Abstract
Acylcarnitines are intermediates of fatty acid and amino acid oxidation found in tissues and body fluids. They are important diagnostic markers for inherited diseases of peroxisomal and mitochondrial oxidation processes and were recently described as biomarkers of complex diseases like the metabolic syndrome. Quantification of acylcarnitine species can become challenging because various species occur as isomers and/or have very low concentrations. Here we describe a new LC-MS/MS method for quantification of 56 acylcarnitine species with acyl-chain lengths from C2 to C18. Our method includes amino acid-derived positional isomers, like methacrylyl-carnitine (2-M-C3:1-CN) and crotonyl-carnitine (C4:1-CN), and odd-numbered carbon species, like pentadecanoyl-carnitine (C15:0-CN) and heptadecanoyl-carnitine (C17:0-CN), occurring at very low concentrations in plasma and tissues. Method validation in plasma and liver samples showed high sensitivity and excellent accuracy and precision. In an application to samples from streptozotocin-treated diabetic mice, we identified significantly increased concentrations of acylcarnitines derived from branched-chain amino acid degradation and of odd-numbered straight-chain species, recently proposed as potential biomarkers for the metabolic syndrome. In conclusion, the LC-MS/MS method presented here allows robust quantification of isomeric acylcarnitine species and extends the palette of acylcarnitines with diagnostic potential derived from fatty acid and amino acid metabolism.
Keywords: diabetes, fatty acid/oxidation, mitochondria, liver, diagnostic tools, liquid chromatography-tandem mass spectroscopy
Acylcarnitines are intermediates in fatty acid and amino acid breakdown generated from the conversion of acyl-CoA species by the action of carnitine acyltransferases (1). The formation of carnitine conjugates is crucial for the transport of activated fatty acids (acyl-CoAs) from the cytosol across the inner mitochondrial membrane into the matrix where fatty acid oxidation takes place. It has now become clear that a net efflux of acylcarnitine species from the mitochondria into the cytosol and ultimately into plasma is particularly important in situations of impaired fatty acid oxidation to prevent accumulation of potentially toxic acyl-CoA intermediates in the mitochondrion (2, 3). How acylcarnitines are released into the extracellular space is not known, but changes in plasma and/or urinary acylcarnitine profiles are used to detect disorders in fatty acid and amino acid oxidation (4). More recently, alterations in plasma acylcarnitine profiles were described as strongly associated with obesity and type 2 diabetes (5–7). A broad spectrum of short-, medium-, and long-chain acylcarnitine species is generated from multiple metabolic routes including various fatty acid and amino acid oxidation pathways. A comprehensive quantification that covers as many as possible of these entities is therefore not only important for proper clinical diagnosis of inherited enzymatic impairments, but also for basic studies of complex diseases such as the metabolic syndrome.
Acylcarnitine profiles are commonly established by direct infusion ESI-MS/MS. This technology, however, does not allow the discrimination of isomeric acylcarnitine species. Furthermore, isobaric matrix interferences can lead to an overestimation of metabolite concentrations producing false positive results, in particular, for compounds with low abundance (8). Although LC-MS/MS methods have been developed for successful separation of isomeric compounds (9–12), the identification of the exact compound in the chromatogram often remains elusive. Additionally, the abundance of many species in biological samples is often too low for robust detection and quantification.
Here we describe a robust and highly sensitive LC-MS/MS method for comprehensive quantification of an array of acylcarnitine species with a special focus on amino acid-derived intermediates, which often occur at very low concentrations and in isomeric forms. Because alterations in plasma odd-numbered fatty acids were recently shown to associate with impaired glucose tolerance and diabetes (13), we also focused on quantification of the corresponding odd-numbered acylcarnitines. The method was validated and applied to plasma and liver samples from streptozotocin (STZ)-induced insulin-deficient mice, which serve as a model of type I diabetes.
MATERIALS AND METHODS
Chemicals, solutions, reference substances, and internal standards
Ammonium acetate, n-butanol (99.7%), thionyl chloride, carnitine hydrochloride, formic acid (LC-MS grade), heptafluorobutyric acid (HFBA) (≥99.5%), acetyl chloride, and acetonitrile (LC-MS grade) were purchased from Sigma-Aldrich (Taufkirchen, Germany). Water (LC-MS grade) and diethyl ether (Baker analyzed) were obtained from J. T. Baker Chemicals (Center Valley, PA). Methanol (LC-MS grade) and trichloroacetic acid were purchased from Merck (Darmstadt, Germany). The internal standard (IS) mixture containing d9-carnitine (0.785 pmol/μl), d3-acetyl-carnitine (0.327 pmol/μl), d3-propionyl-carnitine (0.086 pmol/μl), d3-butyryl-carnitine (0.043 pmol/μl), d9-isovaleryl-carnitine (0.039 pmol/μl), d3-hexanoyl-carnitine (0.046 pmol/μl), d3-octanoyl-carnitine (0.042 pmol/μl), d3-decanoyl-carnitine (0.046 pmol/μl), d3-dodecanoyl-carnitine (0.043 pmol/μl), d3-tetradecanoyl-carnitine (0.041 pmol/μl), d3-hexadecanoyl-carnitine (0.082 pmol/μl), and d3-octadecanoyl-carnitine (0.074 pmol/μl) was obtained from ChromSystems (München, Germany). The free fatty acids used for synthesis of acylcarnitines (crotonoic acid, methacrylic acid, tiglic acid, dimethylacrylic acid, isobutyric acid, 2-methylbutyric acid, (S)-3-hydroxybutyric acid, (R)-3-hydroxybutyric acid, sodium-(S)-β-hydroxyisobutyric acid, 3-hydroxyisovaleric acid, trans-2-methyl-butenoic acid, heptanoic acid, nonanoic acid, undecanoic acid, tridecanoic acid, pentadecanoic acid, heptadecanoic acid, and adipic acid) were obtained from Sigma-Aldrich. Valeryl-carnitine was acquired from LGC-Standards (Wesel, Germany), 15-methylhexadecanoic acid was obtained from Larodan (Solna, Sweden), and 3-methylglutaryl-carnitine was purchased at Avanti Polar Lipids (Alabaster, AL).
Synthesis of reference substances
O-acylated acylcarnitines were synthesized from carnitine chloride and free fatty acids using a modified protocol from Ziegler et al. (14). Briefly, the free fatty acid was incubated with thionyl chloride (15 mg free fatty acid per microliter thionyl chloride) under continuous shaking for 4 h at 70°C to generate fatty acyl chlorides. Carnitine hydrochloride dissolved in trichloroacetic acid was added to the acyl chloride and incubated at 45°C for 18 h. After cooling, the product was precipitated and washed three times in cold diethyl ether. Sodium-(s)-β-hydroxyisobutyric acid was first treated with hydrochloric acid to produce the free fatty acid, followed by evaporation of water at 90°C for 30 min, before incubation with thionyl chloride. For (S)- and (R)-β-hydroxybutyric acid and for (S)-β-hydroxyisobutyric acid, this protocol generated a racemic mixture of both (S)- and (R)- stereoisomers.
Sample preparation and derivatization
Plasma (10 μl) was dissolved in 100 μl ice-cold methanol containing the IS mixture under ultra-sonication to prevent sample aggregation. Dissolved samples were evaporated to dryness in a vacuum concentrator. Tissue samples were first grounded in liquid nitrogen using pestle and mortar. Forty milligrams of tissue were then extracted using 1,800 μl 100% ice-cold methanol and centrifuged at 10,000 g, 4°C for 10 min. IS was added to 200 μl of the supernatant before vacuum-drying the samples.
Acylcarnitines were derivatized to their butyl esters as described by Gucciardi et al. (9). Briefly, 100 μl n-butanol containing 5% v/v acetyl chloride was added to the dried samples, incubated at 60°C for 20 min at 800 rpm (Eppendorf Thermomixer Comfort; Eppendorf, Hamburg, Germany) and subsequently evaporated to dryness. Samples were reconstituted in 100 μl (for plasma samples) or 200 μl (for tissue samples) methanol/water and transferred to glass vials.
LC-MS/MS
The analysis was performed on a triple quadrupole QTRAP5500 LC-MS/MS system (AB Sciex, Framingham, MA), equipped with a 1200 series binary pump, a degasser, and a column oven (Agilent, Santa Clara, CA) connected to a HTC pal autosampler (CTC Analytics, Zwingen, Switzerland). A Turbo V ion spray source operating in positive ESI mode was used for ionization. The source settings were: ion spray voltage, 5,500 V; heater temperature, 600°C; source gas 1, 50 psi; source gas 2, 50 psi; curtain gas, 40 psi; and collision gas (CAD), medium.
