Abstract
Quantitative detection of molecules of interest from biological and environmental samples in a rapid manner, particularly with a relevant concentration range, is imperative to the timely assessment of human diseases and environmental issues. In this work, we employed the microwave-accelerated bioassay (MAB) technique, which is based on the combined use of circular bioassay platforms and microwave heating, for rapid and quantitative detection of Glial Fibrillary Acidic Protein (GFAP) and Shiga like toxin (STX 1). The proof-of-principle use of the MAB technique with the circular bioassay platforms for the rapid detection of GFAP in buffer based on colorimetric and fluorescence readouts was demonstrated with a 900 W kitchen microwave. We also employed the MAB technique with a new microwave system (called the iCrystal system) for the detection of GFAP from mice with brain injuries and STX 1 from a city water stream. Control bioassays included the commercially available gold standard bioassay kits run at room temperature. Our results show that the lower limit of detection (LLOD) of the colorimetric and fluorescence based bioassays for GFAP was decreased by ~1,000 times using the MAB technique and our circular bioassay platforms as compared to the commercially available bioassay kits. The overall bioassay time for GFAP and STX 1 was reduced from 4 hours using commercially available bioassay kits to 10 minutes using the MAB technique.
Keywords: Glial Fibrillary Acidic Protein, Shiga like toxin, ELISA, Microwave-Accelerated Bioassays, Silver Island Films, Circular Bioassay Platform, Metal-Enhanced Fluorescence
2. INTRODUCTION
Biosensors are useful tools for the assessment and monitoring of environmental and healthcare issues caused by biological and chemical attacks in warfare and the accidental release of harmful chemicals and organisms into the water resources and food chain (Aslan and Geddes 2008; Chen Gui 2011; Jones et al. 2002; Kiilerich-Pedersen et al. 2011). Various technologies have been combined to introduce a variety of biosensing devices, which can detect analytes of interest from biological fluids and environmental samples. Most biosensors have successfully utilized enzyme-based techniques to detect target analyte in a mixture of samples (Aslan and Geddes 2008; Aslan et al. 2005b; Aslan et al. 2006; Chen Gui 2011).
Enzyme-linked immunosorbent assay (ELISA) is the most common method used in health care and environmental industry for analyzing biological or environmental samples for specific proteins and autoantibodies, which can confirm the existence of a diseased state or microbial presence (Chen Gui 2011; Geginat et al. 2012; Liu et al. 2009; Mohammed and Aslan 2013a). Since its first publication in 1968, ELISA technique has been modified over the years to face new challenges of low assay sensitivity, assay linearity and cost efficiency (Aslan and Geddes 2008). ELISAs can be used for qualitative detection (such as pregnancy test, in food industry to detect allergens) or quantitative (such as blood glucose level test, HIV test, entero-toxin of E. coli in feces and water samples (Geginat et al. 2012). In this regard, colorimetric-based detection, which gives a visual conformation of the presence of an analyte of interest, is the most widely used detection technique for ELISA based systems (Dixit et al. 2010). Other detection methods, such as fluorescence and chemiluminescence, are routinely used due to the potential improvements in the detection limit of the bioassays compared to colorimetric detection (Aslan et al. 2005a; Aslan and Geddes 2006; Chowdhury et al. 2006a, b; Chowdhury et al. 2007a; Chowdhury et al. 2007b). Despite these latest developments, biosensors still require long processing times, especially for complex biological and environmental samples (Chen Gui 2011).
The Aslan Research group has demonstrated the proof-of-principle use of the MAB technique in conjunction with circular bioassay platforms and 900 W kitchen microwave for a biotin-avidin model bioassay and p53-MDM2 bioassay show significant improvement in LLOD (up to ~100 times) as well as reduction in total assay time (up to ~10 min) in comparison to traditional methods (4–8 hours) (Mohammed and Aslan 2014). The working principle of the MAB technique is based on Microwave-Induced Temperature Gradient (MITG) phenomenon and has been discussed previously (Grell et al. 2013; Mohammed and Aslan 2014). However, these studies were based on direct ELISA method which is qualitative in nature and required for diagnostic relevance. In this paper, we show results of an indirect sandwich-ELISA using two proteins:
GFAP: a protein that has been used as markers for many brain disorders and diseases such as Alexander’s disease (Jany et al. 2013; Messing 2011; Wada et al. 2013), Wernicke’s encephalopathy, Down’s syndrome, schizophrenia and depression (Ilhan-Mutlu et al. 2013; Mayer et al. 2013; Serrano-Pozo et al. 2013). It has been used as an astrocyte marker protein to ascertain any traumatic brain damage and injury after potential nervous system insult caused by stress scenarios (PTSD, shockwave impact) (Kamnaksh et al. 2011; Xia et al. 2013), environmental contaminants or agents (Lotosh et al. 2013; Mondello et al. 2011; Storoni et al. 2011; Wei et al. 2013; Yokobori et al. 2013), and exposure to harmful electromagnetic waves (Carballo-Quintas et al. 2011). GFAP levels are almost constant in the human body and slight increase in quantity may indicate impact of an injury or damage, therefore it is essential to develop a sensitive assay that can detect minute changes in GFAP quantity. Currently, traditional ELISA is used and the LLOD for GFAP is reported to be 1.56 ng/mL.
