Abstract
CDH1, as a tumor suppressor gene, contributes sporadic breast cancer (BC) progression. However, the association between CDH1 hypermethylation and BC, and its clinicopathological significance remains unclear. We conducted a meta-analysis to investigate the relationship between the CDH1 methylation profile and the major clinicopathological features. A detailed literature was searched through the electronic databases PubMed, Web of Science™, and EMBASE™ for related research publications. The data were extracted and assessed by two reviewers independently. Odds ratios (ORs) with corresponding confidence intervals (CIs) were calculated and summarized respectively. The frequency of CDH1 methylation was significantly higher in invasive ductal carcinoma than in normal breast tissues (OR =5.83, 95% CI 3.76–9.03, P<0.00001). CDH1 hypermethylation was significantly higher in estrogen receptor (ER)-negative BC than in ER-positive BC (OR =0.62, 95% CI 0.43–0.87, P=0.007). In addition, we found that the CDH1 was significantly methylated in HER2-negative BC than in HER2-positive BC (OR =0.26, 95% CI 0.15–0.44, P<0.00001). However, CDH1 methylation frequency was not associated with progesterone receptor (PR) status, or with grades, stages, or lymph node metastasis of BC patients. Our results indicate that CDH1 hypermethylation is a potential novel drug target for developing personalized therapy. CDH1 hypermethylation is strongly associated with ER-negative and HER2-negative BC, respectively, suggesting CDH1 methylation status could contribute to the development of novel therapeutic approaches for the treatment of ER-negative or HER2-negative BC with aggressive tumor biology.
Keywords: methylation, estrogen receptor, HER2, triple-negative breast cancer
Introduction
Breast Cancer (BC) is the most common malignancy among women in most western countries.1 The development of BC involves a progression, starting with atypical hyperplasia, followed by intermediate stages until the invasive carcinoma, and finally into metastatic disease. A series of epigenetic and genetic changes contribute to this multistep process of BC onset and progression. Epigenetics refers to heritable changes in gene activity and expression that do not involve changes to underlying DNA sequence and has recently gained significant attention of researchers.2 Epigenetic alterations occur in transformed cells and includes global hypomethylation, focal hypermethylation, histone modifications, and nucleosomal remodeling.3 Aberrant DNA hypermethylation is a commonly observed epigenetic modification in human malignancies, including BC.4–6 The CDH1 gene encodes a transmembrane glycoprotein E-cadherin that maintains Ca2+-dependent cell–cell adhesion in epithelial tissues and therefore plays an important role in tumorigenesis as a tumor suppressor gene.7–10 Alteration in E-cadherin expression has been observed in several types of malignancies, including ovarian cancer,11 prostate cancer,12 hepatocellular carcinoma,13 and BC.14 Recent evidence indicate that reduced E-cadherin expression is associated with pathological features, such as poor differentiation, infiltrative growth, lymph node metastasis, and poor prognosis.15–17 However, the association of between CDH1 promoter hypermethylation and BC, and its clinicopathological significance, remains under investigation. We conducted a meta-analysis to investigate the relationship between CDH1 methylation status in BC and the major clinicopathological features including estrogen receptor (ER), progesterone receptor (PR) and HER2.
Methods
Literature search and selection of studies
We conducted a systematic search of the literature published in English and Chinese prior to November 2014. The electronic databases included PubMed, Web of Science™, and EMBASE™. The search was conducted using the following key words: “CDH1 or E-cadherin methylation” and “breast cancer” or “breast carcinoma” for relative articles. There were 84 articles identified from PubMed, 143 articles from Web of Science, and 101 articles from EMBASE. A total of 328 articles were screened using article titles and abstracts. Reference lists of identified articles were searched manually for further relevant articles (Figure 1).
Figure 1.
Schematic flow diagram for selection of included studies.
