Abstract
A low level of HDL cholesterol (HDL-C) is a common clinical scenario and an important marker for increased cardiovascular risk. Many patients with very low or very high HDL-C have a rare mutation in one of several genes, but identification of the molecular abnormality in patients with extreme HDL-C is rarely performed in clinical practice. We investigated the accuracy and diagnostic yield of a targeted next-generation sequencing (NGS) assay for extreme levels of HDL-C. We developed a targeted NGS panel to capture the exons, intron/exon boundaries, and untranslated regions of 26 genes with highly penetrant effects on plasma lipid levels. We sequenced 141 patients with extreme HDL-C levels and prioritized variants in accordance with medical genetics guidelines. We identified 35 pathogenic and probably pathogenic variants in HDL genes, including 21 novel variants, and performed functional validation on a subset of these. Overall, a molecular diagnosis was established in 35.9% of patients with low HDL-C and 5.2% with high HDL-C, and all prioritized variants identified by NGS were confirmed by Sanger sequencing. Our results suggest that a molecular diagnosis can be identified in a substantial proportion of patients with low HDL-C using targeted NGS.
Keywords: ATP binding cassette transporter A1, diagnostic tools, genetics, genomics, high density lipoprotein, atherosclerosis, molecular diagnosis
A low concentration of plasma HDL cholesterol (HDL-C) is one of the most common lipid abnormalities and an important risk factor for CVD. Low HDL-C predicts increased CVD risk, even among patients with aggressively treated LDL cholesterol (LDL-C) (1–3). HDL-C levels are highly heritable (4) and display both locus and allelic heterogeneity, with multiple variants in several genes leading to very high or very low HDL-C. The genetic architecture of HDL-C is notable among polygenic traits in that rare variants with presumed large effect sizes are present in a substantial proportion of patients with very low or very high HDL-C, usually in the same genes that cause extremely rare Mendelian disorders of HDL-C (5–12). This suggests that it may be possible to establish a molecular diagnosis in many patients with extreme HDL-C levels. However, detection of the specific molecular abnormality in patients with extreme HDL-C is rarely performed outside of specialized research laboratories because of the cost, complexity, and time required to do so, and uncertainty regarding the clinical utility of establishing a molecular diagnosis.
The development of next-generation sequencing (NGS) technologies has created new opportunities for the routine use of sequencing in clinical medicine. Targeted NGS panels have been effectively used for newborn carrier screening and for the diagnosis of inherited cardiomyopathy, hereditary cancers, and other conditions (13), but to date have not been established for disorders of HDL-C. The objective of this study was to evaluate the accuracy and diagnostic yield of a targeted NGS panel to establish the molecular diagnosis of abnormal HDL-C in a range of patients with extreme HDL phenotypes visiting a specialty lipid clinic. Our results indicate that this approach can reliably and accurately identify pathogenic variants in a substantial proportion of patients with low levels of HDL-C.
MATERIALS AND METHODS
Patients
We recruited consecutive patients with HDL-C levels below the 10th percentile or greater than the 90th percentile using age- and gender-adjusted population data from the Lipid Research Clinics (14), hereafter referred to as “extreme HDL-C,” regardless of other lipid parameters, from the Healthy Heart Program Prevention Clinic at St. Paul’s Hospital, Vancouver, Canada, a large specialty lipid clinic serving a multi-ethnic patient population. We did not attempt to exclude patients with potential secondary causes of low HDL-C (e.g., hypertriglyceridemia) or high HDL-C (e.g., alcohol use). Related individuals were not specifically recruited. Patients were excluded if they could not speak English or were unable or unwilling to provide written informed consent. As a positive control, we included patients in whom a molecular diagnosis of a Mendelian disorder of HDL had previously been established by Sanger sequencing (15). Clinical data were abstracted from the patients’ medical records. In cases where multiple sets of laboratory values were available, we used the most recent values. All subjects provided written informed consent. This study was approved by the Clinical Research Ethics Board of the University of British Columbia.
DNA sequencing
DNA was isolated from venous blood samples using DNeasy kits (Qiagen) or saliva using Oragene kits (DNA Genotek) and quantified using the Quant-iT PicoGreen assay (Life Technologies). Sequencing libraries were prepared according to the manufacturer’s instructions (Illumina, San Diego, CA). In-solution hybridization capture was performed with SeqCap EZ library reagents (Roche-Nimblegen). Captured DNA was quantified by the KAPA system (Kapa Biosystems) and diluted to a final concentration of 5 nM. Sequencing was performed on an Illumina MiSeq instrument in 2 × 151 bp mode. Reads were mapped to the reference human genome (hg19) using Burrows-Wheeler alignment tool v0.5.9 in paired read mode (maximal exact match algorithm) and processed following the recommended guidelines by the Genome Analysis Toolkit “Best Practices for Variant Calling” (available at http://www.broadinstitute.org/genome-analysis-toolkit). Variants were called using the Genome Analysis Toolkit v3 Unified Genotyper. We applied the following variant-level and genotype-level quality filters for variant calling: quality by depth (QD) < 2.0; root mean square of mapping quality (MQ) < 40.0; rank sum test of mapping quality (MQRankSum) < −12.5; rank sum test of read position (ReadPosRankSum) < −8.0, <20 times base coverage; <Q20 genotype quality; and allele balance <0.15 for heterozygous calls and <0.85 for homozygous calls. All single nucleotide variants and indels that passed these quality control filters were annotated with the SIFT, Polyphen 2 [HumDiv], Polyphen 2 [HumVar], PROVEAN, and Condel tools, and matched against public databases of variants (1000 Genomes, HapMap, and National Heart, Lung, and Blood Institute exome variant server). We then prioritized variants in HDL genes according to the following criteria: 1) variants that are reported to be disease-causing in the Human Gene Mutation Database (HGMD) (16); 2) disruptive variants [nonsense, splice-site (two nucleotides on either side of the intron/exon boundary) and frameshift] that are novel or rare [minor allele frequency (MAF) <1% in public databases and MAF <4% in study samples]; and 3) novel or rare (MAF <1%) missense variants that were predicted to be deleterious by all of SIFT, Polyphen 2 [HumDiv], Polyphen 2 [HumVar], PROVEAN, and Condel. Variants that met these criteria were validated by bidirectional Sanger sequencing of PCR amplicons (primer sequences available on request).
Biochemical assays
ABCA1 cDNA sequences were cloned into the pCDNA3.1 vector and the W590L and R2200* mutations generated by site-directed mutagenesis. Immunoblotting and immunofluorescence in transiently transfected HEK293 cells were carried out as previously described (15). Cholesterol efflux assays were performed, as previously described (15), in transiently transfected HEK293 cells. Data represent the mean ± SD of at least three independent experiments, each performed in triplicate.
LCAT assays were performed as recommended by the manufacturer (Calbiochem) using 5 μl of human plasma. The fluorescent intensities at 390 nm and 470 nm were measured at time = 0, 2, 4, and 8 h. LCAT activity was calculated as the rate of change in the ratio of fluorescent intensities at 470 nm and 390 nm. Data represent the mean ± SD of three independent experiments, each performed in triplicate.