STX 1: one of the most common environmental toxins. Shigella species are responsible for an estimated 122,800 deaths globally due to bacillary dysentery (Lozano et al. 2012). The cytotoxins produced by STEC are divided into STX1 and STX2 subgroups (Nyholm et al. 2015). STEC causes wide variety of diseases in young children and infants including severe dehydration, dysentery, hemolytic-uremic syndrome (HUS) and hemorrhagic colitis (Franke et al. 1995; Lozano et al. 2012; Ruggenenti et al. 2001). STX1 protein consists of one molecule of subunit A and 5 molecules of subunit B. Subunit A is responsible for the toxicity and subunit B molecules are responsible for binding to the target cell (Lumor et al. 2012; Shimizu et al. 2007).
Current methods for quantification of both these proteins are laborious, time consuming and require elaborate systems that are available only in laboratories of a hospital or biological testing facilities. These tests conducted in hospital or diagnostic laboratory are based on ELISA (Ahadi et al. 2015; Jiang et al. 2014) and RT-PCR (Andrievskaia and Tangorra 2014), that have a turnaround time of 1–3 days, depending upon the availability of manpower and the medical emergency of the patient.
In this work, bioassays were conducted at room temperature using gold standard EIA kits in high-throughput screening plates and using the MAB technique (in 900 W multimode kitchen microwave and the iCrystal system (comprised of a 100 W solid state microwave source and a small mono-mode microwave cavity). In order to facilitate a direct comparison between various currently used methods of assay analysis and their compatibility with the MAB technique, detection of bioassays were carried out with two different methods: 1) colorimetric 2) fluorescence; and data was analyzed for assay linearity, LLOD of assay and enhancement of signal strength.
3. MATERIALS and METHODS
3.1 MATERIALS
Sodium phosphate, o-phenylene diamine HCl (OPD), citric acid, hydrogen peroxide (30%), sulfuric acid 98%, PMMA beads (120,000 Da molecular weight), methyl methacrylate (MMA), N,N-dimethyl-para-toluidine (DMPT), Tween 20 and phosphate buffer saline (PBS), Amersham Enhanced Chemiluminescence kit (ECL) was purchased from GE Healthcare, USA. Deionized water (DI) was obtained from Milli-Q water purifiers at 18.2 MΩ.cm at 25 °C and filtered through 0.22 μm filter.
Glial Fibrillary Acidic Protein (GFAP) standards, anti-GFAP polyclonal antibody (rabbit), goat anti-rabbit polyclonal secondary antibody tagged with HRP and FITC were purchased Abcam, USA, Dako, USA and KPL, USA, respectively. GFAP EIA kits were purchased from Millipore Laboratories, USA. Homogenized Balb/cByJ mouse brain tissue samples in PBS were obtained from Dr. Michael Koban’s laboratory at Morgan State University, Baltimore, MD, USA. STX-1 Shiga toxin standard and anti-STX-1 monoclonal antibody (mouse) was purchased from Toxin Technology, USA and goat anti-mouse polyclonal antibody tagged with HRP and FITC was purchased from KPL, USA. 900 W Frigidaire kitchen microwave (Model No-FCM09Z03KB, 900 W, duty cycle = seconds) was purchased from Walmart Inc., USA. The iCrystal system (Figure S1, Supplementary Material) was previously described.
3.2 METHODS
3.2.1 Preparation of circular PMMA platform
100 g of 120,000 Da (average molecular weight) PMMA beads were dissolved in 300 mL MMA maintained at 80 °C. After solution turns clear, 100 μl of DMPT was added and then the solution was poured in to 5 cm circular polypropylene cups and left at room temperature to polymerize for 7 days. The circular platforms were taken out of the cups and stored in fry environment until use.
3.2.2 Deposition of SNFs
PMMA platforms were subjected to plasma cleaning for 2 min and 10 nm thick SNFs are deposited selectively into the wells using a 21-well polypropylene mask using plasma sputtering technique as described previously (Mohammed and Aslan 2014). The SNFs deposited PMMA platforms (i.e., circular bioassay platforms) are rinsed with deionized water and air dried followed by the attaching of a 21-well silicone isolator.
3.2.3 Preparation of samples for bioassays
GFAP
Adult Balb C mice were sacrificed at post-natal day 90 and their brain cortex tissue was collected and stored in −80°C until further use. The cortex tissue was homogenized in PBS at pH 7 using an ultrasound tissue homogenizer for 30 seconds and subsequently centrifuged at 2°C and 6500 rpm for 10 min. The supernatant was carefully separated out into another Eppendorf tube, partitioned into aliquots and stored at −20°C until further use. The pellet was stored in −80°C freezer for further protein extraction when needed.
STX1
Water samples were collected from a stream flowing into Montebello waste water plant, Baltimore, MD, USA. Samples were spiked with known concentrations of STX1 proteins to investigate feasibility of using MAB technique for environmental samples without any preparation method involved. STX1 protein standard solution was prepared in PBS at pH 7.4 with 5 mM NaCl, 2 mM KCl to replicate serum conditions.