The following inclusion criteria were applied: 1) studies that reported the association between CDH1 methylation and the clinicopathological significance of BC and 2) studies that investigated the frequency of CDH1 methylation in different ER, PR, as well as HER2 statuses of BC. After screening by article titles and abstracts, 25 relevant articles were included for full text review. The exclusion criteria were as follows: 1) reviews; 2) studies in which CDH1 protein expression was investigated; 3) studies in which cell lines or mice were utilized; and 4) studies in which same populations or overlapping data were used. Finally, eleven articles were selected for inclusion in this meta-analysis. The variables from eleven relevant studies were listed in Table 1.
Table 1.
Main characteristics of included studies
| Author | Year | Country | Sample
|
Stage (I + II/III) | Lymph node (0/+) | Grade (I + II/III) | ER status (−/+) | HER2 status (−/+) | |
|---|---|---|---|---|---|---|---|---|---|
| Normal | IDC | ||||||||
| Asiaf et al36 | 2014 | India | 128 | 128 | 89/39 | 79/49 | 108/20 | 39/89 | |
| Karray-Chouayekh et al37 | 2010 | Tunisia | 78 | 21/30 | 36/42 | 61/17 | 31/47 | 51/13 | |
| Hoque et al50 | 2009 | Italy | 12 | 45 | |||||
| Zou et al51 | 2009 | New Zealand | 19 | 20 | |||||
| Sunami et al52 | 2008 | USA | 130 | 102/8 | 76/54 | 65/65 | 78/29 | ||
| Prasad et al38 | 2008 | India | 50 | 15/35 | 27/23 | ||||
| Li et al53 | 2006 | Australia | 193 | 83/71 | 54/134 | 142/30 | |||
| Shinozaki et al27 | 2005 | USA | 10 | 151 | 102/43 | 37/114 | |||
| Caldeira et al54 | 2005 | Brazil | 71 | 53/18 | 53/18 | 45/26 | 63/8 | ||
| Hu et al55 | 2002 | People’s Republic of China | 23 | 18/5 | 15/8 | 5/10 | 10/3 | ||
| Toyooka et al28 | 2002 | USA | 10 | 51 | |||||
Abbreviations: ER, estrogen receptor; IDC, invasive ductal carcinoma.
Data extraction and study assessment
Two independent reviewers (RH and PD) extracted the following data: first author’s name, year of publication, geographical location, sample size of the different histologic categories of BC, grade of BC, stage of BC, and ER, PR, and HER2 status. Any disagreement was resolved by discussion between two authors (RH and PD). If they could not reach a consensus, a third author (YD) was consulted.
Statistics analysis
The meta-analysis was conducted using Review Manager 5.2 (software update; The Nordic Cochrane Centre, Copenhagen, Denmark). Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated. The assessment of statistical heterogeneity was done by using the Cochran’s Q statistic and I 2 tests. If the I 2 value was below 50%, a fixed effects model was used, and if the I 2 value was 50% or greater, a random effects model was used. The analysis was conducted to evaluate the association of the frequency of CDH1 methylation with invasive ductal carcinoma (IDC). Furthermore, we compared the frequency of CDH1 methylation in different ER, PR, and HER2 statuses as well as in different grades and stages. All P-values were two sided, with P<0.05 considered statistically significant. Publication bias was determined using funnel plot analysis.
Results
CDH1 was more frequently methylated in IDCs than in normal breast tissues (pooled OR =5.83, 95% CI 3.76–9.03, z=7.88, P<0.00001, I 2=46%, P=0.10) (Figure 2). CDH1 hypermethylation was significantly higher in ER-negative than in ER-positive patients with BC (OR =0.62, 95% CI 0.43–0.87, z=2.72, P=0.007, I 2=39%, P=0.13) (Figure 3). CDH1 promoter was significantly more methylated in HER2-negative patients than in HER2-positive patients with BC (OR =0.26, 95% CI 0.15–0.44, z=4.97, P<0.00001, I 2=0%, P=0.57) (Figure 4). The methylation frequency was similar between PR-positive and -negative patients with BC (OR =0.76, 95% CI 0.49–1.15, z=1.30, P=0.19, I2=0%, P=0.45) (Figure 5). The frequency of CDH1 hypermethylation was not significantly higher in grade III than in grade I/II (OR =1.46, 95% CI 0.86–2.49, z=1.40, P=0.16, I 2=11%, P=0.35) (Figure 6). The frequency of CDH1 hypermethylation was not significantly changed between late stage and early stage of BC (OR =1.09, 95% CI 0.63–1.88, z=0.30, P=0.77, I 2=41%, P=0.16) (Figure 7). The frequency of CDH1 hypermethylation was similar between BC patients with positive lymph node metastasis and those with negative lymph node metastasis (OR =1.21, 95% CI 0.59–2.47, z=0.53, P=0.60, I 2=71%, P=0.007) (Figure 8).