Chromatographic separation was achieved on a Zorbax Eclipse XDB-C18 column (length 150 mm, internal diameter 3.0 mm, particle size 3.5 μm; Agilent) at a column temperature of 50°C. Eluent A consisted of 0.1% formic acid, 2.5 mM ammonium acetate, and 0.005% HFBA in water. Eluent B consisted of 0.1% formic acid, 2.5 mM ammonium acetate, and 0.005% HFBA in acetonitrile. Gradient elution was performed with the following program: 100% A (0.5 ml/min) for 0.5 min, a linear decrease to 65% A (0.5 ml/min) for 2.5 min, hold for 3 min, a linear decrease to 40% A (0.5 ml/min) for 3.7 min, a linear decrease to 5% A (0.5 ml/min) for 1 min. Elution was then carried out at 100% B (1 ml/min) for 0.5 min, hold for 7.3 min (1.5 ml/min). Re-equilibration was finally performed at 100% A (0.5 m/min) for 4 min. The complete running time of the program was 22 min. Analytes were measured in scheduled multiple reaction monitoring (MRM) (total scan time, 0.5 s; scan time window, 0.5 min). Quadrupoles were working at unit resolution.
Calibration and quantification
Calibration was achieved by spiking plasma and liver samples with different quantities of acylcarnitine standards. A ten-point calibration was performed by addition of increasing amounts of each standard and IS, as described in the sample preparation section. Calibration curves were constructed and fitted by linear regression without weighting. Data analysis was done using Analyst 1.5® software (AB Sciex).
Animal experiment and sample collection
C57BL6/N mice (n = 10), at an age of 12 weeks, were injected intraperitoneally with a single dose of STZ (180 mg/kg body weight dissolved in 0.1 M citric acid buffer) and mice (n = 10) injected intraperitoneally with citric acid buffer served as controls. All mice were fed a standard chow diet (ssniff V1534-0 R/M-H) and were not fasted prior to sampling of blood and organs. Body weight and blood glucose levels were measured daily using tail-vein puncture. After 5 days, mice were anesthetized with isoflurane. Blood was collected into EDTA-coated tubes and was centrifuged for 10 min at 1,200 g and 4°C. Plasma was separated and snap-frozen in liquid nitrogen. Mice were euthanized and liver tissue was collected and immediately snap-frozen in liquid nitrogen. All samples were stored at −80°C until the day of measurement. The research was conducted in conformity with the Public Health Service policy on humane care and use of laboratory animals. All experiments were performed according to the German guidelines for animal care and were approved by the state of Bavaria (Regierung von Oberbayern) ethics committee (reference number 55.2.1-54-2532-22-11).
Statistical analysis
Levels of significance between metabolite concentrations in STZ-treated and control mice were assessed using two-sided independent Student’s t-tests.
RESULTS
Sample extraction and derivatization
Extraction of acylcarnitine species from plasma and tissue samples was achieved using methanol and analytes were derivatized to their butyl esters. Because dicarboxylic acylcarnitines were derivatized at both carboxyl groups (see Fig. 1), we could discriminate various isobaric acylcarnitines, which have different masses as corresponding butyl esters [e.g., hydroxybutyryl-carnitine (C4-OH-CN, nonderivatized m/z 248, butylated m/z 304) and malonyl-carnitine (C3:0-DC-CN, nonderivatized m/z 248, butylated m/z 360)].
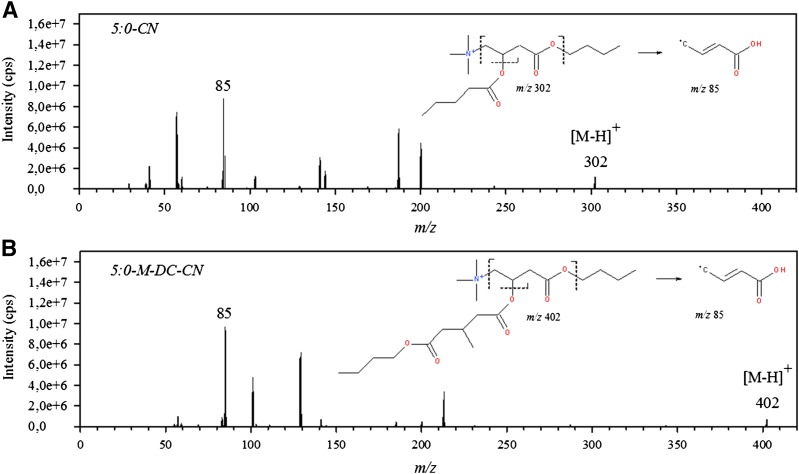
Fig. 1.
Product ion spectra and proposed fragmentation patterns for valeryl-carnitine (C5:0-CN) (A) and 3-methylglutaryl-carnitine (C5-M-DC-CN) (B).
Acylcarnitine fragmentation
Mass spectrometric analysis of acylcarnitine species was performed using ESI in positive mode and product ion spectra were acquired. Butylation of acylcarnitines (especially of dicarboxylic species) increased the ionization efficiency (data not shown). Representative fragmentation patterns for valeryl-carnitine (C5:0-CN; monocarboxylic) and 3-methylglutaryl-carnitine (C5:0-M-DC-CN; dicarboxylic) are shown in Fig. 1. Fragmentation patterns of acylcarnitines were reported to have a prominent fragment ion at m/z 85 in common (15). This fragment ion displayed the highest intensity in our measurements and was thus used for quantification. It is proposed to be formed due to loss of the butyl group(s) (m/z 56), the acyl chain [m/z 102 and m/z 146 for valeryl-carnitine (C5:0-CN) and 3-methylglutaryl-carnitine (C5:0-M-DC-CN), respectively], and the trimethylamine fragment (m/z 59), as initially described by Chace et al. (15) in 1997. A compilation of all acylcarnitine species covered by our method, including the corresponding MS parameters, is shown in Table 1.
TABLE 1.
MS parameters and retention times
| MRM (m/z) | IS | IS MRM (m/z) | Retention Time (min) | DP (V) | EP (V) | CE (V) | CXP (V) | ||
| Free carnitine | C0 | 218.1 → 85.1 | d9-C0 | 227.5 → 85.1 | 5.9 | 61 | 10 | 23 | 15 |
| Hydroxypropionyl-carnitine | C3-OH | 290.1 → 85.1 | d3-C3 | 277.1 → 85.1 | 6.1 | 53 | 10 | 30 | 15 |
| Hydroxybutyryl-carnitine a | C4-OH a | 304.1 → 85.1 | d3-C4 | 291.4 → 85.1 | 6.2 | 55 | 10 | 32 | 15 |
| Hydroxybutyryl-carnitine b | C4-OH b | 304.1 → 85.1 | d3-C4 | 291.4 → 85.1 | 6.3 | 55 | 10 | 32 | 15 |
| 3-Hydroxyisobutyryl-carnitine | 2-M-C3-OH | 304.1 → 85.1 | d3-C4 | 291.4 → 85.1 | 6.4 | 55 | 10 | 32 | 15 |
| Acetyl-carnitine | C2 | 260.5 → 85.1 | d3-C2 | 263.2 → 85.1 | 6.4 | 41 | 10 | 18 | 15 |
| Hydroxyvaleryl-carnitine | C5-OH | 318.1 → 85.1 | d9-C5 | 311 → 85.1 | 6.5 | 58 | 10 | 33 | 15 |
| Propionyl-carnitine | C3:0 | 274.4 → 85.1 | d3-C3 | 277.1 → 85.1 | 6.9 | 46 | 10 | 29 | 15 |
| Hydroxyhexanoyl-carnitine a | C6-OH a | 332.2 → 85.1 | d3-C6 | 319.2 → 85.1 | 7.3 | 61 | 10 | 35 | 15 |
| Crotonyl-carnitine | C4:1 | 286.1 → 85.1 | d3-C4 | 291.4 → 85.1 | 7.3 | 52 | 10 | 29 | 15 |
| Hydroxyhexanoyl-carnitine b | C6-OH b | 332.2 → 85.1 | d3-C6 | 319.2 → 85.1 | 7.5 | 61 | 10 | 35 | 15 |
| Methacrylyl-carnitine | 2-M-C3:1 | 286.1 → 85.1 | d3-C4 | 291.4 → 85.1 | 7.5 | 52 | 10 | 29 | 15 |
| Isobutyryl-carnitine | 2-M-C3:0 | 288.2 → 85.1 | d3-C4 | 291.4 → 85.1 | 7.8 | 46 | 10 | 29 | 15 |