Statistical analyses
Results are presented as mean ± SD. Differences between groups were compared with two-tailed Student’s t-test, or one-way ANOVA for three or more groups, or for comparison between proportions, chi-squared test. Calculations were performed in GraphPad Prism software. P < 0.05 was considered statistically significant.
RESULTS
Sequencing extreme HDL-C patients
We generated a customized NGS panel to capture the exons, intron/exon boundaries, and flanking untranslated regions (UTRs) of 26 genes with known roles in plasma lipid metabolism based on data from the HGMD and manual curation of the literature (Table 1). We included genes in which rare variants are known to cause highly penetrant effects on plasma levels of HDL-C, LDL-C, TG, and lipoprotein (a), as well as on the response to lipid-lowering therapy. We generated a probe library using Nimblegen SeqCap technology to capture these targets.
TABLE 1.
List of genes sequenced
| Phenotype | Gene | MIM Number | Disease | Locus | Number of Exons |
| Low HDL-C | ABCA1 | 600046 | Tangier disease | 9q31.1 | 50 |
| Low HDL-C | APOA1 | 107680 | ApoA-I deficiency | 11q23-q24 | 5 |
| Low HDL-C | APOA2 | 107670 | ApoA-II deficiency | 1q23.3 | 4 |
| Low HDL-C | LCAT | 606967 | Familial LCAT deficiency/fish eye disease | 16q22.1 | 6 |
| Low HDL-C | NPC1 | 607623 | Niemann-Pick disease | 18q11.2 | 27 |
| Low HDL-C | PLTP | 172425 | — | 20q13.12 | 17 |
| High HDL-C | CETP | 118470 | — | 16q21 | 17 |
| High HDL-C | GALNT2 | 602274 | — | 1q41-q42 | 16 |
| High HDL-C | LIPG | 603684 | — | 18q21.1 | 11 |
| High HDL-C | SCARB1 | 601040 | — | 12q24.31 | 13 |
| High LDL-C | ABCG5 | 605459 | Sitosterolemia | 2p21 | 16 |
| High LDL-C | ABCG8 | 605460 | Sitosterolemia | 2p21 | 13 |
| High LDL-C/low LDL-C | APOB | 107730 | FH | 2p24-p23 | 29 |
| High LDL-C | LDLR | 606945 | FH | 19p13.2 | 18 |
| High LDL-C | LDLRAP1 | 605747 | FH | 1p36-p35 | 13 |
| High LDL-C/low LDL-C | PCSK9 | 607786 | FH | 1p32.3 | 12 |
| Low LDL-C | MTTP | 157147 | Abetalipoproteinemia | 4q24 | 19 |
| High TG | APOA5 | 606368 | — | 11q23 | 4 |
| High TG | APOC2 | 608083 | Chlyomicronemia | 19q13.2 | 4 |
| High TG | APOE | 107741 | Dysbetalipoproteinemia | 19q13.2 | 4 |
| High TG | GPIHBP1 | 612757 | — | 8q24.3 | 4 |
| High TG | LMF1 | 611761 | — | 16p13.3 | 20 |
| High TG | LPL | 609708 | Chlyomicronemia | 8p22 | 10 |
| Low TG/high HDL-C | APOC3 | 107720 | — | 11q23.3 | 4 |
| High lipoprotein (a) | LPA | 152200 | — | 6q26 | 40 |
| Statin myopathy | SLCO1B1 | 604843 | — | 12p | 15 |
The MIM number is from the Online Mendelian Inheritance in Man database. FH, familial hypercholesterolemia.
To assess the accuracy and diagnostic yield of this panel, we sequenced 141 patients with extreme levels of HDL-C, including 64 patients with extremely low HDL-C and 77 patients with extremely high HDL-C. This patient population was hypothesized to harbor a substantial percentage of damaging variants in HDL-related genes (5–12). The clinical characteristics of these patients are shown in Table 2. Patients with low HDL-C also had lower total plasma cholesterol and LDL-C, as well as higher TGs, higher BMI, and a higher frequency of diabetes mellitus, consistent with previous observations (17). Most patients in both groups were of self-reported European ancestry.
TABLE 2.
Characteristics of patients with extreme HDL-C
| High HDL-C Patients | Low HDL-C Patients | P | |
| Number | 77 | 64 | — |
| Age (mean ± SD) | 58.4 ± 10 | 53.4 ± 13 | 0.01 |
| Gender (% male) | 26.0 | 79.7 | <0.0001 |
| TC (mean ± SD, mmol/l) | 5.87 ± 1.3 | 4.15 ± 1.2 | <0.0001 |
| HDL-C (mean ± SD, mmol/l) | 2.47 ± 0.4 | 0.66 ± 0.2 | <0.0001 |
| LDL-C (mean ± SD, mmol/l) | 3.06 ± 1.1 | 2.13 ± 0.9 | <0.0001 |
| TG (mean ± SD, mmol/l) | 0.88 ± 0.4 | 3.88 ± 3.4 | <0.0001 |
| ApoA-I (mean ± SD, g/l) | 2.14 ± 0.3 | 1.07 ± 0.2 | <0.0001 |
| BMI (mean ± SD, kg/m2) | 25.0 ± 16 | 29.7 ± 6 | <0.0001 |
| CAD (%) | 6.5 | 12.5 | 0.2 |
| Diabetes mellitus (%) | 0.0 | 27.0 | <0.0001 |
| Current smoker (%) | 4.8 | 14.1 | 0.07 |
| Lipid lowering medication (%) | 59.5 | 62.9 | 0.6 |
| Self-reported ancestry | |||
| European (%) | 81.8 | 71.8 | 0.3 |
| Asian (%) | 11.7 | 14.1 | 0.3 |
| Other/not reported (%) | 6.5 | 14.1 | 0.3 |
TC, total cholesterol; CAD, coronary artery disease. P values represent two-tailed t-test for comparison of means and chi-squared test for comparison of proportions.
We performed high-throughput sequencing of this cohort to a mean depth of 1,559 ± 290 reads per base after quality filtering. Ninety-three percent of target bases were covered by 20 or more reads. Ninety-four percent of targeted exons had ≥50% of bases covered with an average of 15 or more reads. The mean GC (guanine-cytosine) content of exons with <50% of bases covered with 15 or more reads was 70.8 ± 4% compared with 52.0 ± 8% for exons with ≥50% of bases covered with 15 or more reads, suggesting that local genomic features interfered with the capture of these loci.
We identified 668 variants that met quality filtering criteria, including 357 exonic variants, 37 intronic variants, 212 3′UTR variants, 37 5′UTR variants, 18 variants downstream of the 3′UTR, and 7 variants upstream of 5′UTR. We then prioritized these variants (supplementary Fig. 1) based on established medical genetics recommendations (18), as follows. Variants known to be disease-causing for abnormalities of HDL-C in HGMD were designated as “pathogenic.” Novel or rare variants (MAF <1% in public databases and MAF <4% in study samples) in HDL-related genes were designated as “probably pathogenic” if they resulted in a premature stop, indel, or splice site alteration, or for missense variants, if they were predicted to be deleterious by five different variant prediction algorithms (SIFT, Polyphen 2 [HumDiv], Polyphen 2 [HumVar], PROVEAN, and Condel).