3.2.4 Bioassay preparation
GFAP and STX 1 assay protocol was based on steps suggested by EIA kit manufacturers, however, with additional use of secondary antibody to enhance signal intensity. A standard curve was constructed with proteins with varying concentration between 0.001 ng/mL to 100 ng/mL for 900 W microwave experiments and for the iCrystal system experiments. In order to prevent damage to the samples, microwave power (900 W) was applied in two short durations of 10 seconds each with 2 seconds interval in between. In case of the iCrystal system, continuous microwave power (100 W) was applied for 1 min.
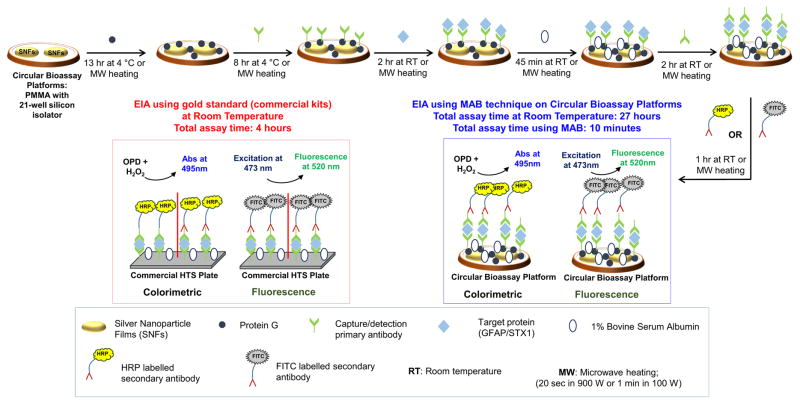
Scheme 1 depicts various steps for GFAP bioassays conducted on circular bioassay platforms and using commercial EIA kits. Detailed description of each step can be found in the Supplementary Material. The total assay time for commercially available kits is 4 hours as recommended by the vendor, inclusive of protein incubation for 90 min, blocking step (using manufacturer supplied super block BSA) for 30 min, primary antibody incubation for 75 min. and secondary antibody incubation for 45 min. We note that the experiments for all diluted and undiluted samples were repeated at least 10 times, and average values of each data point was plotted with the standard deviation for 10 repetitions of each sample.
Scheme 1.
Schematic depiction of GFAP and STX1 bioassays on 21-well circular bioassay platforms and using commercial EIA kits with colorimetric and fluorescence detection methods.
3.2.5 Detection of bioassay
For colorimetric detection, substrate solution was freshly prepared using 4 mg OPD in 10 mL pH 5 buffer (solution of sodium dibasic monophosphate and citric acid) and 4 μL hydrogen peroxide (30% v/v). A 20 μL portion of the substrate was incubated in each well for 2 minutes at room temperature after which the solutions were transferred from the wells to a HTS plate with 200 μL of 2.5 M sulfuric acid in each well as stopping solution. The HTS plate was read using Agilent UV-Vis plate reader between 400–600 nm with 200 μL of sulfuric acid solution as blank. The raw data was collected and normalized (to absorbance scale of 1) along with calculating standard deviation and plotted.
For fluorescence detection, FITC-tagged antibody was used instead of the enzyme complex. After immobilization of FITC-tagged antibody, 10–15 μL of deionized water was added to each well and excitation was carried out using 473 nm blue laser at 45° angle to the platform. The detection of emitted light from the enzyme complex in the well was captured using an Ocean optics flex fluorimeter at 90° to the platform.
3.2.6 Data analysis
Each protein concentration was run in triplicates using separate bioassay platforms and the data was collected at 95% confidence limit. Data was plotted and treated for 4-parameter fit using Sigma Plot statistical software.
4. RESULTS and DISCUSSION
4.1. Rapid and Sensitive Detection of GFAP
The proof-of-principle use of the MAB technique with circular bioassay platforms for the rapid and sensitive detection of a model protein (i.e., GFAP) in buffer was demonstrated with a 900 W kitchen microwave and a comparison of the results to a commercially available bioassay was made (Figure S2–S3, Supplementary Material). These observations clearly demonstrate a significant improvement in the LLOD and linearity of bioassays along with reduction of the background signal and false positives using the MAB technique powered by a 900 W multimode microwave. The total time for room temperature bioassays was found to be 27 hours (including assay preparation time), which was reduced to 10 minutes using the MAB technique, without any alterations to the bioassay protocol.
Despite of these advantages, the use of kitchen microwave (i.e., a multimode system) results in the overheating of bioassay platform and in rapid evaporation of sample solutions, which were to be expected due to the uneven heating pattern as shown in previous publications (Aslan 2010; Aslan and Geddes 2005). Moreover, microwave heating using a kitchen microwave resulted in the deformation of the circular bioassay platforms due to prolonged heating in the multiple steps of the bioassays, which subsequently resulted in sparking of the SNFs (data not shown for the sake of brevity). The samples hitherto tested were based in buffer, which does not necessarily replicate the conditions of a real biological samples that may contain numerous constituents along with the molecules of interest. Subsequently, it is important to evaluate the use of the iCrystal system and the circular bioassay platforms for the quantitative detection of target proteins from actual biological samples.
