. Acta. 1076: 23–28. [DOI] [PubMed] [Google Scholar]
- 45.Myher J. J., Kuksis A., Pind S. 1989. Molecular species of glycerophospholipids and sphingomyelins of human erythrocytes: improved method of analysis. Lipids. 24: 396–407. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen J. T., Faergeman N. J., Kristiansen K., Knudsen J. 1994. Acyl-CoA-binding protein (ACBP) can mediate intermembrane acyl-CoA transport and donate acyl-CoA for beta-oxidation and glycerolipid synthesis. Biochem. J. 299: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y., Cronan J. E. 2011. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol. Microbiol. 80: 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu K., Zhang Y. M., Rock C. O. 2009. Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J. Biol. Chem. 284: 34880–34888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.