Thirty-five variants in 38 individuals met these prioritization criteria, including 29 variants in low HDL-C genes and 6 variants in high HDL-C genes (Table 3). Among the 35 patients with prioritized variants, 10 patients (28.5%) had >1 variant. Variants in the ABCA1 gene were most frequent, followed by variants in LCAT. No variants that met our prioritization criteria were identified in APOA2, GALNT2, APOC3, or PLTP. Twenty-one of the prioritized variants we identified were novel, including ten novel variants in ABCA1, three novel variants in LCAT, two novel variants in each of APOA1, NPC1, LIPG, and one novel variant in each of CETP and SCARB1 (Table 3).
TABLE 3.
Pathogenic and probably pathogenic variants identified in patients with extreme HDL-C
| Ref. Seq. | Nuc. Change | Amino Acid Change | rsID/Novel | HGMD | Functional Prediction | MAF | Phenotype | |
| Low HDL genes | ||||||||
| ABCA1 | NM_005502 | c.1196T>C | p.Val399Ala | rs9282543:G | DM | Tolerated | 0.002 | Low |
| ABCA1 | NM_005502 | c.1755C>A | p.Asp585Glu | Novel | — | Damaging | — | Low |
| ABCA1 | NM_005502 | c.1769G>T | p.Trp590Leu | Novel | DM | Damaging | — | Low |
| ABCA1 | NM_005502 | c.1913G>A | p.Arg638Gln | Novel | DM | Damaging | — | Low |
| ABCA1 | NM_005502 | c.2328G>C | p.Lys776Asn | rs138880920 | DP | Damaging | 0.003 | High |
| ABCA1 | NM_005502 | c.2540C>T | p.Pro847Leu | Novel | — | Damaging | — | Low |
| ABCA1 | NM_005502 | c.3338delT | p.Gln980Serfs*9 | Novel | — | — | — | Low |
| ABCA1 | NM_005502 | c.3121C>G | p.Leu1041Val | rs192935024 | — | Damaging | 0.001 | High |
| ABCA1 | NM_005502 | c.3541C>A | p.Ala1046Asp | rs141021096 | DM | Damaging | — | Low |
| ABCA1 | NM_005502 | c.3946C>T | p.Ser1181Phe | rs76881554 | DM | Damaging | 0.0006 | High |
| ABCA1 | NM_005502 | c.4449delC | p.Leu1484Cysfs*17 | Novel | — | — | — | Low |
| ABCA1 | NM_005502 | c.4939C>T | p.Thr1512M | Novel | DM | Damaging | — | Low |
| ABCA1 | NM_005502 | c.5039G>A | p.Arg1680Gln | rs150125857 | DM | Damaging | 0.0002 | Low |
| ABCA1 | NM_005502 | c.5550G>T | p.Tryp1699Cys | rs146934490 | DM | Damaging | — | Low |
| ABCA1 | NM_005502 | c.5449C>T | p.Arg1817* | Novel | — | Damaging | — | Low |
| ABCA1 | NM_005502 | c.5702_5703dupGA | p.Ile1902Glufs*12 | Novel | — | — | — | Low |
| ABCA1 | NM_005502 | c.7002C>T | p.Arg2200* | Novel | — | Damaging | — | Low |
| ABCA1 | NM_005502 | c.6730G>A | p.Val2244Ile | rs144588452 | DM | Tolerated | 0.0004 | Low |
| APOA1 | NM_000039 | c.138C>T | p.Arg34* | Novel | — | Damaging | — | Low |
| APOA1 | NM_000039 | c.382T>A | p.Lys131Met | rs4882 | — | Damaging | — | Low |
| APOA1 | NM_000039 | c.391_393del | p.Lys131del | Novel | — | — | — | Low |
| LCAT | NM_000229 | c.451C>T | p.Thr147Ile | rs121908050 | DM | Damaging | — | Low |
| LCAT | NM_000229 | c.487G>A | p.Arg159Gln | Novel | DM | Damaging | Low | |
| LCAT | NM_000229 | c.694T>A | p.Ser232Thr | rs4986970 | DM | Tolerated | 0.0084 | Both |
| LCAT | NM_000229 | c.1043C>A | p.Thr348Ile | Novel | — | Damaging | — | Low |
| LCAT | NM_000229 | c.1103G>T | p.Gly368Val | Novel | DM | Damaging | — | Low |
| NPC1 | NM_000271 | c.665A>G | p.Asn222Ser | rs55680026 | DM | Tolerated | 0.003 | Both |
| NPC1 | NM_000271 | c.3308G>T | p. Gly1012Cys | Novel | — | Damaging | Low | |
| NPC1 | NM_000271 | c.3689T>C | p.Leu1230Ser | Novel | — | Damaging | — | High |
| High HDL genes | ||||||||
| CETP | NM_000078 | c.118+1-118+4delGTAA | splice site | Novel | DM | — | — | High |
| CETP | NM_000078 | c.1376A>G | p.Asp459Gly | rs2303790 | DM | Damaging | 0.0062 | High |
| LIPG | NM_006033 | c.716T>C | p.Ile239Thr | Novel | DM | Damaging | — | High |
| LIPG | NM_006033 | c.1069G>T | p.Glu273* | Novel | — | Damaging | High | |
| LIPG | NM_006033 | c.1426C>T | p.Arg476Trp | rs117623631 | DM | Damaging | 0.003 | High |
| SCARB1 | NM_001082959 | c.715G>A | p.Gly239Arg | Novel | — | Damaging | — | Low |
MAF is based on 1000 Genomes CEU (Utah residents with ancestry from northern and western Europe) data. Functional prediction based on SIFT. DM, disease-causing mutation in HGMD; DP, disease-associated polymorphism in HGMD; Ref. Seq., reference sequence; Nuc., nulceotide; rsID, reference SNP identification. Phenotype refers to whether the variant was detected in a patient (or patients) with low HDL-C, high HDL-C, or in both groups of patients.
Of note, three patients with extremely low HDL-C were found to harbor homozygous or compound heterozygous mutations in the APOA1, ABCA1, and LCAT genes (Table 4). This included a 43-year-old female subject [patient identification number (ID) 114] with extremely low HDL-C (0.1 mmol/l) and ApoA-I (0.05 g/l), found to be homozygous for a novel premature truncation in the APOA1 gene (p.Arg34*); a 21-year-old male subject (patient ID 18301) with near undetectable HDL-C (0.1 mmol/l) found to be compound heterozygous for one known disease-causing mutation in ABCA1 (p.Trp590Leu) and one novel premature truncation in ABCA1 (p.Arg2200*); and a 21-year-old male subject (patient ID 34) with extremely low HDL-C (0.1 mmol/l) and less severely reduced ApoA-I levels (0.9 g/l), found to be homozygous for a novel predicted deleterious missense variant in LCAT (p.Thr348Ile), as well as heterozygous for a previously identified variant in ABCA1 (p.Val2244Ile) (19). These results established molecular diagnoses of the rare Mendelian disorders ApoA-I deficiency, Tangier disease, and familial LCAT deficiency, respectively, in these three individuals. Another individual (patient ID 35) who is the sibling of patient ID 34 and had a less dramatic reduction in HDL-C (0.5 mmol/l) was found to be heterozygous for the same LCAT variant (p.Thr348Ile), establishing a gene-dosage relationship for this variant with HDL-C levels.