| Butyryl-carnitine | C4:0 | 288.2 → 85.1 | d3-C4 | 291.4 → 85.1 | 8 | 46 | 10 | 29 | 15 |
| Tiglyl-carnitine | 2-M-C4:1 | 300.2 → 85.1 | d9-C5 | 311 → 85.1 | 8.2 | 55 | 10 | 31 | 15 |
| 3-Methylcrotonyl-carnitine | 3-M-C4:1 | 300.2 → 85.1 | d9-C5 | 311 → 85.1 | 8.4 | 55 | 10 | 31 | 15 |
| 2-Methyl-butyryl-carnitine | 2-M-C4:0 | 302.1 → 85.1 | d9-C5 | 311 → 85.1 | 9.5 | 46 | 10 | 29 | 15 |
| Isovaleryl-carnitine | 3-M-C4:0 | 302.1 → 85.1 | d9-C5 | 311 → 85.1 | 9.6 | 46 | 10 | 29 | 15 |
| Valeryl-carnitine | C5:0 | 302.1 → 85.1 | d9-C5 | 311 → 85.1 | 9.7 | 46 | 10 | 29 | 15 |
| Fumaryl-carnitine | C4:1-DC | 372.5 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 9.9 | 56 | 10 | 27 | 15 |
| Malonyl-carnitine | C3-DC | 360.2 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 10.3 | 55 | 10 | 32 | 15 |
| Succinyl-carnitine | C4-DC | 374.2 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 10.8 | 56 | 10 | 33 | 15 |
| Hexanoyl-carnitine | C6:0 | 316.4 → 85.1 | d3-C6 | 319.2 → 85.1 | 10.9 | 56 | 10 | 27 | 15 |
| Methyl-malonyl-carnitine | C3-DC-M | 374.2 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.1 | 58 | 10 | 33 | 15 |
| Glutaryl-carnitine | C5-DC | 388.3 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.1 | 61 | 10 | 35 | 15 |
| Glutaconyl-carnitine | C5:1-DC | 386.3 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.2 | 60 | 10 | 35 | 15 |
| Adipyl-carnitine | C6-DC | 402.3 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.3 | 63 | 10 | 37 | 15 |
| Methylglutaryl-carnitine | C5-M-DC | 402.3 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.4 | 63 | 10 | 37 | 15 |
| Heptanoyl-carnitine | C7:0 | 330.2 → 85.1 | d3-C6 | 319.2 → 85.1 | 11.5 | 60 | 10 | 30 | 15 |
| Pimelyl-carnitine | C7-DC | 416.3 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 11.7 | 66 | 10 | 39 | 15 |
| Octanoyl-carnitine | C8:0 | 344.1 → 85.1 | d3-C8 | 347.4 → 85.1 | 11.9 | 66 | 10 | 33 | 15 |
| Decenoyl-carnitine | C10:1 | 370.2 → 85.1 | d3-C10 | 375.2 → 85.1 | 12.1 | 68 | 10 | 40 | 15 |
| Nonanoyl-carnitine | C9:0 | 358.2 → 85.1 | d3-C8 | 347.4 → 85.1 | 12.1 | 66 | 10 | 39 | 15 |
| Decanoyl-carnitine | C10:0 | 372.2 → 85.1 | d3-C10 | 375.2 → 85.1 | 12.3 | 56 | 10 | 37 | 15 |
| Iso-undecanoyl-carnitine | Iso-C11:0 | 386.3 → 85.1 | d3-C10 | 375.2 → 85.1 | 12.4 | 65 | 10 | 40 | 15 |
| Undecanoyl-carnitine | C11:0 | 386.3 → 85.1 | d3-C10 | 375.2 → 85.1 | 12.5 | 65 | 10 | 40 | 15 |
| Tetradecadienyl-carnitine | C14:2 | 424.3 → 85.1 | d3-C14 | 431.2 → 85.1 | 12.6 | 78 | 10 | 47 | 15 |
| Dodecanoyl-carnitine | C12:0 | 400.3 → 85.1 | d3-C12 | 403.2 → 85.1 | 12.7 | 73 | 10 | 44 | 15 |
| Hydroxyhexadecenyl-carnitine | C16:1-OH | 470.3 → 85.1 | d3-C16 | 459.2 → 85.1 | 12.8 | 87 | 10 | 53 | 15 |
| Isotridecanoyl-carnitine | Iso-C13:0 | 414.3 → 85.1 | d3-C12 | 403.2 → 85.1 | 13 | 80 | 10 | 45 | 15 |
| Tridecanoyl-carnitine | C13:0 | 414.3 → 85.1 | d3-C12 | 403.2 → 85.1 | 13.1 | 80 | 10 | 45 | 15 |
| Hexadecadienyl-carnitine | C16:2 | 452.3 → 85.1 | d3-C16 | 459.2 → 85.1 | 13.1 | 83 | 10 | 51 | 15 |
| Tetradecanoyl-carnitine | C14:0 | 428.3 → 85.1 | d3-C14 | 431.2 → 85.1 | 13.4 | 86 | 10 | 45 | 15 |
| Hydroxyhexadecanoyl-carnitine a | C16-OH a | 472.3 → 85.1 | d3-C16 | 459.2 → 85.1 | 13.4 | 87 | 10 | 53 | 15 |
| Hydroxyhexadecanoyl-carnitine b | C16-OH b | 472.3 → 85.1 | d3-C16 | 459.2 → 85.1 | 13.5 | 87 | 10 | 53 | 15 |
| Hydroxyoctadecenyl-carnitine | C18:1-OH | 498.4 → 85.1 | d3-C18 | 487.5 → 85.1 | 13.5 | 92 | 10 | 57 | 15 |
| Hexadecenyl-carnitine | C16:1 | 454.3 → 85.1 | d3-C16 | 459.2 → 85.1 | 13.6 | 84 | 10 | 51 | 15 |
| Isopentadecanoyl-carnitine | iso-C15:0 | 442.3 → 85.1 | d3-C14 | 431.2 → 85.1 | 13.6 | 85 | 10 | 48 | 15 |
| Pentadecanoyl-carnitine | C15:0 | 442.3 → 85.1 | d3-C14 | 431.2 → 85.1 | 13.7 | 85 | 10 | 48 | 15 |
| Octadecadienyl-carnitine | C18:2 | 480.3 → 85.1 | d3-C18 | 487.5 → 85.1 | 13.7 | 89 | 10 | 54 | 15 |
| Hexadecanoyl-carnitine | C16:0 | 456.5 → 85.1 | d3-C16 | 459.2 → 85.1 | 14.3 | 84 | 10 | 51 | 15 |
| Octadecenyl-carnitine | C18:1 | 482.3 → 85.1 | d3-C18 | 487.5 → 85.1 | 14.3 | 89 | 10 | 55 | 15 |
| Isoheptadecanoyl-carnitine | Iso-C17:0 | 470.4 → 85.1 | d3-C16 | 459.2 → 85.1 | 14.7 | 90 | 10 | 57 | 15 |
| Heptadecanoyl-carnitine | C17:0 | 470.4 → 85.1 | d3-C16 | 459.2 → 85.1 | 14.8 | 90 | 10 | 57 | 15 |
| Dodecadioyl-carnitine | C12-DC | 486.4 → 85.1 | d6-C5-DC | 394.2 → 85.1 | 15.6 | 86 | 10 | 45 | 15 |
| Octadecanoyl-carnitine | C18:0 | 484.4 → 85.1 | d3-C18 | 487.5 → 85.1 | 15.6 | 96 | 10 | 63 | 15 |
DP, declustering potential; EP, entrance potential; CE, collision energy; CXP, collision cell exit potential.
Chromatographic separation and identification of isomeric acylcarnitine species
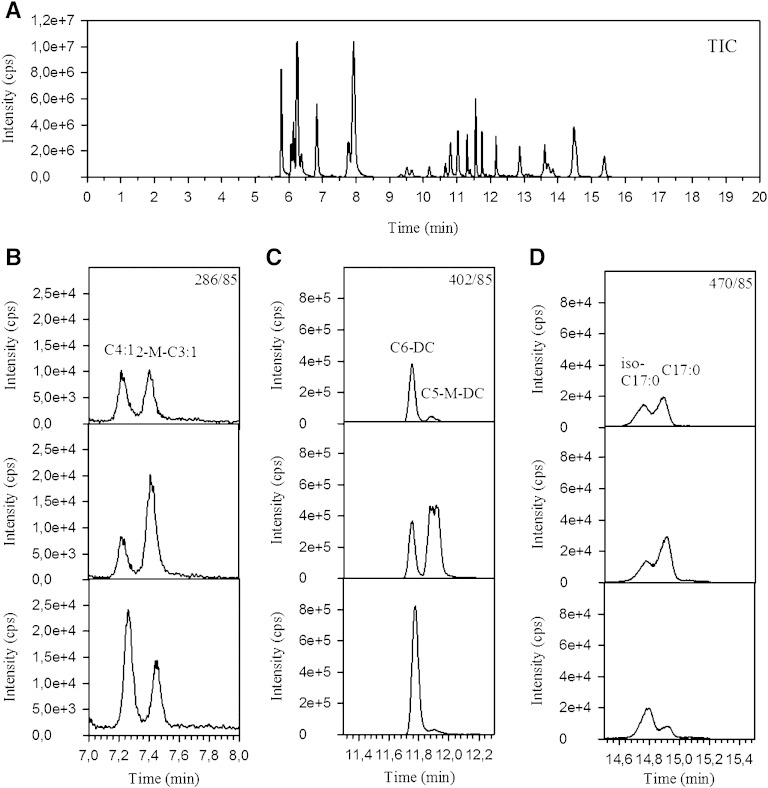
Because all acylcarnitines form a similar product ion (m/z 85) and many compounds are isobaric or occur as positional isomers, appropriate chromatographic separation is essential. Chromatographic separation was established using a C18-reversed phase HPLC column (length, 15 cm; internal diameter, 3.0 mm; particle size, 3.5 μm). We favored an HPLC column over an ultra-performance LC column, because larger sample volumes of complex matrices were injected. Peaks were well shaped, sharp (∼0.1 min; Fig. 2), and well separated. A coelution of analyte and IS was achieved, which is important to compensate for matrix effects and varying ionization efficiencies during gradient elution (16). The addition of HFBA to the eluents as an ion-pairing reagent (17) improved peak separation and peak sharpness. Addition of ammonium acetate enhanced ionization efficiency. Figure 2A shows a typical chromatogram (total ion count) derived from a liver sample containing acylcarnitines ranging from 0 to 18 carbon atoms. Retention times increased with the number of carbons and decreased with the number of double bonds. Because >50 mass transitions were required to quantify all metabolites, data were acquired in the scheduled MRM mode to yield more and shorter scan cycles per peak, to facilitate peak integration and data analysis.