Figure 2.
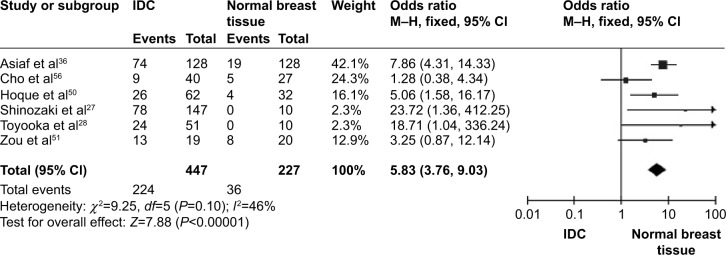
Forest plot for CDH1 hypermethylation in IDC and normal breast tissue.
Abbreviations: CI, confidence interval; IDC, invasive ductal carcinoma; M–H, Mantel–Haenszel.
Figure 3.
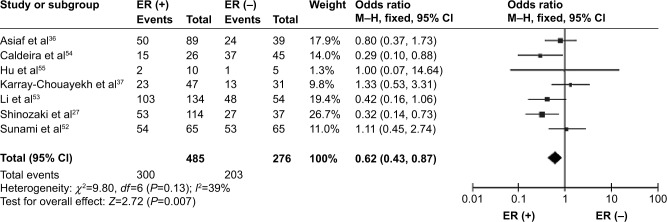
Forest plot for CDH1 hypermethylation in ER-positive and -negative BC.
Abbreviations: BC, breast cancer; CI, confidence interval; ER, estrogen receptor; M–H, Mantel–Haenszel.
Figure 4.
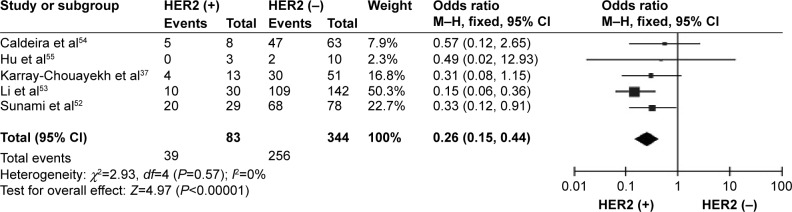
Forest plot for CDH1 hypermethylation in HER2-positive and -negative BC.
Abbreviations: BC, breast cancer; CI, confidence interval; M–H, Mantel–Haenszel.
Figure 5.
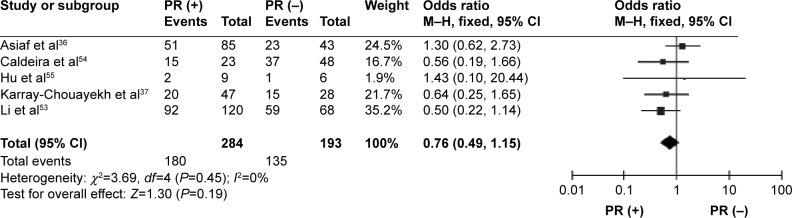
Forest plot for CDH1 hypermethylation in PR-positive and -negative BC.
Abbreviations: BC, breast cancer; CI, confidence interval; PR, progesterone receptor; M–H, Mantel–Haenszel.
Figure 6.