TABLE 4.
Individuals with a new molecular diagnosis of HDL
| Patient ID | Gender | Age | TC | HDL-C | LDL-C | TG | ApoA-I | Gene | Type | Variant |
| Low HDL-C patients | ||||||||||
| 114 | F | 43 | 3.48 | 0.10 | 2.30 | 2.49 | 0.05 | APOA1 | Homo. | p.Arg34* |
| 18301 | M | 21 | 1.80 | 0.10 | 1.36 | 0.85 | 0.27 | ABCA1 | Cmpd. het. | p.Arg2200* |
| pTrp590Leu | ||||||||||
| 34 | F | 21 | 2.30 | 0.10 | 1.70 | 1.10 | 0.90 | LCAT | Homo. | p.Thr348Ile |
| ABCA1 | Het. | p.Val2244Ile | ||||||||
| 13 | M | 63 | 5.18 | 0.33 | 3.73 | 2.44 | 1.04 | ABCA1 | Het. | p.Arg638Gln |
| 96 | M | 59 | 3.70 | 0.36 | – | 4.06 | 0.89 | LCAT | Het. | p.Arg159Gln |
| 41 | M | 62 | 3.87 | 0.39 | 2.28 | 2.65 | 1.15 | LCAT | Het. | p.Ala165Thr |
| ABCA1 | Het. | p.Arg1680Gln | ||||||||
| 28 | M | 41 | 3.10 | 0.40 | 1.30 | 3.00 | 0.60 | APOA1 | Het. | p.Lys131del |
| LCAT | Het. | p.Ser232Thr | ||||||||
| 4 | F | 67 | 3.15 | 0.44 | 1.71 | 2.21 | 0.62 | ABCA1 | Het. | p.Arg1817* |
| 21 | M | 61 | 4.70 | 0.50 | 2.50 | 3.70 | 1.00 | ABCA1 | Het. | p.Asp585Glu |
| 91 | M | 62 | 3.40 | 0.50 | 1.80 | 2.40 | 0.76 | ABCA1 | Het. | p.Pro847Leu |
| 35 | M | 26 | 4.00 | 0.50 | 2.70 | 1.70 | 1.14 | LCAT | Het. | p.Thr348Ile |
| ABCA1 | Het. | p.Val2244Ile | ||||||||
| 87 | M | 49 | 3.20 | 0.52 | 1.60 | 2.20 | 0.98 | ABCA1 | Het. | p.Glu1902Ilefs*12 |
| SCARB1 | Het. | p.Gly239Arg | ||||||||
| 117 | F | 58 | 6.63 | 0.59 | 5.30 | 1.61 | 1.12 | ABCA1 | Cmpd. het. | p.Thr1512Met |
| p.Gln980Serfs*9 | ||||||||||
| 9 | F | 40 | 4.90 | 0.60 | – | 8.70 | 1.13 | LCAT | Het. | p.Ser232Thr |
| 90 | M | 48 | 3.60 | 0.60 | 2.30 | 1.60 | 1.11 | LCAT | Het. | p.Gly368Val |
| 20 | M | 56 | 1.83 | 0.65 | 0.79 | 0.85 | 1.14 | ABCA1 | Het. | p.Val399Ala |
| 115 | F | 54 | 4.20 | 0.66 | 2.40 | 2.50 | 1.30 | ABCA1 | Cmpd. het. | p.Trp1699Cys |
| p.Ala1046Asp | ||||||||||
| LCAT | Het. | p.Ser232Thr | ||||||||
| 139 | M | 60 | 3.43 | 0.66 | 1.16 | 3.55 | 1.16 | ABCA1 | Het. | p.Val399Ala |
| 134 | M | 71 | 3.40 | 0.70 | 2.00 | 1.50 | 0.99 | LCAT | Het. | p.Thr147Ile |
| 14 | M | 31 | 6.60 | 0.70 | 4.40 | 3.20 | 1.01 | LCAT | Het. | p.Ser232Thr |
| 57 | M | 35 | 4.10 | 0.70 | 2.90 | 1.00 | 0.96 | APOA1 | Het. | p.Lys131Met |
| 53 | M | 47 | 4.16 | 0.71 | 2.49 | 2.12 | 0.93 | ABCA1 | Het. | p.Cys1484Leufs*17 |
| 66 | F | 53 | 3.80 | 1.00 | 2.40 | 0.80 | 1.11 | ABCA1 | Het. | p.Trp590Leu |
| High HDL-C patients | ||||||||||
| 36 | F | 77 | 6.80 | 3.15 | 3.40 | 0.69 | — | LIPG | Cmpd. het. | p.Ile239Thr |
| p.Arg476Trp | ||||||||||
| 48 | F | 61 | 5.10 | 2.20 | 2.20 | 1.47 | 2.38 | CETP | Het. | p.Asp459Gly |
| ABCA1 | Het. | p.Leu1041Val | ||||||||
| 44 | M | 71 | 5.10 | 2.20 | 2.20 | 2.00 | 1.97 | CETP | Het. | Splice site |
| 100 | M | 89 | 5.46 | 2.13 | 2.87 | 1.01 | 1.69 | LIPG | Het. | p.Glu273* |
TC, total cholesterol; Het., heterozygous; Cmpd. het., compound heterozygous; Homo., homozygous.
One individual with extremely high HDL-C (3.2 mmol/l) was compound heterozygous for two known disease-causing variants in LIPG, p.Arg476Trp, and p.Ile239Thr (20). One individual of self-reported Japanese ancestry with high HDL-C (1.9 mmol/l) was heterozygous for p.Asp459Gly in CETP, a known high HDL-C-causing variant that is present in 2–3% of individuals of Asian ancestry, but absent in individuals of European ancestry (http://www.1000genomes.org). One patient with low HDL-C (0.52 mmol/l) was heterozygous for both a novel 2 bp insertion in ABCA1 leading to premature protein truncation (p.Glu1902Ilefs*1912) and a novel predicted damaging mutation in SCARB1 (p.Gly239Arg). This observation is consistent with previous data that mutations in ABCA1 are dominant to the effects of mutations in SCARB1 (21).
The distribution of prioritized alleles in genes known to cause low HDL-C was highly skewed toward patients with low HDL-C, with 44/50 (88%) of such alleles occurring in patients with low HDL-C and only 6/50 (12%) in patients with high HDL-C. Similarly, 83% of prioritized alleles in genes known to cause high HDL-C were observed in patients with high HDL-C. Among patients with low HDL-C, 34/35 (97.1%) of the prioritized alleles observed were in genes known to cause low HDL-C, and only 2.9% were in genes known to cause high HDL-C. These observations suggest that our prioritization criteria were effective, in general, at identifying variants with penetrant effects on the lipid phenotypes in question. A notable exception is the NPC1 gene, in which we detected pathogenic or probably pathogenic variants in two individuals with low HDL-C (3.1% of low HDL-C patients) and two individuals with high HDL-C (2.6% of high HDL-C patients), all in the heterozygous state, suggesting that these variants may not contribute to the differences in HDL-C in these patients.