Fig. 2.
Total ion count (TIC) chromatogram (A) and MRM chromatograms of exemplified spiking experiments (B–D).
Various acylcarnitine isomer species were expected to result from known amino acid and fatty acid breakdown pathways (18). Indeed, chromatographic separation generated multiple peaks for various m/z transitions [e.g., 2-methylbutyryl-carnitine (2-M-C4:0-CN), 3-methylbutyryl-carnitine (3-M-C4:0-CN), and valeryl-carnitine (C5:0-CN)]. For peak identification, matrix samples were spiked with individual acylcarnitine reference compounds as shown in Fig. 2B–D. In general, for saturated acylcarnitines, branched-chain forms elute earlier than straight-chain forms [e.g., isobutyryl-carnitine (2-M-C3:0-CN) < butyryl-carnitine (C4:0-CN)]. This was also true for odd-numbered acylcarnitines, for which two peaks were generally found (see Fig. 2D). In contrast, for monounsaturated and dicarboxylic acylcarnitines, branched-chain forms eluted later [e.g., crotonyl-carnitine (C4:1-CN) < methacrylyl-carnitine (2-M-C3:1-CN), see Fig. 2B, C]. Furthermore, hydroxy-acylcarnitines [hydroxybutyryl-carnitine (C4-OH-CN), hydroxyhexanoyl-carnitine (C6-OH-CN), hydroxyhexadecanoyl-carnitine (C16-OH-CN)], displayed two peaks. This was in accordance with previous reports suggesting the presence of S- and R-stereoisomers (9, 11, 19).
Calibration and quantification
For quantification of metabolite concentrations and to compensate for variation in sample preparation and ionization efficiency, an acylcarnitine standard mixture containing 13 deuterium-labeled acylcarnitine species was added to the samples (Table 2). Calibration lines were generated by addition of different concentrations of acylcarnitine species to a pool of liver and plasma samples. The ratio between analyte and IS was used for quantification (“stable isotope dilution”). For those analytes without corresponding IS, analyte-IS pairs were selected according to the closest retention times and the best accuracies for these analyte-IS allocations [e.g., nonanoyl-carnitine (C9:0-CN) to d3-octanoyl-carnitine (d3-C8:0-CN)]. For all analytes and matrices, calibration curves were linear and correlation coefficients (r2) were greater than 0.99 (Table 2). Limits of detection (LODs) were determined as the signal-to-noise ratio of three, and limits of quantitation (LOQs) were calculated as the triple fold of the LOD and were sufficient enough for quantification of acylcarnitine species in plasma and liver samples (Table 2).
TABLE 2.
Calibration data
| Calibration Data (Plasma Matrix) | LOD (pmol/μl) | LOQ (pmol/μl) | Calibration Data (Liver Matrix) | LOD (pmol/mg w.w.) | LOQ (pmol/mg w.w.) | |||||||
| Calibration Range (pmol/μl) | IS (pmol/μl) | Slope | R2 | Calibration Range (pmol/mg w.w.) | IS Added (pmol/mg w.w.) | Slope | Correlation Coefficient (R2) | |||||
| C0 | 1.5–500 | 7.70 | 0.28 | 0.999 | 0.089 | 0.266 | 1.5–1,000 | 180.55 | 0.0088 | 0.990 | 0.275 | 0.824 |
| C2 | 0.375–125 | 3.21 | 0.34 | 0.999 | 0.038 | 0.113 | 0.375–250 | 75.21 | 0.0099 | 0.999 | 0.074 | 0.222 |
| C3 | 0.075–25 | 0.84 | 1.37 | 0.999 | 0.008 | 0.024 | 0.075–50 | 19.78 | 0.0389 | 0.999 | 0.003 | 0.008 |
| C4 | 0.075–25 | 0.42 | 3.40 | 0.999 | 0.008 | 0.023 | 0.075–50 | 9.89 | 0.098 | 0.999 | 0.008 | 0.011 |
| 2-M-C3 | 0.075–25 | 0.42 | 3.49 | 0.999 | 0.007 | 0.021 | 0.075–50 | 9.89 | 0.101 | 0.999 | 0.004 | 0.012 |
| C5 | 0.075–25 | 0.38 | 5.67 | 0.999 | 0.006 | 0.018 | 0.075–50 | 8.97 | 0.159 | 0.999 | 0.003 | 0.009 |
| 3-M-C4 | 0.075–25 | 0.38 | 5.95 | 0.999 | 0.007 | 0.021 | 0.075–50 | 8.97 | 0.158 | 0.999 | 0.003 | 0.009 |
| C6 | 0.075–25 | 0.45 | 3.18 | 0.999 | 0.013 | 0.038 | 0.075–50 | 10.58 | 0.098 | 0.999 | 0.008 | 0.024 |
| C8 | 0.075–25 | 0.41 | 2.27 | 0.997 | 0.006 | 0.020 | 0.075–50 | 9.66 | 0.071 | 0.999 | 0.002 | 0.006 |
| C10 | 0.075–25 | 0.45 | 2.43 | 0.993 | 0.009 | 0.028 | 0.075–50 | 10.58 | 0.083 | 0.999 | 0.005 | 0.014 |
| C12 | 0.15–50 | 0.42 | 2.45 | 0.992 | 0.009 | 0.027 | 0.15–100 | 9.89 | 0.093 | 0.999 | 0.004 | 0.012 |
| C14 | 0.15–50 | 0.40 | 2.56 | 0.996 | 0.009 | 0.028 | 0.15–100 | 9.43 | 0.104 | 0.999 | 0.003 | 0.009 |
| C16 | 0.15–50 | 0.80 | 1.80 | 0.998 | 0.002 | 0.007 | 0.15–100 | 18.86 | 0.064 | 0.999 | 0.002 | 0.005 |
| C18 | 0.15–50 | 0.73 | 1.31 | 0.994 | 0.021 | 0.065 | 0.15–100 | 17.02 | 0.049 | 0.999 | 0.021 | 0.063 |
Calibration lines were generated by plotting the area ratios of analyte/IS against the spiked concentrations. Concentrations in liver are given per milligram of liver wet weight. w.w., wet weight.
Method validation and sample stabilities
The accuracy of the method was tested in three liver and three plasma samples by spiking with acylcarnitine standards in concentrations covering the whole calibration range. Accuracies between 81 and 108% were found for the liver matrix and between 89 and 120% for the plasma matrix (Table 3), except for the lowest levels of free carnitine in liver tissues for which concentrations were overestimated. Method reproducibility was examined by determining intra- and inter-day precisions in pools of plasma and liver samples (Table 4). With a few exceptions, intra- and inter-day coefficients of variation (CVs) for liver and plasma samples were below 10% for high-abundant acylcarnitines (>0.01 pmol/mg tissue; >0.01 pmol/μl plasma) and between 10 and 20% for low-abundant acylcarnitines. Recoveries were calculated as the difference between area ratios of prespiked samples (addition of standards before extraction) and postspiked samples (addition of standards to the methanol extract). Recoveries between 59 and 99% were found (supplementary Table 1). Matrix effects were determined by comparing the peak areas of acylcarnitine species spiked in matrix samples (three plasma and three liver samples) and corresponding amounts dissolved in methanol at two levels (supplementary Tables 2, 3). The area ratios of spiked acylcarnitine species in plasma samples ranged between 110 and 120% compared with nonmatrix samples. Area ratios of spiked acylcarnitine species in liver were between 85 and 122% compared with nonmatrix samples, with a few exceptions for short-chain species.