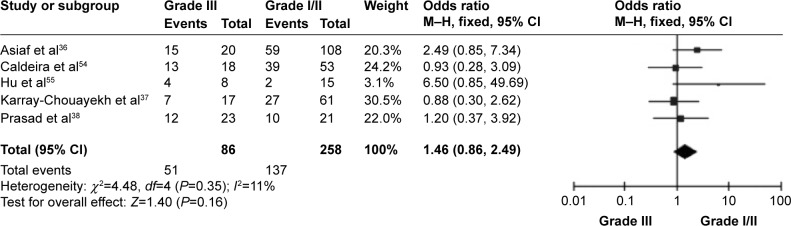
Forest plot for CDH1 hypermethylation in different grades of BC.
Abbreviations: BC, breast cancer; CI, confidence interval; M–H, Mantel–Haenszel.
Figure 7.
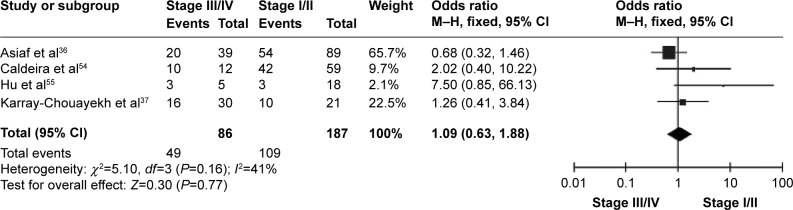
Forest plot for CDH1 hypermethylation in different stages of BC.
Abbreviations: BC, breast cancer; CI, confidence interval; M–H, Mantel–Haenszel.
Figure 8.
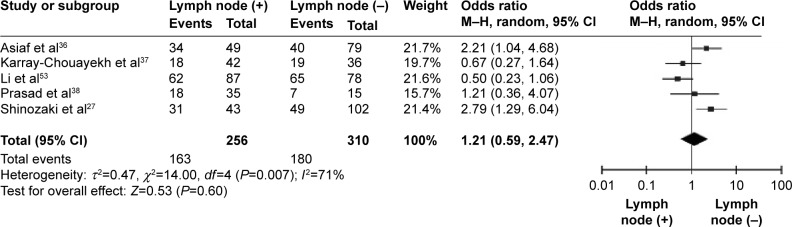
Forest plot for CDH1 hypermethylation in lymph node-positive and -negative metastasis of BC.
Abbreviations: BC, breast cancer; CI, confidence interval; M–H, Mantel–Haenszel.
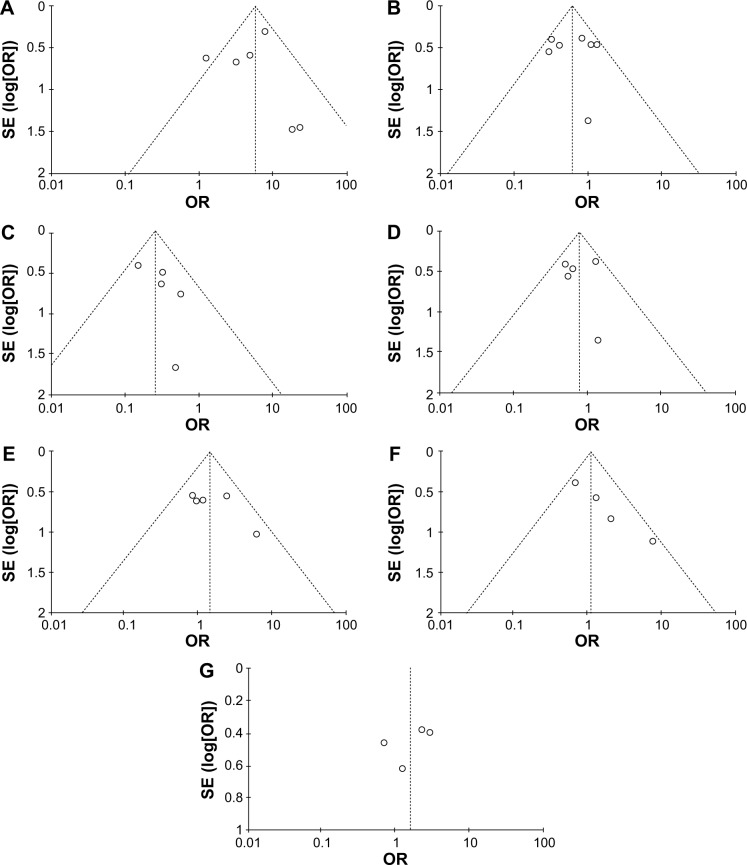
The assessment of quality of the included articles was done using the Newcastle–Ottawa Quality Assessment Scale.18 This scale allocates a maximum of 9 points for the quality of selection, comparability, exposure, and outcomes for study participants. The Newcastle–Ottawa scale scores ranged from 0 to 9 (a score of 7 or more indicates a good quality). Of the studies, three scored 9 points, three scored 8 points, and five scored 7 points. Hence, the studies were of a relatively high quality (data not shown). We further conducted analyses to determine the result stability in the choice of a single study; the finding suggested no single study had the effect on the pooled ORs, indicating the stability of our analyses. There were no publication biases in the meta-analysis according to the largely symmetric funnel plots (Figure 9).
Figure 9.
Funnel plots for publication bias.
Notes: (A) CDH1 methylation in IDC and normal breast tissue; (B) CDH1 methylation in ER-positive and -negative BC; (C) CDH1 methylation in HER2-positive and -negative BC; (D) CDH1 methylation in PR-positive and -negative BC; (E) CDH1 methylation in different grades of BC; (F) CDH1 hypermethylation in different stages of BC; and (G) CDH1 hypermethylation in lymph node-positive and -negative metastasis of BC.
Abbreviations: BC, breast cancer; ER, estrogen receptor; IDC, invasive ductal carcinoma; PR, progesterone receptor; OR, odds ratio.
Discussion