We performed Sanger sequencing of PCR amplicons to confirm the 35 prioritized variants, present as 50 genotypes. All 50 of these genotypes were bi-directionally confirmed by Sanger sequencing, indicating that the NGS assay had 100% concordance with the gold standard (95% confidence interval 92.9–100.0%). We also assessed the reproducibility of the NGS assay by performing technical replication of one sample in two independent NGS runs. After quality filtering was performed, we observed 100% concordance in quality filtered variant calls between runs.
Diagnostic yield for extreme HDL-C
We considered a molecular diagnosis to have been made if a patient had a pathogenic or probably pathogenic variant detected in a gene known to cause the phenotype present in that patient (i.e., in a low HDL-C gene in a patient with low HDL-C and vice versa). We excluded variants in NPC1 from this analysis because of the observation that a similar proportion of patients with high versus low HDL-C had variants in this gene. Overall, a molecular diagnosis was established in 23 out of 64 patients with low HDL-C (35.9%) and 4 out of 77 patients with high HDL-C (5.2%) (Table 4). The percentage of patients in whom a molecular diagnosis was made did not appreciably change when we excluded patients of non-European ancestry [patients with a molecular diagnosis 15/46 (32.6%) with low HDL-C and 3/63 (4.8%) with high HDL-C], suggesting that these results were not substantially affected by population stratification.
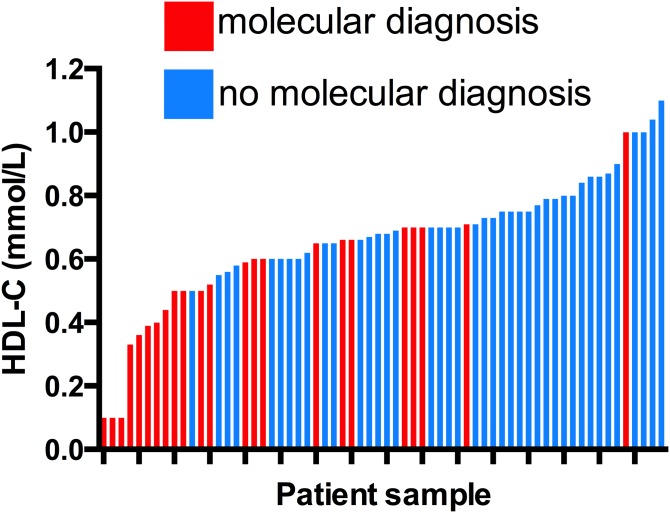
Within the low HDL-C group of patients, HDL-C levels were significantly lower among those in whom a molecular diagnosis was made compared with those in whom no molecular diagnosis was made (0.51 ± 0.2 mmol/l vs. 0.74 ± 0.1 mmol/l, P < 0.0001) (Fig. 1). Similarly, within the low HDL-C group, patients in whom a molecular diagnosis was made had lower TG levels (2.44 ± 1.6 mmol/l vs. 4.52 ± 3.9 mmol/l, P = 0.02) and a trend toward a lower prevalence of type 2 diabetes (13.0% vs. 36.6%, P = 0.08), compared with those in whom no molecular diagnosis was made. In contrast, among patients with high HDL-C, the HDL-C level did not differ between those with or without a molecular diagnosis (2.42 ± 0.5 mmol/l vs. 2.47 ± 0.4 mmol/l, P = 0.8).
Fig. 1.
HDL-C levels in patients with or without a molecular diagnosis of low HDL. Each patient is represented by one column in order of increasing HDL-C level from left to right. Red columns indicate patients in whom a molecular diagnosis of low HDL-C was made by NGS, and blue columns indicate those in whom no molecular diagnosis of low HDL-C was made.
Identification of known mutations
To assess the sensitivity of our assay to detect previously identified mutations, we sequenced six patients with known disease-causing rare mutations in the ABCA1 gene previously identified by Sanger sequencing (15). This included five individuals heterozygous for missense mutations in ABCA1 and one individual compound heterozygous for mutations in ABCA1. All seven genotypes were correctly identified by NGS. However, it is noteworthy that one of these mutations, c.4176-11T>G, previously shown to be pathogenic (15), would not have passed conservative prioritization criteria based on its distance from the intron/exon junction (11 nucleotides upstream of the exon start site).
Functional significance of pathogenic variants
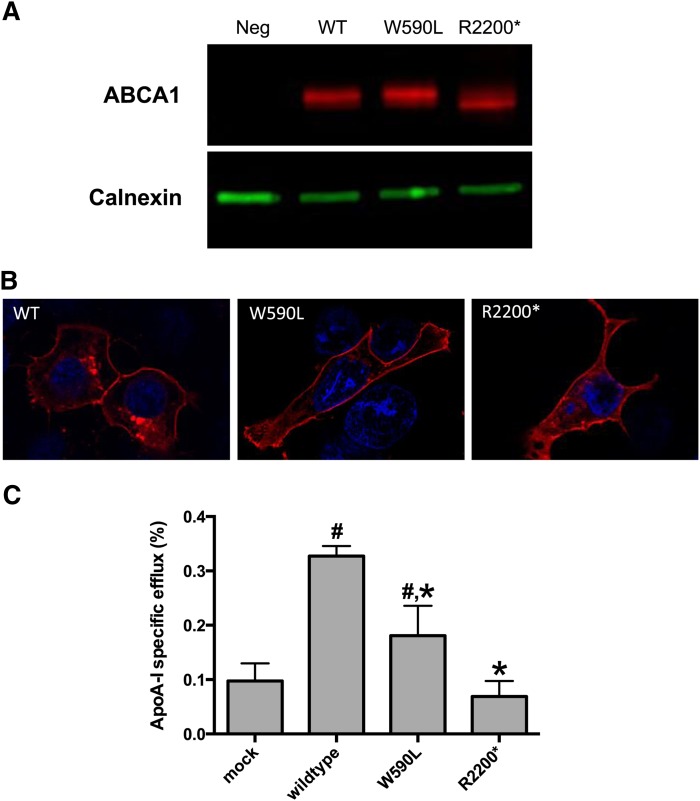
To investigate the functional significance of variants identified by our prioritization approach, we performed functional validation of a subset of these variants. We generated the ABCA1 p.Trp590Leu and p.Arg2200* variants, identified in patient ID 18301, and transiently expressed them in HEK293 cells. Both variants displayed similar levels of protein expression by Western blot (Fig. 2A) and similar patterns of subcellular localization by immunofluorescence (Fig. 2B), as compared with wild-type ABCA1. We then tested the ability of cells transfected with these variants to efflux radiolabeled cholesterol in response to ApoA-I. The p.Arg2200* variant displayed complete abrogation of ApoA-I-dependent cholesterol efflux activity, whereas the p.Trp590Leu variant displayed an ∼50% reduction in efflux compared with wild-type ABCA1 (Fig. 2C), indicating that these two variants represent complete and partial loss-of-function alleles, respectively. Similar results have been reported previously for another mutation in ABCA1 (p.Trp590Ser) that affects the same amino acid position (22).
Fig. 2.