TABLE 3.
Accuracies
| Liver Spiked (pmol/mg w.w.) | Accuracy (%) | Plasma Spiked (pmol/μl) | Accuracy (%) | |
| C0 | 30 | 261.8 | 20 | 119.7 |
| 150 | 87.4 | 150 | 95.5 | |
| 500 | 89 | 300 | 97.8 | |
| C2 | 7.5 | 106.7 | 5 | 116.1 |
| 37.5 | 102.7 | 37.5 | 97.7 | |
| 125 | 97.1 | 75 | 97.9 | |
| C3:0 | 1.5 | 104.2 | 1 | 116.3 |
| 7.5 | 98.1 | 7.5 | 94.2 | |
| 25 | 98.7 | 15 | 97.8 | |
| C4:0 | 1.5 | 96.2 | 1 | 115.5 |
| 7.5 | 101 | 7.5 | 92.2 | |
| 25 | 101 | 15 | 93.3 | |
| 2-M-C3:0 | 1.5 | 82.9 | 1 | 120.0 |
| 7.5 | 97.2 | 7.5 | 93.3 | |
| 25 | 96.4 | 15 | 93.9 | |
| 3-M-C4:0 | 1.5 | 84.3 | 1 | 113.9 |
| 7.5 | 94.3 | 7.5 | 89.5 | |
| 25 | 94.2 | 15 | 96.0 | |
| C5:0 | 1.5 | 84.1 | 1 | 108.3 |
| 7.5 | 95.8 | 7.5 | 91.8 | |
| 25 | 94.1 | 15 | 97.6 | |
| C6:0 | 1.5 | 90.8 | 1 | 115.4 |
| 7.5 | 100.8 | 7.5 | 102.6 | |
| 25 | 102 | 15 | 100.6 | |
| C8:0 | 1.5 | 90.3 | 1 | 110.1 |
| 7.5 | 105 | 7.5 | 104.7 | |
| 25 | 107.9 | 15 | 100.9 | |
| C10:0 | 1.5 | 86.3 | 1 | 109.8 |
| 7.5 | 99.3 | 7.5 | 103.6 | |
| 25 | 98.1 | 15 | 106.6 | |
| C12:0 | 3 | 87.2 | 2 | 108.7 |
| 15 | 95.8 | 15 | 107.5 | |
| 50 | 96.5 | 30 | 105.7 | |
| C14:0 | 3 | 89.3 | 2 | 119.1 |
| 15 | 91.7 | 15 | 109.2 | |
| 50 | 90 | 30 | 105.8 | |
| C16:0 | 3 | 81.7 | 2 | 112.3 |
| 15 | 90.3 | 15 | 102.4 | |
| 50 | 92.4 | 30 | 102.1 | |
| C18:0 | 3 | 82.5 | 2 | 107.8 |
| 15 | 99.6 | 15 | 106.0 | |
| 50 | 98.5 | 30 | 104.0 |
The displayed accuracy is the average of the assayed concentration (corrected by endogenous levels of plasma and liver samples) in percent of the actual spiked concentration. Concentrations in liver are given per milligram of liver wet weight. w.w., wet weight.
TABLE 4.
Intra- and inter-day precisions
| Name | Intra-day Liver (n = 5) | Inter-day Liver (n = 5) | Intra-day Plasma (n = 5) | Inter-day Plasma (n = 5) | ||||
| Mean ± SD (pmol/mg w.w.) | CV (%) | Mean ± SD (pmol/mg w.w.) | CV (%) | Mean ± SD (pmol/μl) | CV (%) | Mean ± SD (pmol/μl) | CV (%) | |
| C0 | 230.91 ± 17.61 | 7.6 | 228.47 ± 9.35 | 4.1 | 34.99 ± 3.29 | 9.4 | 33.34 ± 2.01 | 6.1 |
| C3-OH | 0.0098 ± 0.001 | 14.6 | 0.013 ± 0.001 | 13.2 | — | — | — | — |
| C4-OH a | 1.62 ± 0.099 | 6.1 | 1.71 ± 0.075 | 4.4 | 0.051 ± 0.01 | 12.7 | 0.049 ± 0.01 | 15.4 |
| C4-OH b | 3.89 ± 0.21 | 5.5 | 4.09 ± 0.10 | 2.5 | 0.10 ± 0.01 | 11.7 | 0.098 ± 0.01 | 7.8 |
| 2-M-C3-OH | 0.17 ± 0.006 | 4.2 | 0.17 ± 0.009 | 5.5 | 0.062 ± 0.00 | 14.2 | 0.059 ± 0.00 | 8.3 |
| C2 | 60.01 ± 5.23 | 8.7 | 61.26 ± 2.34 | 3.8 | 16.73 ± 1.34 | 8.0 | 15.34 ± 0.69 | 4.6 |
| C5-OH | 0.68 ± 0.056 | 8.3 | 0.67 ± 0.015 | 2.3 | 0.067 ± 0.00 | 10.6 | 0.067 ± 0.00 | 14.3 |
| C3:0 | 12.73 ± 1.31 | 10.3 | 12.69 ± 0.48 | 3.8 | 0.52 ± 0.05 | 9.7 | 0.51 ± 0.04 | 9.7 |
| C6-OH a | 0.14 ± 0.013 | 9.8 | 0.14 ± 0.003 | 2.2 | 0.005 ± 0.00 | 10.4 | 0.005 ± 0.00 | 14.5 |
| C4:1 | 0.0046 ± 0.000 | 8.4 | 0.0046 ± 0.000 | 11.5 | 0.003 ± 0.00 | 11.0 | 0.003 ± 0.00 | 11.4 |
| C6-OH b | 0.051 ± 0.004 | 9.3 | 0.051 ± 0.002 | 4.0 | 0.005 ± 0.00 | 12.3 | 0.005 ± 0.00 | 15.2 |
| 2-M-C3:1 | 0.010 ± 0.001 | 11.9 | 0.0099 ± 0.001 | 13.2 | — | — | — | — |
| 2-M-C3:0 | 1.29 ± 0.10 | 8.0 | 1.35 ± 0.094 | 7.0 | 0.16 ± 0.01 | 9.5 | 0.15 ± 0.01 | 10.3 |
| C4:0 | 5.34 ± 0.45 | 8.5 | 5.40 ± 0.22 | 4.1 | 0.45 ± 0.04 | 8.9 | 0.43 ± 0.03 | 9.3 |
| 2-M-C4:1 | 0.011 ± 0.001 | 17.8 | 0.011 ± 0.000 | 7.3 | 0.005 ± 0.00 | 8.9 | 0.005 ± 0.00 | 12.9 |
| 3-M-C4:1 | 0.0053 ± 0.000 | 13.8 | 0.0049 ± 0.000 | 13.8 | — | — | — | — |
| 2-M-C4:0 | 0.72 ± 0.060 | 8.4 | 0.70 ± 0.026 | 3.8 | 0.054 ± 0.00 | 12.3 | 0.054 ± 0.00 | 12.4 |
| 3-M-C4:0 | 0.18 ± 0.018 | 10.1 | 0.18 ± 0.008 | 5.0 | 0.070 ± 0.00 | 9.7 | 0.070 ± 0.00 | 11.3 |
| C5:0 | 1.56 ± 0.17 | 10.8 | 1.56 ± 0.072 | 4.6 | 0.011 ± 0.00 | 11.4 | 0.011 ± 0.00 | 11.0 |
| C4:1-DC | 0.024 ± 0.002 | 11.4 | 0.025 ± 0.002 | 12.2 | 0.001 ± 0.00 | 12.7 | 0.001 ± 0.00 | 14.2 |
| C3-DC | 4.61 ± 0.44 | 9.5 | 4.78 ± 0.23 | 4.7 | 0.039 ± 0.00 | 11.1 | 0.038 ± 0.00 | 11.0 |
| C4-DC | 1.51 ± 0.16 | 10.6 | 1.49 ± 0.039 | 2.6 | 0.029 ± 0.00 | 8.8 | 0.029 ± 0.00 | 9.3 |
| C6:0 | 1.33 ± 0.13 | 10.0 | 1.31 ± 0.059 | 4.6 | 0.068 ± 0.00 | 10.9 | 0.066 ± 0.00 | 10.3 |
| C3-DC-M | 0.11 ± 0.010 | 9.6 | 0.11 ± 0.010 | 10.0 | 0.010 ± 0.00 | 18.6 | 0.011 ± 0.00 | 16.8 |
| C5-DC | 5.87 ± 0.53 | 9.0 | 5.84 ± 0.13 | 2.3 | 0.032 ± 0.00 | 11.7 | 0.030 ± 0.00 | 11.0 |
| C5:1-DC | 0.91 ± 0.16 | 17.1 | 0.92 ± 0.074 | 8.1 | 0.017 ± 0.00 | 13.2 | 0.020 ± 0.00 | 12.1 |
| C6-DC | 4.92 ± 0.53 | 10.9 | 4.81 ± 0.20 | 4.2 | — | — | — | — |
| C5-M-DC | 0.095 ± 0.014 | 15.3 | 0.097 ± 0.009 | 9.3 | 0.043 ± 0.00 | 10.7 | 0.039 ± 0.00 | 8.1 |