In order to compare the current “gold standard” methods and the MAB technique for rapid and sensitive detection of biologically relevant proteins, additional experiments were also carried out using commercially available EIA kits and the MAB technique. The manufacturer’s recommended protocol was employed in the MAB technique with modifications as specified in the Methods section. Brian cortex tissues of a minimum of three mice were homogenized and evaluated for total GFAP content. Due to the lower bioassay sensitivity of the MAB technique reported in Figure S2 and S3, the standard curve for the MAB was constructed using lower protein concentrations and the unknown samples were subsequently diluted further i.e., 1,000 and 10,000 times.
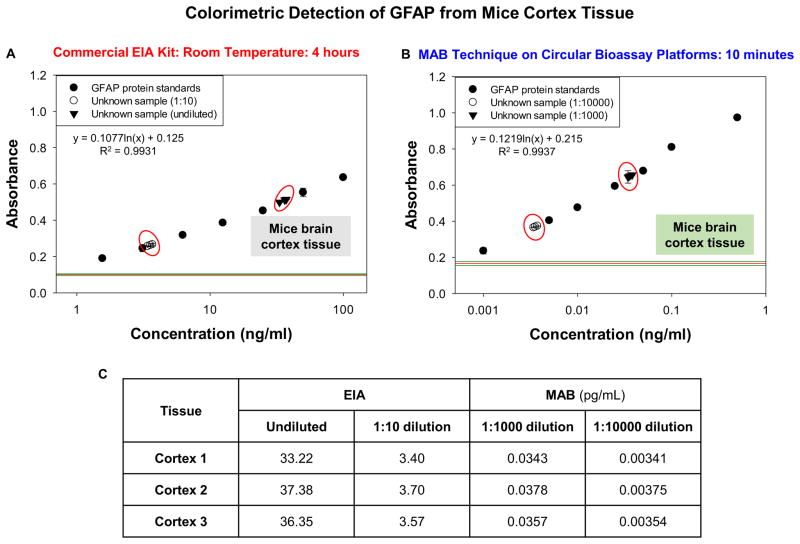
Figure 1 shows the absorbance values obtained from the colorimetric GFAP bioassay conducted using the commercially available EIA kit and using the MAB technique. The LLOD from the commercial EIA kit was observed to be 1.56 ng/mL and the assay linearity was in the range of 1.56 ng/mL to 100 ng/mL (Figure 1A) as suggested by the manufacturer. Figure 1B shows the absorbance data for GFAP bioassay conducted using the MAB technique and the iCrystal system. LLOD for the MAB bioassay was 0.001 ng/mL and linearity of bioassay was in the range of 0.001 ng/mL to 0.5 ng/mL The data points from the standard curve indicate the sensitivity of the MAB technique (~1,000 times) in comparison to the EIA kits and the background was also found to be significantly lower than the background observed with gold standard despite using 1/1000th of concentration for constructing standard curve. The overall time taken for the completion of bioassay was reduced to 10 minutes with the MAB technique as compared to gold standard. Figure 1C shows the concentrations of the unknown samples calculated from the plots (Figures 1A and 1B). With the use of EIA kits the concentrations of unknown samples without dilution was found to be 33.22 ng/mL for cortex tissue 1, 37.38 ng/mL for cortex tissue 2 and 36.35 ng/mL for cortex tissue 3. On 1:10 dilution of the unknown samples, the concentration was found to be 3.40 ng/mL for cortex tissue 1, 3.70 ng/mL for cortex tissue 2 and 3.57 ng/mL for cortex tissue 3. With the MAB technique, the concentrations of unknown samples diluted 1:1000 times were 0.0343 ng/mL for cortex tissue 1, 0.0378 ng/mL for cortex tissue 2 and 0.0357 ng/mL for cortex tissue 3. In experiments with further dilution to 1:10000, the concentrations were found to be 0.00341 ng/mL for cortex tissue 1, 0.00375 ng/mL for cortex tissue 2 and 0.00354 ng/mL for cortex tissue 3.
Figure 1.
Colorimetric data obtained from GFAP bioassays conducted at room temperature using commercial EIA kit (A) and with the MAB technique (1 min MW heating for each step) (B). The unknown concentration values for all dilutions used calculated from the plot (C). GFAP samples were collected from mice brain cortex tissue. The MAB technique employed the iCrystal system.