Functional characterization of variants in ABCA1. A: Western blot analysis showing normal protein expression of wild-type and mutant ABCA1. The two ABCA1 variants, as well as wild-type ABCA1, were generated in vitro and expressed in HEK293 cells. Cell lysates were probed with antibodies to ABCA1 and calnexin (as loading control). Neg, negative control. B: Immunofluorescence results showing normal subcellular localization of wild-type, W590L, and R2200* ABCA1 proteins. C: HEK293 cells were transiently transfected with wild-type, W590L, and R2200* ABCA1, and cholesterol efflux measured in the presence or absence of ApoA-I. # Indicates a statistically significant difference compared with mock transfected and * indicates a statistically significant difference compared with wild-type ABCA1 at P < 0.05 using a one-way ANOVA.
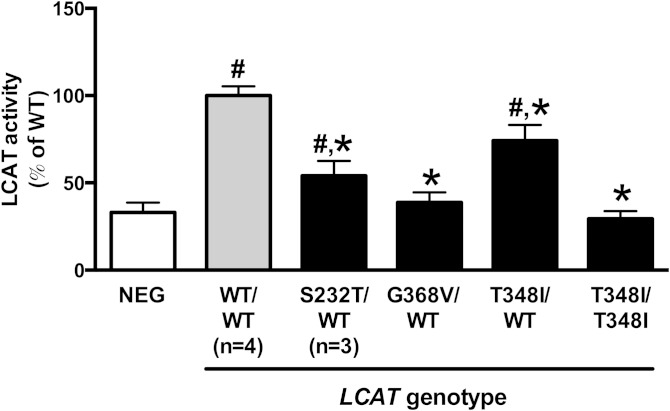
We measured LCAT activity in plasma from six individuals with variants in the LCAT gene compared with four patients with wild-type LCAT and similar HDL-C levels. This included the novel variant p.Thr348Ile, observed in one patient in the homozygous state and in one patient in the heterozygous state. We also included the p.Ser232Thr and p.Gly368Val variants, both of which have been previously reported to cause low HDL-C (5, 23), but have not been functionally characterized. We observed p.Ser232Thr in three patients and p.Gly368Val in one patient, all in the heterozygous state. Relative to patients with wild-type LCAT and similar HDL-C levels, heterozygotes for either p.Ser232Thr or p.Thr348Ile variants had significant reductions in plasma LCAT activity, with some residual activity (Fig. 3). The patient homozygous for p.Thr348Ile, as well as the patient heterozygous for p.Gly368Val, had complete abrogation of LCAT activity with residual activity that was not significantly different than the negative control sample. These data indicate that patients with these variants in LCAT have impaired plasma LCAT activity, consistent with the concept that these variants represent loss-of-function alleles.
Fig. 3.
LCAT activity in plasma from patients with wild-type or variant LCAT genes. LCAT activity was determined in plasma using a fluorescent assay. Values represent the mean ± SD of triplicate samples from individual patients unless otherwise indicated. # Indicates a statistically significant difference compared with negative control and * indicates a statistically significant difference compared with wild-type, both at P < 0.05 using a one-way ANOVA.
DISCUSSION
We have described a targeted NGS panel for molecular diagnosis of disorders of lipid metabolism. We assessed the performance of this panel in a population of patients with extreme HDL-C levels without previously identified mutations, as well as a collection of patients with known rare mutations that impact HDL-C. The major findings of this study are: 1) that this panel is highly accurate and has high analytic sensitivity to identify pathogenic and probably pathogenic variants that impact HDL-C; 2) the report of several novel variants in HDL-related genes with associated clinical phenotypes and functional validation of a subset of these; and 3) that a molecular diagnosis can be established in a substantial percentage of patients with extremely low HDL-C visiting a specialty lipid clinic.
Our results showed high analytic accuracy of this assay to detect pathogenic variants. It is noteworthy that while our assay accurately detected a known pathogenic mutation in the ABCA1 gene (c.4176-11T>G) (15) with high levels of confidence (Q > 30, 3,188 depth of coverage), the location of this mutation, 11 nucleotides 5′ of the start of an exon, means that it would have been excluded by most conservative prioritization approaches that generally identify splice site variants two nucleotides up- or downstream of the intron/exon junction. This example highlights the ongoing challenge of determining pathogenicity by computational approaches.
We showed that the NGS assay could successfully identify the molecular causes of rare Mendelian disorders of HDL-C, namely, ApoA-I deficiency, Tangier disease, and familial LCAT deficiency. Notably, none of these three patients displayed the classical physical examination findings associated with these conditions (i.e., enlarged yellow tonsils in Tangier disease or corneal opacities in familial LCAT deficiency). This suggests that the sequencing-based approach described here may be more sensitive to identify patients with rare Mendelian disorders of HDL-C compared with an approach based on classical stigmata of these conditions. Previously, we have employed Sanger sequencing to identify pathogenic mutations in patients with Mendelian disorders of HDL-C (15). For large genes such as ABCA1 (50 exons), this approach is costly, labor-intensive, time-consuming, and generally requires sequential testing of candidate genes if no pathogenic mutation is identified in the initial screening. In contrast, the targeted resequencing strategy we describe here allows processing of multiple samples and multiple genes in parallel, thus substantially reducing both the cost and time on a per-patient and per-gene basis.
While our study was designed primarily to detect known pathogenic variants, it is noteworthy that 21 of the prioritized variants we detected were novel. Importantly, we confirmed functionality of some of these variants by demonstrating either reduced activity in vitro or reduced enzyme activity ex vivo. These results highlight the research and discovery potential of targeted gene panels for both known disease genes and important candidate genes. In this regard, we included NPC1 as a candidate gene based on the observation that patients with the autosomal recessive disorder, Niemann-Pick disease, have reduced levels of plasma HDL-C (24–26). Our results showing a similar proportion of patients with low versus high HDL-C carrying predicted damaging variants in NPC1 (3.1% vs. 2.6%) do not support a role for heterozygous variants in this gene causing low HDL-C in the general population. For this reason, we did not consider the finding of a pathogenic or probably pathogenic variant in NPC1 to constitute a molecular diagnosis of low HDL-C. Larger sample sizes and functional testing may help to clarify the impact of rare heterozygous variants in NPC1 on HDL-C levels.
One previous study evaluated the use of a targeted NGS panel for lipid disorders in patients with abnormal TG or LDL-C levels and in patients with lipodystrophy and maturity-onset diabetes of the young (27). That study also reported a high level of analytic accuracy in detecting known mutations (95.2%). The mean depth of coverage achieved by that assay (345.1×) was substantially less than was achieved by our assay (1,559×), which relates to the smaller genomic capture region of our assay. Together with these previous results on disorders of TG and LDL-C, the current study, demonstrating high accuracy to identify pathogenic variants causing abnormalities of HDL-C, suggest that targeted NGS panels can accurately identify pathogenic variants across the entire range of clinical dyslipidemias.