The CDH1 gene encodes E-cadherin protein, which is a 120 kDa glycoprotein consisting of an extracellular domain of five tandem repeated domains, a cytoplasmic domain, and a single transmembrane domain.19,20 The extracellular domain binds to cadherin on adjacent cells and forms cell–cell adhesion, whereas the cytoplasmic domain binds to catenin and activates the Wnt signaling cascade.21 CDH1 hypermethylation is one of the mechanisms that cause gene silencing and results in reduced E-cadherin expression. Lack of E-cadherin leads to the dysfunction of the cell–cell adhesion system, resulting in a loss of cell–cell adhesion, release of cytoplasmic β-catenin, increase in Wnt signaling, and increased tumor invasiveness.22 CDH1 hypermethylation and its reduced expression have been observed in several of malignancies, including gastric, ovarian, lung, and BC.23–26 In the present study, we compared the frequency of CDH1 hypermethylation between IDC and normal breast tissues. Our results showed CDH1 promoter hypermethylation was significantly higher in IDC than in normal breast tissue, which is in agreement with previous studies.27,28 There was a 5.83 times increased risk to BC in the subjects with methylated CDH1 promoter. The major treatments of BC include chemotherapy, hormone therapy, and target therapy, and drug target therapy has been gaining more attention recently. CDH1, as a suppressor gene, is a potential novel drug target, and its hypermethylation could be reversed through demethylation. Inhibitors of DNA methylation (DNMTis), such as 5-Aza-CdR and 5-fluoro-2′-deoxycytidine, have been applied to human lung cancer and BC cells, and currently 5-fluoro-2′-deoxycytidine is in clinical trials for the treatment of BC and other solid tumors.29–31 Lopes et al induced E-cadherin expression in triple-negative MDA-MB-231 cells with 1α,25(OH)2D3, through CDH1 promoter demethylation.32 They observed that 1α,25(OH)2D3 promoted differentiation of MDA-MB-231 cells by inducing de novo E-cadherin expression, an effect that was time- and dose-dependent. These preclinical studies showed the therapeutic potential of restoration of the tumor suppressor gene through epigenetic modulation, which may decrease the aggressiveness of BC. Therefore, CDH1 hypermethylation is a potential novel drug target in the development of personalized therapy.