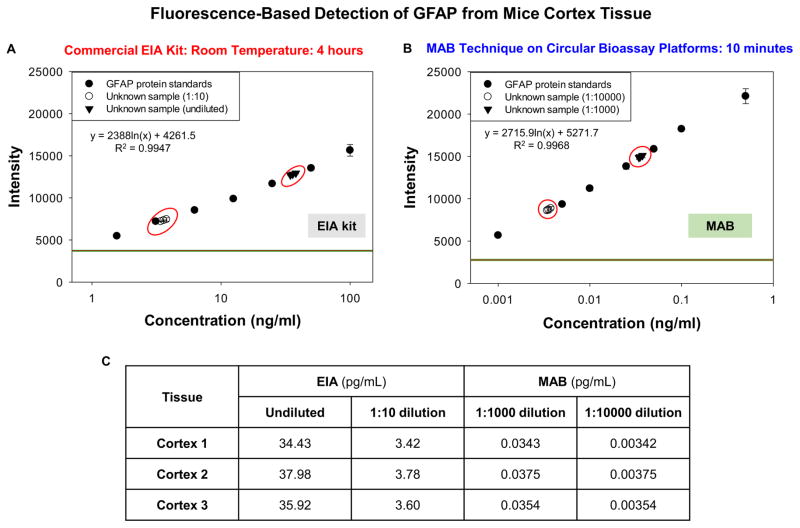
Figure 2 shows the fluorescence emission data of GFAP bioassay conducted using the commercially available EIA kit at room temperature and using the MAB technique. The LLOD for room temperature bioassay was 1.56 ng/mL and linearity of the bioassay was in the range of 1.56 ng/mL to 100 ng/mL with R2 value of 0.9947 (Figure 2A). For bioassay conducted using the MAB technique, LLOD value was found to be 0.001 ng/mL and the linearity of the bioassay was in the range of 0.001 ng/mL to 0.5 ng/mL with R2 value of 0.9968. Figure 2C shows the concentrations of the unknown samples calculated from the standard curve plots. From the room temperature plot, the concentrations of the undiluted samples was calculated to be 34.43 ng/mL for cortex tissue 1, 37.98 ng/mL for cortex tissue 2 and 35.92 ng/mL for cortex tissue 3. On dilution of this sample by 10 times (1:10), the concentrations were calculated to be 3.42 ng/mL for cortex tissue 1, 3.78 ng/mL for cortex tissue 2 and 3.60 ng/mL for cortex tissue 3. With the standard curve of the bioassay obtained by using the MAB technique, the concentrations of unknown samples after 1:1000 dilution were 0.0343 ng/mL for cortex tissue 1, 0.0375 ng/mL for cortex tissue 2 and 0.0354 ng/mL for cortex tissue 3.
Figure 2.
Fluorescence emission data obtained from GFAP bioassays conducted at room temperature using commercial EIA kit (A) and with the MAB technique (1 min MW heating for each step) (B) The unknown concentration values calculated for all dilutions used from the plot (C). GFAP samples were collected from mice brain cortex tissue. The MAB technique employed the iCrystal system
4.2. Rapid and Sensitive Detection of STX 1
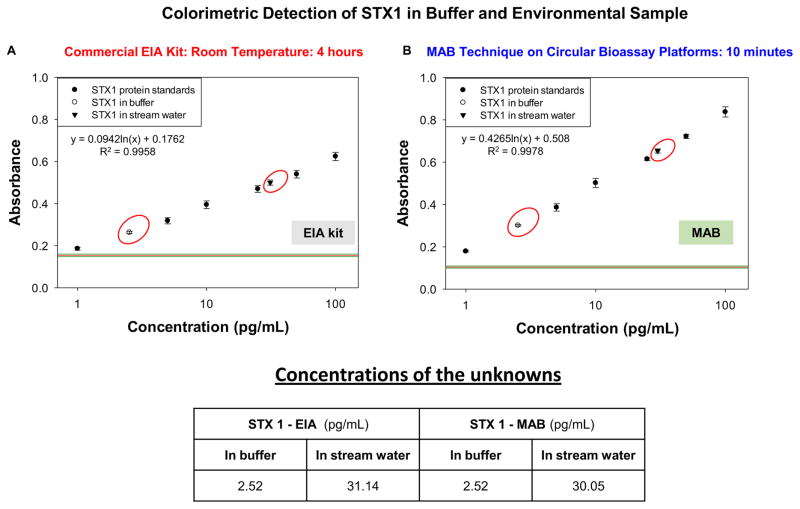
Figure 3 shows the absorbance values obtained from STX 1 bioassays conducted using EIA kits at room temperature and the MAB technique. LLOD of room temperature bioassay using EIA kits was 1 pg/mL and linearity of bioassay was in the range of 1 pg/mL to 100 pg/mL with R2 value of 0.9958 (Figure 3A). Similarly, LLOD of 1 pg/mL was observed for bioassay conducted using MAB technique and its linearity was in the range of 1 pg/mL to 100 pg/mL (Figure 3B). The concentration of STX 1 protein in buffer sample was calculated from the standard curves and found to be 2.52 pg/mL using both room temperature and the MAB bioassay. Concentration of STX1 protein in stream water was found to be 31.14 pg/mL by EIA kit and 30.05 by MAB bioassay. In both these methods, no significant differences were found in terms of assay sensitivity, background and assay linearity. However, it should be noted that EIA kits used for these experiments (both colorimetric and fluorescence) were modified for signal enhancement, which is not the case with commercially available kits.
Figure 3.
Colorimetric data obtained from STX1 bioassays conducted at room temperature using commercial EIA kit (A) and with the MAB technique (1 min MW heating for each step) (B). The concentration values for all solutions used calculated from the plot (C). STX1 was detected in buffer and an environmental sample. The MAB technique employed the iCrystal system.