We identified a pathogenic or probably pathogenic variant in 35.9% of patients with low HDL-C, suggesting a high diagnostic yield in this patient population. Notably, among patients with low HDL-C, the HDL-C level was significantly lower among those in whom a pathogenic variant was identified compared with those in whom no such variant was identified (Fig. 1). This suggests that identification of a molecular defect in patients with low HDL-C may identify a more extreme subpopulation of patients, and likewise, that the diagnostic yield will be highest among patients with the lowest levels of HDL-C. Indeed, had we included only patients with an HDL-C of less than or equal to 0.7, 0.6, or 0.5 mmol/l, the molecular diagnosis rate would have increased to 51.2, 65.2, or 91.7%, respectively. This further indicates that these established HDL genes account for the vast majority of cases of extremely low HDL-C, and that there are likely to be few additional major genes causing this phenotype that remain to be discovered. The observation that patients with low HDL-C in whom no pathogenic variant was identified had higher TG levels and a trend toward a higher prevalence of diabetes mellitus is consistent with the concept that the low HDL-C phenotype in these patients may be secondary to other metabolic factors, rather than being primarily due to a major genetic cause.
In contrast to the low HDL-C group, the diagnostic yield in patients with extremely high HDL-C was lower in our cohort (5.2%), suggesting that there may be additional unknown genetic or nongenetic factors that impact this phenotype, or alternatively, that a greater proportion of such patients have multiple variants in genes with more modest effect sizes that may not have been detected by our approach (12).
There are several limitations of our study. Our assay was primarily designed to detect single nucleotide variants and small indels. As such, larger indels or complex rearrangements may not have been detected. As bioinformatics approaches to identifying structural and copy number variants improve, it is expected that detection of this category of variant will increase. Importantly, out of the 200 curated mutations in the ABCA1 gene, only 6 (3%) are large indels or complex rearrangements, suggesting that the majority of pathogenic variants will be detected by our assay. Second, despite applying stringent prioritization criteria, it is not possible to conclusively determine the pathogenicity of novel variants by computational methods alone. This is particularly the case for missense variants, and it is likely that some of the novel missense variants detected in our study that were predicted to be damaging are in fact benign. We used a relatively stringent bioinformatics approach, by requiring that five different computational algorithms all agreed in their predictions that a novel missense variant was deleterious, for us to classify it as probably pathogenic. Notwithstanding the limitations of bioinformatics approaches to predict functionality, our functional validation of a subset of novel variants provides confidence that many of the variants detected in this study are functional. In addition, the prioritized variants showed strong enrichment with the associated phenotype: 88% of prioritized alleles in low HDL genes were in patients with low HDL-C. In contrast, analysis of all rare coding variants in low HDL-C genes without prioritization displayed a more equal distribution: 60% of rare nonprioritized coding alleles in low HDL genes were in patients with low HDL-C. This indicates that our prioritization approach selected variants with penetrant effects on these lipid phenotypes. Lastly, we biochemically characterized only a subset of the variants detected. While our functional data is consistent with the concept that many of the variants we detected do impair the function of the encoded proteins, we did not perform exhaustive functional characterization of all novel variants detected. This also reflects the reality of clinical practice, in which novel variants are frequently discovered but functional characterization may not be possible.
Identifying the molecular cause of HDL-C or other lipid or lipoprotein parameters provides diagnostic closure and allows rapid cascade screening in the family members of affected individuals. In patients with mutations causing elevated LDL-C, introduction of a molecular diagnostic program has been shown to significantly increase the appropriate use of lipid-lowering therapy (28, 29). Similarly, identification of molecular abnormalities in patients with low HDL-C could guide personalized cardiovascular prevention and risk-reduction strategies, and may improve patient compliance with therapeutic lifestyle modification or pharmacological interventions. Moreover, targeted therapies for specific disorders of HDL metabolism are in development (30, 31), and the identification of specific molecular defects in patients with abnormal HDL-C levels may identify candidates for these future therapies.
In summary, we have described a novel targeted NGS panel for disorders of lipid metabolism. Our validation study in patients with extreme levels of HDL-C indicates that this assay is highly accurate for the identification of pathogenic variants and that the diagnostic yield of such an approach is high, particularly among patients with very low HDL-C. Targeted sequencing approaches hold the promise to refine diagnostic categories and may lead to new opportunities for personalized treatment and prevention strategies in patients with abnormalities of plasma lipid levels.
Supplementary Material
Acknowledgments
The authors thank the patients who participated in this study.
Footnotes
Abbreviations:
- HDL-C
- HDL cholesterol
- HGMD
- Human Gene Mutation Database
- ID
- identification number
- LDL-C
- LDL cholesterol
- MAF
- minor allele frequency
- NGS
- next-generation sequencing
- UTR
- untranslated region
This work was supported by a project grant from the British Columbia Clinical Genomics Network (to L.R.B. and J.F.), by the Canadian Institutes of Health Research (to G.A.F., MOP-119418), and by the National University of Singapore and the Biomedical Research Council of the Agency for Science, Technology, and Research of Singapore. L.R.B. is the recipient of a clinician-scientist new investigator grant from the National Medical Research Council of Singapore.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Yusuf S., Hawken S., Ounpuu S., Dans T., Avezum A., Lanas F., McQueen M., Budaj A., Pais P., Varigos J., et al. 2004. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 364: 937–952. [DOI] [PubMed] [Google Scholar]
- 2.Boekholdt S. M., Arsenault B. J., Hovingh G. K., Mora S., Pedersen T. R., Larosa J. C., Welch K. M., Amarenco P., Demicco D. A., Tonkin A. M., et al. 2013. Levels and changes of HDL cholesterol and apolipoprotein A-I in relation to risk of cardiovascular events among statin-treated patients: a meta-analysis. Circulation. 128: 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barter P., Gotto A. M., LaRosa J. C., Maroni J., Szarek M., Grundy S. M., Kastelein J. J., Bittner V., Fruchart J. C.; Treating to New Targets Investigators. 2007. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N. Engl. J. Med. 357: 1301–1310. [DOI] [PubMed] [Google Scholar]