| C7:0 | 0.19 ± 0.026 | 14.0 | 0.20 ± 0.013 | 6.8 | 0.001 ± 0.00 | 19.9 | 0.001 ± 0.00 | 17.0 |
| C7-DC | 2.59 ± 0.27 | 10.3 | 2.53 ± 0.10 | 4.1 | 0.011 ± 0.00 | 9.1 | 0.010 ± 0.00 | 10.8 |
| C8:0 | 0.19 ± 0.017 | 8.9 | 0.20 ± 0.006 | 3.2 | 0.009 ± 0.00 | 9.0 | 0.008 ± 0.00 | 12.5 |
| C10:1 | — | — | — | — | 0.004 ± 0.00 | 11.8 | 0.007 ± 0.00 | 19.5 |
| C9:0 | 0.038 ± 0.003 | 10.3 | 0.038 ± 0.002 | 5.7 | — | — | — | — |
| C10:0 | 0.10 ± 0.010 | 10.9 | 0.092 ± 0.008 | 9.2 | 0.010 ± 0.00 | 13.6 | 0.009 ± 0.00 | 14.9 |
| Iso-C11:0 | 0.0094 ± 0.000 | 17.7 | 0.0086 ± 0.000 | 3.1 | — | — | — | — |
| C11:0 | 0.0055 ± 0.000 | 18.6 | 0.0060 ± 0.001 | 12.7 | — | — | — | — |
| C14:2 | 0.047 ± 0.002 | 5.1 | 0.044 ± 0.005 | 12.5 | 0.001 ± 0.00 | 10.9 | 0.001 ± 0.00 | 19.9 |
| C12:0 | 0.090 ± 0.011 | 13.2 | 0.082 ± 0.002 | 3.1 | 0.015 ± 0.00 | 19.9 | 0.013 ± 0.00 | 19.8 |
| C16:1-OH | 0.031 ± 0.013 | 44.6 | 0.033 ± 0.004 | 12.5 | — | — | — | — |
| Iso-C13:0 | 0.0035 ± 0.000 | 20.0 | 0.0039 ± 0.000 | 14.2 | 0.000 ± 2.37 | 7.9 | 0.000 ± 3.54 | 12.6 |
| C13:0 | 0.0035 ± 0.000 | 18.4 | 0.0039 ± 0.000 | 14.2 | 0.001 ± 0.00 | 18.8 | 0.001 ± 0.00 | 16.8 |
| C16:2 | 0.089 ± 0.009 | 10.3 | 0.087 ± 0.003 | 4.4 | 0.011 ± 0.00 | 18.4 | 0.010 ± 0.00 | 25.3 |
| C14:0 | 0.22 ± 0.021 | 9.5 | 0.21 ± 0.009 | 4.3 | 0.048 ± 0.00 | 16.5 | 0.044 ± 0.00 | 19.6 |
| C16-OH a | 0.050 ± 0.005 | 10.3 | 0.049 ± 0.002 | 4.7 | 0.003 ± 0.00 | 19.0 | 0.010 ± 0.00 | 24.7 |
| C16-OH b | 0.0098 ± 0.001 | 13.4 | 0.0098 ± 0.001 | 11.1 | 0.002 ± 0.00 | 16.3 | 0.002 ± 0.00 | 23.7 |
| C18:1-OH | 0.11 ± 0.022 | 19.5 | 0.13 ± 0.013 | 10.6 | — | — | — | — |
| C16:1 | 0.54 ± 0.053 | 10.0 | 0.57 ± 0.021 | 3.8 | 0.049 ± 0.00 | 18.9 | 0.045 ± 0.01 | 31.1 |
| Iso-C15:0 | 0.011 ± 0.001 | 15.2 | 0.030 ± 0.001 | 3.8 | 0.001 ± 0.00 | 18.7 | 0.000 ± 0.00 | 20.0 |
| C15:0 | 0.012 ± 0.000 | 6.5 | 0.011 ± 0.001 | 9.6 | 0.002 ± 0.00 | 15.2 | 0.002 ± 0.00 | 13.5 |
| C18:2 | 1.45 ± 0.18 | 12.6 | 1.50 ± 0.049 | 3.3 | 0.051 ± 0.01 | 20.9 | 0.051 ± 0.01 | 38.7 |
| C16:0 | 1.40 ± 0.12 | 8.3 | 1.35 ± 0.050 | 3.7 | 0.18 ± 0.03 | 19.5 | 0.17 ± 0.03 | 18.5 |
| C18:1 | 2.67 ± 0.25 | 9.4 | 2.74 ± 0.045 | 1.7 | 0.11 ± 0.02 | 22.6 | 0.11 ± 0.03 | 36.9 |
| Iso-C17:0 | 0.019 ± 0.001 | 9.8 | 0.018 ± 0.000 | 4.4 | 0.001 ± 0.00 | 16.3 | 0.001 ± 0.00 | 17.4 |
| C17:0 | 0.029 ± 0.004 | 14.2 | 0.025 ± 0.001 | 6.9 | 0.001 ± 0.00 | 16.1 | 0.001 ± 0.00 | 14.0 |
| C12-DC | 0.0048 ± 0.000 | 18.2 | 0.0045 ± 0.001 | 24.1 | — | — | — | — |
| C18:0 | 0.91 ± 0.081 | 9.0 | 0.93 ± 0.038 | 4.1 | 0.028 ± 0.00 | 17.8 | 0.027 ± 0.00 | 19.2 |
Intra- and inter-day precisions were assessed by calculating average concentrations and CVs from five individual plasma and liver samples. Concentrations in liver are given per milligram of liver wet weight. w.w., wet weight.
To assess sample stability in plasma, samples were stored at room temperature for 1 h before extraction and peak areas were compared with samples that were directly extracted. Acylcarnitines in plasma remained stable over 1 h at room temperature. For liver tissue, thawing of samples in three freeze-thaw cycles strongly affected the abundance of various analytes, but postpreparative storage of plasma and liver samples at −20°C showed a high stability of most analytes. See supplementary Table 4 for a summary on plasma and liver sample stabilities.
Method application
The developed method was applied to plasma and liver samples from STZ-treated insulin-deficient and healthy control mice. STZ causes pancreatic β-cell destruction resulting in insulin-deficient type I diabetes (20). STZ-treated animals showed very low circulating insulin levels accompanied by a dramatic weight loss and a severe increase in blood glucose levels 5 days after treatment (see supplementary Table 5).
We found that concentration ranges of acylcarnitine species in plasma and liver were in good agreement with levels reported previously (21), with highest concentrations for saturated short-chain species [acetyl-carnitine (C2:0-CN, ∼20%), propionyl-carnitine (C3:0-CN, ∼5%), and butyryl-carnitine (C4:0-CN, ∼2%)] and lowest concentrations for odd-numbered medium- and long-chain species [undecanoyl-carnitine (C11:0-CN), tridecanoyl-carnitine (C13:0-CN), and pentadecanoyl-carnitine (C15:0-CN)]. Whereas almost all acylcarnitine species could be found in liver samples, the concentrations in plasma were generally far lower and a number of species (especially monounsaturated species like crotonyl- and 3-methyl-crotonyl-carnitine) could not be detected.