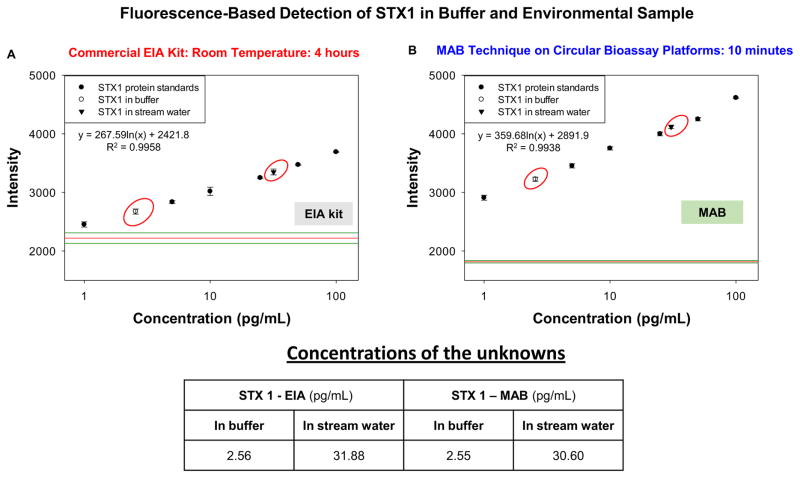
Figure 4 shows fluorescence emission data of STX 1 bioassays conducted at room temperature with EIA kits and using the MAB technique. As observed with colorimetric method (Figures 3A and 3B), the LLOD and linearity of both these methods were found to be same. However, there was significant signal enhancement observed with MAB technique due to the plasmon resonance effect (Aslan 2010; Aslan and Geddes 2005). The concentration of STX 1 protein in buffer was found to be 2.56 pg/mL with EIA kits and 2.55 pg/mL with the MAB technique. In the case of the water sample taken from a stream near Morgan State University, STX 1 concentration with EIA kits was found to be 31.88 pg/mL and 30.60 pg/mL with the MAB technique (Figure 4C). Time taken for completion of STX 1 bioassay using EIA kits was 28 hours including assay preparation whereas using the MAB technique the same bioassay was completed in 10 minutes. These results also highlight that the MAB technique afford for improved bioassay sensitivity for the detection of target proteins from unprocessed samples (brain tissue and lake water), unlike traditional methods that give false positives resulting in inaccurate analyte quantification.
Figure 4.
Fluorescence emission data obtained from STX1 bioassays conducted at room temperature using commercial EIA kit (A) and with the MAB technique (1 min MW heating for each step) (B) The concentration values calculated for all solutions used from the plot (C). STX1 was detected in buffer and an environmental sample. The MAB technique employed the iCrystal system.
It is important to compare the results obtained in this study with the commercially available gold-standard ELISA. Table 1 shows comparison between MAB technique using the iCrystal system and currently used traditional methods. In comparison to traditional quantitative methods, bioassays carried out using the MAB technique in the iCrystal system were found to be more sensitive (up to ~10-fold decrease in the LLOD as the lowest reported LLOD for biological samples and up to ~1,000-fold for GFAP) and rapid (~10 min). We have also shown the advantage of using the iCrystal system over the 900 W kitchen microwave, where mono-mode microwave heating in the iCrystal system resulted in significant decrease of LLOD (~1,000-fold for GFAP) and reduced the background signal obtained due to non-specific binding along with “zero” loss of samples or platforms due to overheating. The percentage variation between the concentrations of all diluted and undiluted samples after 10 repetitions were found to be less than 0.1%, indicating the consistency and reproducibility of the MAB technique using circular bioassay platform and the iCrystal system.
Table 1.
Comparison of various quantitative assessment methods currently used in diagnostic laboratories
| Method | LLOD | Assay linearity | Total assay time | On-site analysis |
|---|---|---|---|---|
| Traditional ELISA | 10 pg/ml* | pg/ml to ng/ml | 4–8 hours | No |
| PCR-ELISA | 1 pg/ml | 1–500 pg/ml | 3–8 hours | No |
| ELISPOT | Cell count (pg/ml) | NA | 6 hours | No |
| MAB Technique | 1 pg/ml | 1–100 pg/ml | 10 minutes | Yes |
5. CONCLUSIONS
The successful application of the MAB technique using our circular bioassay platforms for the rapid and sensitive detection of two target molecules based on colorimetric and fluorescence detection schemes in a kitchen microwave and the iCrystal system demonstrates the wide applicability of our MAB technique. The bioassays conducted for proteins were completed in less than 10 minutes using the MAB technique and circular bioassay platforms, which is significantly faster (up to ~30 times) than the identical bioassays carried out at room temperature that required several hours to complete. Rapid processing times of critical biological and environmental samples provided with the MAB technique can help to initiate remedial measures for patients with life threatening internal injuries and safety and recovery procedures for the environment of interest.
Supplementary Material
Highlights.
Circular bioassay platform and microwave heating was used in a biosensing scheme.