Epithelial-to-mesenchymal transition (EMT) is an important process in embryonic development and also plays a particularly important role during tumor progression.33 Loss of E-cadherin induces EMT, and the nonpolarized mesenchymal cells are highly motile and invasive, therefore allowing tumor invasion and metastatic spread.34,35 Previous reports showed inconsistent results of the association between CDH1 hypermethylation and clinicopathologic parameters in BC, due to small sample size and different ethnicity.36,37 We pooled eleven studies and included 940 patients in the present study. Our data showed that CDH1 hypermethylation was not significantly associated with grade, stage, or lymph node metastasis in BC, with a high-power sample compared with previous studies.37,38
About 70% of BCs are ER-positive.39 The ER is responsible for estrogen-induced mitogenic signaling in epithelial cells in the breast and plays a crucial role in BC onset and progression. When estradiol (E2) binds to ER, the ER undergoes conformational changes and forms dimers, which attract coactivators and corepressors. This complex regulates hundreds of genes involved in a variety of processes, including proliferation, differentiation, survival, invasion, and metastasis.40,41 The ER status of breast provides prognostic information but more importantly, is a predictor of response to endocrine therapy.42 We investigated CDH1 methylation status in ER-positive versus -negative BC in the present study, showing significantly higher frequency of CDH1 hypermethylation in ER-negative BC than in ER-positive BC. Interestingly, a few studies reported that loss of β-catenin membranous expression and nuclear accumulation of β-catenin are associated with ER-negative status and reduced CDH1 expression,43,44 along with aggressive tumor phenotype and poor patient outcome in BC.44,45 CDH1 methylation is one of the mechanisms to regulate E-cadherin expression in the ER-negative subtype of BC. In addition, HER2 is a transmembrane tyrosine kinase receptor, located on chromosome 17q21. HER2, encoded by the ERBB2/HER2 oncogene, belongs to a family of epidermal growth factor receptors (EGFRs) structurally related to EGFR.46 This oncogene is overexpressed in 20%–30% of BC and has been recognized as a marker of poor prognosis, including increased metastasis potential and decreased overall survival.47 Inhibition of HER2 activity with antibodies has been applied to treat HER2-positive BC.48 In present study, the frequency of CDH1 hypermethylation was significantly higher in HER2-negative BC than in HER2-positive BC. Our results showed that CDH1 hypermethylation was increased in HER2-negative and ER-negative BCs, respectively, but was not significantly associated with PR status. In the future, CDH1 hypermethylation should be investigated in triple-negative BCs, which are HER2-negative, ER-negative, and PR-negative. As we know, about 85% of triple-negative BCs are basal-like subtypes, which are associated with poor clinical outcome.49 CDH1 hypermethylation could contribute to the development of a new treatment strategy for triple-negative BC, especially in the basal-like subtypes. Finally, our study only selected the published articles and did not include some relevant unpublished papers, which may have resulted in certain publication bias. Therefore, the results should be interpreted carefully.