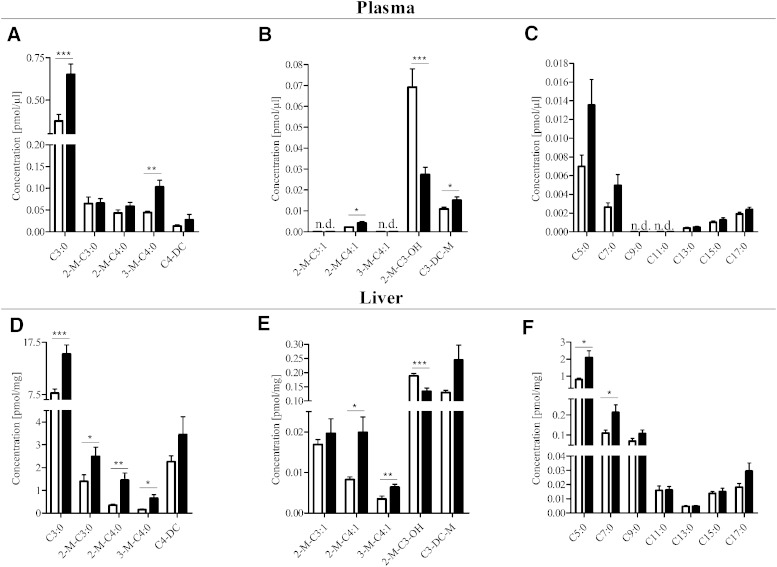
Comparison of acylcarnitine concentrations between diabetic and healthy mice revealed strong increases in the concentrations of various branched-chain amino acid-derived compounds in plasma and liver of diabetic mice (Fig. 3A–D). These included leucine-derived isovaleryl-carnitine (3-M-C4-CN) and 3-methylcrotonyl-carnitine (3-M-C4:1-CN) and isoleucine-derived 2-methylbutyryl-carnitine (2-M-C4-CN) and 2-methylcrotonyl-carnitine (2-M-C4:1-CN), as well as propionyl-carnitine (C3-CN) and methylmalonyl-carnitine (C3-M-DC-CN). In contrast, 3-hydroxyisobutyryl-carnitine (2-M-C3-OH-CN), which is derived from valine breakdown, showed a marked decrease, both in plasma and liver. Furthermore, elevated levels of various straight-chain odd-numbered acylcarnitines were observed in diabetic mice, most strongly in liver samples (Fig. 3E, F), whereas decreased levels were found for hexadecenoyl-carnitine (C16:1-CN) in plasma and liver and tetradecanoyl-carnitine (C14:0-CN) in liver. Supplementary Table 6 provides concentrations of all acylcarnitine species quantified in liver and plasma of healthy and diabetic mice in our study.
Fig. 3.
Levels of acylcarnitine species in plasma and liver tissue samples obtained from STZ-treated animals and controls determined with our LC-MS/MS method. A, D: High-abundant branched-chain amino acid-derived species. B, E: Low-abundant branched-chain amino acid derived species. C, F: Odd-numbered species. Concentrations in liver are given per milligram of liver wet weight. White and black bars represent metabolite concentrations in controls and diabetic animals, respectively. *P < 0.05, **P < 0.01, and ***P < 0.005.
DISCUSSION
Acylcarnitines have recently gained considerable interest as markers for impairments in fatty acid and amino acid oxidation associated with the metabolic syndrome (5–7). Moreover, the metabolites were proposed to have predictive quality for disease initiation and progression (5). Various acylcarnitines occur as positional isomers and many species often appear at very low concentrations. Even though several LC-MS/MS methods have been described for quantification of isomeric acylcarnitine species (9–12), a comprehensive method that covers low-abundant species and includes odd-numbered acylcarnitines is lacking.
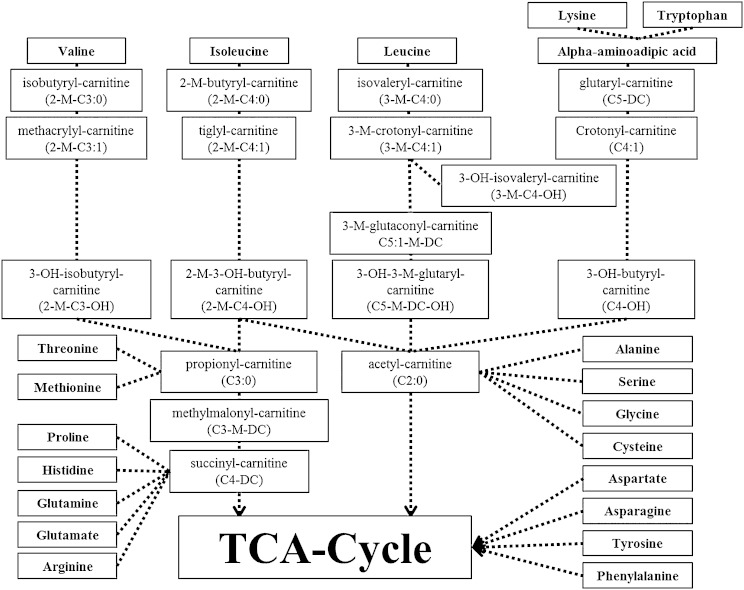
Here we describe a sensitive and robust method for quantification of individual isomeric acylcarnitine species, including low-abundant amino acid-derived forms (see Fig. 4). In addition, our method includes acylcarnitines with odd-numbered acyl-chain lengths, which gained increased attention recently as metabolite markers in disease states (5, 22, 23).
Fig. 4.
Scheme displaying amino acid breakdown pathways and acylcarnitine species that can be formed from acyl-CoA intermediates.
We applied ESI in positive mode for ionization. Collision-induced dissociation produced an intense product ion at m/z 85 for all analytes, which was described to be a specific product ion of acylcarnitine fragmentation (15). LC with a C18-reversed phase HPLC column allowed the chromatographic separation of isomeric acylcarnitine species, and by spiking with reference compounds, we could subsequently assign the individual isomers to the respective peaks. Proper chromatographic separation is mandatory because positional isomers have similar mass transitions [e.g., crotonyl-carnitine (C4:1-CN) and methacrylyl-carnitine (2-M-C3:1-CN), m/z 286 → m/z 85]. In addition, quantification of acylcarnitine concentrations in a direct infusion approach is challenged by potential interfering matrix components which lead to an overestimation of analyte concentrations (9). Although a separation of underivatized acylcarnitine isomers has been described (11), we applied a butylation to achieve an enhanced sensitivity, particularly for dicarboxylic acylcarnitines, as described previously (24). To further improve chromatographic separation and to enhance peak shapes, the ion pairing agent, HFBA, was added to the eluents in very low concentrations (0.005%). HFBA was favored over the stronger trifluoroacetic acid, which is more commonly used (9), because trifluoroacetic acid causes very strong ion suppression (25). We could achieve baseline separation of all acylcarnitine species including positional isomers, and all peaks were well shaped. The total running time of 22 min is in the range of other reported methods (9–11).
Calibration was performed by stable isotope dilution. A coelution of standards and available ISs was achieved, which is important for compensation of matrix effects and differences in ionization efficiency (16). LODs and LOQs were sufficient to determine analytes in plasma and/or liver (supplementary Table 6). In addition, we observed that matrix effects of liver tissue are comparable to other tissue types such as muscle, heart, or kidney tissue, and thus suggest that the method can be applied to other tissues to determine analyte concentrations in these matrices (data not shown). Importantly, extraction of increased tissue quantities (>40 mg) does not improve method performance, because this leads to increased noise and reduced signal-to-noise ratios. In general, for comparison of analyte concentrations between samples, it is recommended to extract similar quantities of tissue to reduce variation resulting from matrix effects.
To evaluate LC-MS/MS method performance, we validated our method in plasma and liver samples as described previously (26). Precisions showed CVs <15% for high-abundant metabolites and CVs <20% for low-abundant compounds. Accuracies displayed CVs of ±10% with a few exceptions. This is a major improvement over previously published methods, for which up to 30% variation was reported in plasma (9), or for which no measures were reported at all (11). We observed that plasma samples were stable when stored for 1 h at room temperature prior to preparation. Furthermore, most acylcarnitine species in plasma and liver showed a high postpreparative stability.
We applied our method to plasma and liver samples of STZ-treated insulin-deficient mice. STZ is selectively taken up into the pancreas and causes β-cell toxicity via alkylation, resulting in β-cell necrosis (20). STZ-treated mice showed very low circulating insulin levels paralleled by a strong increase in blood glucose and a dramatic decrease in body weight (see supplementary Table 5).
We found that, particularly, the branched-chain acylcarnitines, as intermediates of branched-chain amino acid degradation, were increased in plasma and liver, and among them, most strongly, propionyl-carnitine (C3:0-CN), 2-methylbutyryl-carnitine (2-M-C4:0-CN), and isovaleryl-carnitine (3-M-C4:0-CN), as well as 2-methylcrotonyl-carnitine (2-M-C4:1-CN) and 3-methylcrotonyl-carnitine (3-M-C4:1-CN). Interestingly, we also detected elevated levels of the 5-carbon straight-chain valeryl-carnitine (C5:0-CN), for which the metabolic origin is obscure. Increases in plasma levels of 3-carbon-chain and 5-carbon-chain acylcarnitines derived from breakdown of branched-chain amino acids, as determined by a direct infusion MS/MS approach, were recently reported as important predictors for type 2 diabetes (5). Furthermore, while concentrations of 3-hydroxybutyryl-carnitine (C4-OH-CN) were strongly increased (by ∼40%), in diabetic mice, probably as a result of increased ketone body production (27), levels of 3-hydroxyisobutyryl-carnitine (2-M-C3-OH-CN), an intermediate in valine breakdown, displayed a strong decrease (by ∼50%) in both plasma and liver. For a correct interpretation of changes in these different pathways, it is thus necessary to include LC separation prior to MS for individual quantification of these metabolites. Similarly, while using direct infusion MS/MS, Mihalik et al. (6) reported C4-dicarboxyl-carnitine as a marker for glucolipotoxicity in type 2 diabetes, but did not discriminate between succinyl-carnitine (C4-DC-CN) and methylmalonyl-carnitine (C3-M-DC-CN), which are interconverted via methylmalonyl-CoA mutase. With our method, we found that only levels of methylmalonyl-carnitine (C3-M-DC-CN) were significantly increased in diabetic mice, while alterations in succinyl-carnitine (C4-DC-CN) concentrations remained insignificant. Finally, we found elevated levels of several odd-numbered acylcarnitines in diabetic animals, most significantly the short-chain species, propionyl-carnitine (C3-CN), valeryl-carnitine (C5-CN), and heptanoyl-carnitine (C7-CN) in liver. Interestingly, odd-numbered fatty acids were recently proposed as potential markers of the metabolic syndrome (22). The origin and cause for these alterations in disease states is not known.