Glial Fibrillary Acidic Protein was detected in buffer and from mice brain tissue samples
Shiga like toxin was detected in buffer and from environmental samples
The overall bioassay time was reduced from 27 hours to 10 minutes
Lower limit of detection of bioassays was reduced up to 1,000-fold
Acknowledgments
This work was supported by National Institute of Biomedical Imaging and Bioengineering, award number 5-K25EB007565-05. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or NIH. We thank Dr. Michael Koban and Lalith S. Naidu for their help with mice samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahadi R, et al. Diagnostic Value of Serum Levels of GFAP, pNF-H, and NSE Compared With Clinical Findings in Severity Assessment of Human Traumatic Spinal Cord Injury. Spine (Phila Pa 1976) 2015;40(14):E823–830. doi: 10.1097/BRS.0000000000000654. [DOI] [PubMed] [Google Scholar]
- Andrievskaia O, Tangorra E. Detection of bovine central nervous system tissues in rendered animal by-products by one-step real-time reverse transcription PCR assay. J Food Prot. 2014;77(12):2088–2097. doi: 10.4315/0362-028X.JFP-14-223. [DOI] [PubMed] [Google Scholar]
- Aslan K. Rapid whole blood bioassays using microwave-accelerated metal-enhanced fluorescence. NanoBio Med Eng. 2010;2(1):1. doi: 10.5101/nbe.v2i1.p1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan K, et al. Metal-enhanced fluorescence from plastic substrates. J Fluoresc. 2005a;15(2):99–104. doi: 10.1007/s10895-005-2515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan K, Geddes CD. Microwave-accelerated metal-enhanced fluorescence: platform technology for ultrafast and ultrabright assays. Anal Chem. 2005;77(24):8057–8067. doi: 10.1021/ac0516077. [DOI] [PubMed] [Google Scholar]
- Aslan K, Geddes CD. Microwave-accelerated metal-enhanced fluorescence (MAMEF): application to ultra fast and sensitive clinical assays. J Fluoresc. 2006;16(1):3–8. doi: 10.1007/s10895-005-0026-z. [DOI] [PubMed] [Google Scholar]
- Aslan K, Geddes CD. New tools for rapid clinical and bioagent diagnostics: microwaves and plasmonic nanostructures. Analyst. 2008;133(11):1469–1480. doi: 10.1039/b808292h. [DOI] [PubMed] [Google Scholar]
- Aslan K, et al. Metal-enhanced fluorescence: an emerging tool in biotechnology. Curr Opin Biotechnol. 2005b;16(1):55–62. doi: 10.1016/j.copbio.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan K, et al. Microwave-Accelerated Metal-Enhanced Fluorescence (MAMEF) with silver colloids in 96-well plates: Application to ultra fast and sensitive immunoassays, High Throughput Screening and drug discovery. J Immunol Methods. 2006;312(1–2):137–147. doi: 10.1016/j.jim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Carballo-Quintas M, et al. A study of neurotoxic biomarkers, c-fos and GFAP after acute exposure to GSM radiation at 900 MHz in the picrotoxin model of rat brains. Neurotoxicology. 2011;32(4):478–494. doi: 10.1016/j.neuro.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Chen Gui XDaDC. Nanotechnological Advances in Biosensors. Nano Biomedicine and Engineering. 2011;3(4) [Google Scholar]
- Chowdhury MH, et al. Metal-enhanced chemiluminescence. J Fluoresc. 2006a;16(3):295–299. doi: 10.1007/s10895-006-0082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury MH, et al. Metal-enhanced chemiluminescence: Radiating plasmons generated from chemically induced electronic excited states. Appl Phys Lett. 2006b;88(17):173104. doi: 10.1063/1.2195776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury MH, et al. Computational study of fluorescence scattering by silver nanoparticles. J Opt Soc Am B. 2007a;24(9):2259–2267. doi: 10.1364/JOSAB.24.002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury MH, et al. First Observation of Surface Plasmon-Coupled Chemiluminescence (SPCC) Chem Phys Lett. 2007b;435(1–3):114–118. doi: 10.1016/j.cplett.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit CK, et al. Development of a high sensitivity rapid sandwich ELISA procedure and its comparison with the conventional approach. Anal Chem. 2010;82(16):7049–7052. doi: 10.1021/ac101339q. [DOI] [PubMed] [Google Scholar]
- Franke S, et al. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J Clin Microbiol. 1995;33(12):3174–3178. doi: 10.1128/jcm.33.12.3174-3178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat G, et al. Evaluation of third-generation ELISA and a rapid immunochromatographic assay for the detection of norovirus infection in fecal samples from inpatients of a German tertiary care hospital. Eur J Clin Microbiol Infect Dis. 2012;31(5):733–737. doi: 10.1007/s10096-011-1366-z. [DOI] [PubMed] [Google Scholar]
- Grell TA, et al. Microwave-accelerated surface modification of plasmonic gold thin films with self-assembled monolayers of alkanethiols. Langmuir. 2013;29(43):13209–13216. doi: 10.1021/la402455x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhan-Mutlu A, et al. High plasma-GFAP levels in metastatic myxopapillary ependymoma. J Neurooncol. 2013;113(3):359–363. doi: 10.1007/s11060-013-1134-2. [DOI] [PubMed] [Google Scholar]