- 4.Goode E. L., Cherny S. S., Christian J. C., Jarvik G. P., de Andrade M. 2007. Heritability of longitudinal measures of body mass index and lipid and lipoprotein levels in aging twins. Twin Res. Hum. Genet. 10: 703–711. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., Hobbs H. H. 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 305: 869–872. [DOI] [PubMed] [Google Scholar]
- 6.Tietjen I., Hovingh G. K., Singaraja R., Radomski C., McEwen J., Chan E., Mattice M., Legendre A., Kastelein J. J., Hayden M. R. 2012. Increased risk of coronary artery disease in Caucasians with extremely low HDL cholesterol due to mutations in ABCA1, APOA1, and LCAT. Biochim. Biophys. Acta. 1821: 416–424. [DOI] [PubMed] [Google Scholar]
- 7.Frikke-Schmidt R., Nordestgaard B. G., Stene M. C., Sethi A. A., Remaley A. T., Schnohr P., Grande P., Tybjaerg-Hansen A. 2008. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 299: 2524–2532. [DOI] [PubMed] [Google Scholar]
- 8.Kiss R. S., Kavaslar N., Okuhira K., Freeman M. W., Walter S., Milne R. W., McPherson R., Marcel Y. L. 2007. Genetic etiology of isolated low HDL syndrome: incidence and heterogeneity of efflux defects. Arterioscler. Thromb. Vasc. Biol. 27: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 9.Berge K. E., Leren T. P. 2010. Mutations in APOA-I and ABCA1 in Norwegians with low levels of HDL cholesterol. Clin. Chim. Acta. 411: 2019–2023. [DOI] [PubMed] [Google Scholar]
- 10.Candini C., Schimmel A. W., Peter J., Bochem A. E., Holleboom A. G., Vergeer M., Dullaart R. P., Dallinga-Thie G. M., Hovingh G. K., Khoo K. L., et al. 2010. Identification and characterization of novel loss of function mutations in ATP-binding cassette transporter A1 in patients with low plasma high-density lipoprotein cholesterol. Atherosclerosis. 213: 492–498. [DOI] [PubMed] [Google Scholar]
- 11.Alrasadi K., Ruel I. L., Marcil M., Genest J. 2006. Functional mutations of the ABCA1 gene in subjects of French-Canadian descent with HDL deficiency. Atherosclerosis. 188: 281–291. [DOI] [PubMed] [Google Scholar]
- 12.Motazacker M. M., Peter J., Treskes M., Shoulders C. C., Kuivenhoven J. A., Hovingh G. K. 2013. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 33: 1521–1528. [DOI] [PubMed] [Google Scholar]
- 13.Rehm H. L. 2013. Disease-targeted sequencing: a cornerstone in the clinic. Nat. Rev. Genet. 14: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heiss G., Johnson N. J., Reiland S., Davis C. E., Tyroler H. A. 1980. The epidemiology of plasma high-density lipoprotein cholesterol levels. The Lipid Research Clinics Program Prevalence Study. Summary. Circulation. 62: IV116–IV136. [PubMed] [Google Scholar]
- 15.Brunham L. R., Kang M. H., Van Karnebeek C., Sadananda S. N., Collins J. A., Zhang L. H., Sayson B., Miao F., Stockler S., Frohlich J., et al. 2015. Clinical, biochemical, and molecular characterization of novel mutations in ABCA1 in families with Tangier disease. JIMD Rep. 18: 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson P. D., Mort M., Ball E. V., Howells K., Phillips A. D., Thomas N. S., Cooper D. N. 2009. The Human Gene Mutation Database: 2008 update. Genome Med. 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juren A. J., Sarwal G., Al-Sarraf A., Vrablik M., Chan D., Humphries K. H., Frohlich J. J. 2013. Low prevalence of type 2 diabetes mellitus among patients with high levels of high-density lipoprotein cholesterol. J. Clin. Lipidol. 7: 194–198. [DOI] [PubMed] [Google Scholar]
- 18.Richards C. S., Bale S., Bellissimo D. B., Das S., Grody W. W., Hegde M. R., Lyon E., Ward B. E.; Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. 2008. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 10: 294–300. [DOI] [PubMed] [Google Scholar]
- 19.Probst M. C., Thumann H., Aslanidis C., Langmann T., Buechler C., Patsch W., Baralle F. E., Dallinga-Thie G. M., Geisel J., Keller C., et al. 2004. Screening for functional sequence variations and mutations in ABCA1. Atherosclerosis. 175: 269–279. [DOI] [PubMed] [Google Scholar]
- 20.Khetarpal S. A., Edmondson A. C., Raghavan A., Neeli H., Jin W., Badellino K. O., Demissie S., Manning A. K., DerOhannessian S. L., Wolfe M. L., et al. 2011. Mining the LIPG allelic spectrum reveals the contribution of rare and common regulatory variants to HDL cholesterol. PLoS Genet. 7: e1002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunham L. R., Tietjen I., Bochem A. E., Singaraja R. R., Franchini P. L., Radomski C., Mattice M., Legendre A., Hovingh G. K., Kastelein J. J., et al. 2011. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin. Genet. 79: 575–581. [DOI] [PubMed] [Google Scholar]
- 22.Singaraja R. R., Visscher H., James E. R., Chroni A., Coutinho J. M., Brunham L. R., Kang M. H., Zannis V. I., Chimini G., Hayden M. R. 2006. Specific mutations in ABCA1 have discrete effects on ABCA1 function and lipid phenotypes both in vivo and in vitro. Circ. Res. 99: 389–397. [DOI] [PubMed] [Google Scholar]
- 23.Haase C. L., Tybjaerg-Hansen A., Qayyum A. A., Schou J., Nordestgaard B. G., Frikke-Schmidt R. 2012. LCAT, HDL cholesterol and ischemic cardiovascular disease: a Mendelian randomization study of HDL cholesterol in 54,500 individuals. J. Clin. Endocrinol. Metab. 97: E248–E256. [DOI] [PubMed] [Google Scholar]
- 24.Tängemo C., Weber D., Theiss S., Mengel E., Runz H. 2011. Niemann-Pick Type C disease: characterizing lipid levels in patients with variant lysosomal cholesterol storage. J. Lipid Res. 52: 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garver W. S., Jelinek D., Meaney F. J., Flynn J., Pettit K. M., Shepherd G., Heidenreich R. A., Vockley C. M., Castro G., Francis G. A. 2010. The National Niemann-Pick Type C1 Disease Database: correlation of lipid profiles, mutations, and biochemical phenotypes. J. Lipid Res. 51: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi H. Y., Karten B., Chan T., Vance J. E., Greer W. L., Heidenreich R. A., Garver W. S., Francis G. A. 2003. Impaired ABCA1-dependent lipid efflux and hypoalphalipoproteinemia in human Niemann-Pick type C disease. J. Biol. Chem. 278: 32569–32577. [DOI] [PubMed] [Google Scholar]
- 27.Johansen C. T., Dube J. B., Loyzer M. N., Macdonald A., Carter D. E., McIntyre A. D., Cao H., Wang J., Robinson J. F., Hegele R. A. 2014. LipidSeq: a next-generation clinical resequencing panel for monogenic dyslipidemias. J. Lipid Res. 55: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huijgen R., Kindt I., Verhoeven S. B., Sijbrands E. J., Vissers M. N., Kastelein J. J., Hutten B. A. 2010. Two years after molecular diagnosis of familial hypercholesterolemia: majority on cholesterol-lowering treatment but a minority reaches treatment goal. PLoS One. 5: e9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umans-Eckenhausen M. A., Defesche J. C., van Dam M. J., Kastelein J. J. 2003. Long-term compliance with lipid-lowering medication after genetic screening for familial hypercholesterolemia. Arch. Intern. Med. 163: 65–68. [DOI] [PubMed] [Google Scholar]
- 30.Stoekenbroek R. M., van den Bergh Weerman M. A., Hovingh G. K., Potter van Loon B. J., Siegert C. E., Holleboom A. G. 2013. Familial LCAT deficiency: from renal replacement to enzyme replacement. Neth. J. Med. 71: 29–31. [PubMed] [Google Scholar]
- 31.Alphacore Pharma, LLC. 2012. Effect of ACP-501 on Safety, Tolerability, Pharmacokinetics and Pharmacodynamics in Subjects with Coronary Artery Disease. In Clinicaltrials.gov [Internet]. National Library of Medicine (US), Bethesda, MD. Accessed June 5, 2015, at https://clinicaltrials.gov/show/NCT01554800.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.