Conclusion
In conclusion, CDH1 hypermethylation was shown to be significantly higher in IDC than in normal breast tissue, indicating that CDH1 is a potential novel drug target in the development of personalized therapy. CDH1 hypermethylation is strongly associated with ER-negative and HER2-negative BC, respectively, suggesting CDH1 methylation status could contribute to the development of novel therapeutic approaches of ER-negative and or HER2-negative BC, which is a subtype of BC with poor patient outcome.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Cancer Epigenetics for the 21st Century: What’s Next? Genes Cancer. 2011;2(6):604–606. doi: 10.1177/1947601911423096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atalay C. Epigenetics in breast cancer. Exp Oncol. 2013;35(4):246–249. [PubMed] [Google Scholar]
- 4.Ghavifekr Fakhr M, Farshdousti Hagh M, Shanehbandi D, Baradaran B. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;2013:317569. doi: 10.1155/2013/317569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5(3):301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gall TM, Frampton AE. Gene of the month: E-cadherin (CDH1) J Clin Pathol. 2013;66(11):928–932. doi: 10.1136/jclinpath-2013-201768. [DOI] [PubMed] [Google Scholar]
- 8.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4(2):118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 9.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 10.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1(6):a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy P, Liu L, Ren C, et al. Formation of E-cadherin-mediated cell-cell adhesion activates AKT and mitogen activated protein kinase via phosphatidylinositol 3 kinase and ligand-independent activation of epidermal growth factor receptor in ovarian cancer cells. Mol Endocrinol. 2005;19(10):2564–2578. doi: 10.1210/me.2004-0342. [DOI] [PubMed] [Google Scholar]
- 12.Grant CM, Kyprianou N. Epithelial mesenchymal transition (EMT) in prostate growth and tumor progression. Transl Androl Urol. 2013;2(3):202–211. doi: 10.3978/j.issn.2223-4683.2013.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai B, Yan HX, Liu SQ, Chen L, Wu MC, Wang HY. Reduced expression of E-cadherin/catenin complex in hepatocellular carcinomas. World J Gastroenterol. 2008;14(37):5665–5673. doi: 10.3748/wjg.14.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shargh SA, Sakizli M, Khalaj V, et al. Downregulation of E-cadherin expression in breast cancer by promoter hypermethylation and its relation with progression and prognosis of tumor. Med Oncol. 2014;31(11):250. doi: 10.1007/s12032-014-0250-y. [DOI] [PubMed] [Google Scholar]
- 15.Oka H, Shiozaki H, Kobayashi K, et al. Correlation between E-cadherin expression and metastasis in human breast cancer: preliminary report. Nihon Geka Gakkai Zasshi. 1992;93(1):105. Japanese. [PubMed] [Google Scholar]
- 16.Yoshida R, Kimura N, Harada Y, Ohuchi N. The loss of E-cadherin, alpha- and beta-catenin expression is associated with metastasis and poor prognosis in invasive breast cancer. Int J Oncol. 2001;18(3):513–520. [PubMed] [Google Scholar]
- 17.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153(2):333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 19.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299(3):551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 20.Blaschuk OW, Sullivan R, David S, Pouliot Y. Identification of a cadherin cell adhesion recognition sequence. Dev Biol. 1990;139(1):227–229. doi: 10.1016/0012-1606(90)90290-y. [DOI] [PubMed] [Google Scholar]
- 21.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debies MT, Welch DR. Genetic basis of human breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2001;6(4):441–451. doi: 10.1023/a:1014739131690. [DOI] [PubMed] [Google Scholar]
- 23.Sadot E, Simcha I, Shtutman M, Ben-Ze’ev A, Geiger B. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc Natl Acad Sci U S A. 1998;95(26):15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura G, Yin J, Wang S, et al. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst. 2000;92(7):569–573. doi: 10.1093/jnci/92.7.569. [DOI] [PubMed] [Google Scholar]
- 25.Zheng SY, Hou JY, Zhao J, Jiang D, Ge JF, Chen S. Clinical outcomes of downregulation of E-cadherin gene expression in non-small cell lung cancer. Asian Pac J Cancer Prev. 2012;13(4):1557–1561. doi: 10.7314/apjcp.2012.13.4.1557. [DOI] [PubMed] [Google Scholar]
- 26.Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275(4):2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- 27.Shinozaki M, Hoon DS, Giuliano AE, et al. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11(6):2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 28.Toyooka KO, Toyooka S, Maitra A, et al. Establishment and validation of real-time polymerase chain reaction method for CDH1 promoter methylation. Am J Pathol. 2002;161(2):629–634. doi: 10.1016/S0002-9440(10)64218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beumer JH, Parise RA, Newman EM, et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU) Cancer Chemother Pharmacol. 2008;62(2):363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