- Jany PL, et al. GFAP expression as an indicator of disease severity in mouse models of Alexander disease. ASN Neuro. 2013;5(1):e00109. doi: 10.1042/AN20130003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SH, et al. Effect of hypothermia therapy on serum GFAP and UCH-L1 levels in neonates with hypoxic-ischemic encephalopathy. Zhongguo Dang Dai Er Ke Za Zhi. 2014;16(12):1193–1196. [PubMed] [Google Scholar]
- Jones DA, et al. Microwave heating applications in environmental engineering—a review. Resources, Conservation and Recycling. 2002;34:75–90. [Google Scholar]
- Kamnaksh A, et al. Factors affecting blast traumatic brain injury. J Neurotrauma. 2011;28(10):2145–2153. doi: 10.1089/neu.2011.1983. [DOI] [PubMed] [Google Scholar]
- Kiilerich-Pedersen K, et al. Polymer Based Biosensors for Pathogen Diagnostics Environmental Biosensors. 2011:193–212. [Google Scholar]
- Liu Y, et al. Microchip-based ELISA strategy for the detection of low-level disease biomarker in serum. Anal Chim Acta. 2009;650(1):77–82. doi: 10.1016/j.aca.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Lotosh NG, et al. Autoantibodies to neuron-specific proteins S100, GFAP, MBP and NGF in the serum of rats with streptozotocin-induced diabetes. Bull Exp Biol Med. 2013;155(1):48–51. doi: 10.1007/s10517-013-2077-5. [DOI] [PubMed] [Google Scholar]
- Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumor SE, et al. Comparison of the presence of Shiga toxin 1 in food matrices as determined by an enzyme-linked immunosorbent assay and a biological activity assay. J Food Prot. 2012;75(6):1036–1042. doi: 10.4315/0362-028X.JFP-11-372. [DOI] [PubMed] [Google Scholar]
- Mayer CA, et al. Blood levels of glial fibrillary acidic protein (GFAP) in patients with neurological diseases. PLoS One. 2013;8(4):e62101. doi: 10.1371/journal.pone.0062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing A. Physiology and pathophysiology of neuroglia. Henry Stewart Talks; London: 2011. Astrocytes and Alexander disease the first, but not last, primary astrocyte disease; p. 1. streaming video file. [Google Scholar]
- Mohammed M, Aslan K. Design and Proof-of-Concept Use of a Circular PMMA Platform with 16-Well Sample Capacity for Microwave-Accelerated Bioassays. Nano Biomed Eng. 2013 Jan;5(1):10–19. doi: 10.5101/nbe.v5i1.p20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed M, Aslan K. Rapid and Sensitive Detection of p53 Based on DNA-Protein Binding Interactions Using Silver Nanoparticle Films and Microwave Heating. Nano Biomed Eng. 2014;6(3):76–84. doi: 10.5101/nbe.v6i3.p76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello S, et al. Neuronal and glial markers are differently associated with computed tomography findings and outcome in patients with severe traumatic brain injury: a case control study. Crit Care. 2011;15(3):R156. doi: 10.1186/cc10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm O, et al. Hybrids of Shigatoxigenic and Enterotoxigenic Escherichia coli (STEC/ETEC) Among Human and Animal Isolates in Finland. Zoonoses Public Health. 2015 doi: 10.1111/zph.12177. [DOI] [PubMed] [Google Scholar]
- Ruggenenti P, et al. Thrombotic microangiopathy, hemolytic uremic syndrome, and thrombotic thrombocytopenic purpura. Kidney Int. 2001;60(3):831–846. doi: 10.1046/j.1523-1755.2001.060003831.x. [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A, et al. A phenotypic change but not proliferation underlies glial responses in Alzheimer disease. Am J Pathol. 2013;182(6):2332–2344. doi: 10.1016/j.ajpath.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, et al. Receptor affinity, stability and binding mode of Shiga toxins are determinants of toxicity. Microb Pathog. 2007;43(2–3):88–95. doi: 10.1016/j.micpath.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Storoni M, et al. The use of serum glial fibrillary acidic protein measurements in the diagnosis of neuromyelitis optica spectrum optic neuritis. PLoS One. 2011;6(8):e23489. doi: 10.1371/journal.pone.0023489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, et al. Familial adult-onset Alexander disease with a novel mutation (D78N) in the glial fibrillary acidic protein gene with unusual bilateral basal ganglia involvement. J Neurol Sci. 2013;331(1–2):161–164. doi: 10.1016/j.jns.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Wei P, et al. Serum GFAP autoantibody as an ELISA-detectable glioma marker. Tumour Biol. 2013;34(4):2283–2292. doi: 10.1007/s13277-013-0770-7. [DOI] [PubMed] [Google Scholar]
- Xia L, et al. FGF2 blocks PTSD symptoms via an astrocyte-based mechanism. Behav Brain Res. 2013;256:472–480. doi: 10.1016/j.bbr.2013.08.048. [DOI] [PubMed] [Google Scholar]
- Yokobori S, et al. Biomarkers for the clinical differential diagnosis in traumatic brain injury--a systematic review. CNS Neurosci Ther. 2013;19(8):556–565. doi: 10.1111/cns.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.