In summary, we have developed a sensitive and robust method which allows quantification of a large number of acylcarnitine species, including isomeric forms derived from a variety of metabolic pathways in amino acid and fatty acid oxidation. Compared with existing methods, we expanded the palette of analytes with novel acylcarnitine species, including low-abundant odd-numbered forms. Our LC-MS/MS method was validated in plasma and liver samples from mice and was applied to samples from STZ-treated type I diabetic mice. These mice display increased concentrations of BCAA-derived and odd-numbered acylcarnitine species, which were recently proposed as potential biomarkers for the metabolic syndrome.
Supplementary Material
Acknowledgments
The authors thank Mena Eidens for excellent participation in conduction of mouse experiments and sample collection.
Footnotes
Abbreviations:
- CN
- carnitine
- CV
- coefficient of variation
- HFBA
- heptafluorobutyric acid
- IS
- internal standard
- LOD
- limit of detection
- LOQ
- limit of quantitation
- MRM
- multiple reaction monitoring
- STZ
- streptozotocin
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Ramsay R. R., Gandour R. D., van der Leij F. R. 2001. Molecular enzymology of carnitine transfer and transport. Biochim. Biophys. Acta. 1546: 21–43. [DOI] [PubMed] [Google Scholar]
- 2.Ventura F. V., Ijlst L., Ruiter J., Ofman R., Costa C. G., Jakobs C., Duran M., Tavares de Almeida I., Bieber L. L., Wanders R. J. 1998. Carnitine palmitoyltransferase II specificity towards beta-oxidation intermediates–evidence for a reverse carnitine cycle in mitochondria. Eur. J. Biochem. 253: 614–618. [DOI] [PubMed] [Google Scholar]
- 3.Violante S., Ijlst L., Ruiter J., Koster J., van Lenthe H., Duran M., de Almeida I. T., Wanders R. J., Houten S. M., Ventura F. V. 2013. Substrate specificity of human carnitine acetyltransferase: Implications for fatty acid and branched-chain amino acid metabolism. Biochim. Biophys. Acta. 1832: 773–779. [DOI] [PubMed] [Google Scholar]
- 4.Shekhawat P. S., Matern D., Strauss A. W. 2005. Fetal fatty acid oxidation disorders, their effect on maternal health and neonatal outcome: impact of expanded newborn screening on their diagnosis and management. Pediatr. Res. 57: 78R–86R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., et al. 2009. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mihalik S. J., Goodpaster B. H., Kelley D. E., Chace D. H., Vockley J., Toledo F. G., DeLany J. P. 2010. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring). 18: 1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams S. H., Hoppel C. L., Lok K. H., Zhao L., Wong S. W., Minkler P. E., Hwang D. H., Newman J. W., Garvey W. T. 2009. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J. Nutr. 139: 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdenur J. E., Chamoles N. A., Guinle A. E., Schenone A. B., Fuertes A. N. 1998. Diagnosis of isovaleric acidaemia by tandem mass spectrometry: false positive result due to pivaloylcarnitine in a newborn screening programme. J. Inherit. Metab. Dis. 21: 624–630. [DOI] [PubMed] [Google Scholar]
- 9.Gucciardi A., Pirillo P., Di Gangi I. M., Naturale M., Giordano G. 2012. A rapid UPLC-MS/MS method for simultaneous separation of 48 acylcarnitines in dried blood spots and plasma useful as a second-tier test for expanded newborn screening. Anal. Bioanal. Chem. 404: 741–751. [DOI] [PubMed] [Google Scholar]
- 10.Maeda Y., Ito T., Suzuki A., Kurono Y., Ueta A., Yokoi K., Sumi S., Togari H., Sugiyama N. 2007. Simultaneous quantification of acylcarnitine isomers containing dicarboxylic acylcarnitines in human serum and urine by high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 799–806. [DOI] [PubMed] [Google Scholar]
- 11.Peng M., Fang X., Huang Y., Cai Y., Liang C., Lin R., Liu L. 2013. Separation and identification of underivatized plasma acylcarnitine isomers using liquid chromatography-tandem mass spectrometry for the differential diagnosis of organic acidemias and fatty acid oxidation defects. J. Chromatogr. A. 1319: 97–106. [DOI] [PubMed] [Google Scholar]
- 12.Minkler P. E., Stoll M. S., Ingalls S. T., Yang S., Kerner J., Hoppel C. L. 2008. Quantification of carnitine and acylcarnitines in biological matrices by HPLC electrospray ionization-mass spectrometry. Clin. Chem. 54: 1451–1462. [DOI] [PubMed] [Google Scholar]
- 13.Menni C., Fauman E., Erte I., Perry J. R., Kastenmuller G., Shin S. Y., Petersen A. K., Hyde C., Psatha M., Ward K. J., et al. 2013. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 62: 4270–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegler H. J., Bruckner P., Binon F. 1967. O-acylation of dl-carnitine chloride. J. Org. Chem. 32: 3989–3991. [DOI] [PubMed] [Google Scholar]
- 15.Chace D. H., Hillman S. L., Van Hove J. L., Naylor E. W. 1997. Rapid diagnosis of MCAD deficiency: quantitative analysis of octanoylcarnitine and other acylcarnitines in newborn blood spots by tandem mass spectrometry. Clin. Chem. 43: 2106–2113. [PubMed] [Google Scholar]
- 16.Ecker J. 2012. Profiling eicosanoids and phospholipids using LC-MS/MS: principles and recent applications. J. Sep. Sci. 35: 1227–1235. [DOI] [PubMed] [Google Scholar]
- 17.Frolov A., Hoffmann R. 2008. Separation of Amadori peptides from their unmodified analogs by ion-pairing RP-HPLC with heptafluorobutyric acid as ion-pair reagent. Anal. Bioanal. Chem. 392: 1209–1214. [DOI] [PubMed] [Google Scholar]
- 18.Schooneman M. G., Vaz F. M., Houten S. M., Soeters M. R. 2013. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 62: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soeters M. R., Serlie M. J., Sauerwein H. P., Duran M., Ruiter J. P., Kulik W., Ackermans M. T., Minkler P. E., Hoppel C. L., Wanders R. J., et al. 2012. Characterization of D-3-hydroxybutyrylcarnitine (ketocarnitine): an identified ketosis-induced metabolite. Metabolism. 61: 966–973. [DOI] [PubMed] [Google Scholar]
- 20.Lenzen S. 2008. The mechanisms of alloxan- and streptozotocin-induced diabetes. Diabetologia. 51: 216–226. [DOI] [PubMed] [Google Scholar]
- 21.Primassin S., Ter Veld F., Mayatepek E., Spiekerkoetter U. 2008. Carnitine supplementation induces acylcarnitine production in tissues of very long-chain acyl-CoA dehydrogenase-deficient mice, without replenishing low free carnitine. Pediatr. Res. 63: 632–637. [DOI] [PubMed] [Google Scholar]
- 22.Wang-Sattler R., Yu Z., Herder C., Messias A. C., Floegel A., He Y., Heim K., Campillos M., Holzapfel C., Thorand B., et al. 2012. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 8: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiehn O., Garvey W. T., Newman J. W., Lok K. H., Hoppel C. L., Adams S. H. 2010. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One. 5: e15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Jesús V. R., Chace D. H., Lim T. H., Mei J. V., Hannon W. H. 2010. Comparison of amino acids and acylcarnitines assay methods used in newborn screening assays by tandem mass spectrometry. Clin. Chim. Acta. 411: 684–689. [DOI] [PubMed] [Google Scholar]
- 25.Annesley T. M. 2003. Ion suppression in mass spectrometry. Clin. Chem. 49: 1041–1044. [DOI] [PubMed] [Google Scholar]
- 26.Sterz K., Scherer G., Ecker J. 2012. A simple and robust UPLC-SRM/MS method to quantify urinary eicosanoids. J. Lipid Res. 53: 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackshear P. J., Alberti K. G. 1974. Experimental diabetic ketoacidosis. Sequential changes of metabolic intermediates in blood, liver, cerebrospinal fluid and brain after acute insulin deprivation in the streptozotocin-diabetic rat. Biochem. J. 138: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.