- 30.Gowher H, Jeltsch A. Mechanism of inhibition of DNA methyltransferases by cytidine analogs in cancer therapy. Cancer Biol Ther. 2004;3(11):1062–1068. doi: 10.4161/cbt.3.11.1308. [DOI] [PubMed] [Google Scholar]
- 31.Newman EM, Morgan RJ, Kummar S, et al. A phase I, pharmacokinetic, and pharmacodynamic evaluation of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine, administered with tetrahydrouridine. Cancer Chemother Pharmacol. 2015;75(3):537–546. doi: 10.1007/s00280-014-2674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes N, Carvalho J, Durães C, et al. 1Alpha, 25-dihydroxyvitamin D3 induces de novo E-cadherin expression in triple-negative breast cancer cells by CDH1-promoter demethylation. Anticancer Res. 2012;32(1):249–257. [PubMed] [Google Scholar]
- 33.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154(1):8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 34.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 36.Asiaf A, Ahmad ST, Aziz SA, et al. Loss of expression and aberrant methylation of the CDH1 (E-cadherin) gene in breast cancer patients from Kashmir. Asian Pac J Cancer Prev. 2014;15(15):6397–6403. doi: 10.7314/apjcp.2014.15.15.6397. [DOI] [PubMed] [Google Scholar]
- 37.Karray-Chouayekh S, Trifa F, Khabir A, et al. Aberrant methylation of RASSF1A is associated with poor survival in Tunisian breast cancer patients. J Cancer Res Clin Oncol. 2010;136(2):203–210. doi: 10.1007/s00432-009-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad CP, Mirza S, Sharma G, et al. Epigenetic alterations of CDH1 and APC genes: relationship with activation of Wnt/beta-catenin pathway in invasive ductal carcinoma of breast. Life Sci. 2008;83(9–10):318–325. doi: 10.1016/j.lfs.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Tokunaga E, Hisamatsu Y, Tanaka K, et al. Molecular mechanisms regulating the hormone sensitivity of breast cancer. Cancer Sci. 2014;105(11):1377–1383. doi: 10.1111/cas.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renoir JM, Marsaud V, Lazennec G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem Pharmacol. 2013;85(4):449–465. doi: 10.1016/j.bcp.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2(2):101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 43.Kashiwagi S, Yashiro M, Takashima T, et al. Significance of E-cadherin expression in triple-negative breast cancer. Br J Cancer. 2010;103(2):249–255. doi: 10.1038/sj.bjc.6605735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geyer FC, Lacroix-Triki M, Savage K, et al. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24(2):209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 45.Nakopoulou L, Mylona E, Papadaki I, et al. Study of phospho-beta-catenin subcellular distribution in invasive breast carcinomas in relation to their phenotype and the clinical outcome. Mod Pathol. 2006;19(4):556–563. doi: 10.1038/modpathol.3800562. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto T, Ikawa S, Akiyama T, et al. Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature. 1986;319(6050):230–234. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- 47.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 48.Hudis CA. Trastuzumab – mechanism of action and use in clinical practice. N Engl J Med. 2007;357(1):39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 49.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 50.Hoque MO, Prencipe M, Poeta ML, et al. Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2694–2700. doi: 10.1158/1055-9965.EPI-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou D, Yoon HS, Perez D, Weeks RJ, Guilford P, Humar B. Epigenetic silencing in non-neoplastic epithelia identifies E-cadherin (CDH1) as a target for chemoprevention of lobular neoplasia. J Pathol. 2009;218(2):265–272. doi: 10.1002/path.2541. [DOI] [PubMed] [Google Scholar]
- 52.Sunami E, Shinozaki M, Sim MS, et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10(3):R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006;237(2):272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu XC, Loo WT, Chow LW. E-cadherin promoter methylation can regulate its expression in invasive ductal breast cancer tissue in Chinese woman. Life Sci. 2002;71(12):1397–1404. doi: 10.1016/s0024-3205(02)01843-x. [DOI] [PubMed] [Google Scholar]
- 56.Cho YH, Yazici H, Wu HC, et al. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30(7):2489–2496. [PMC free article] [PubMed] [Google Scholar]