Abstract
DNA barcoding coupled high resolution melting (Bar-HRM) is an emerging method for species discrimination based on DNA dissociation kinetics. The aim of this work was to evaluate the suitability of different primer sets, derived from selected DNA regions, for Bar-HRM analysis of species in Croton (Euphorbiaceae), one of the largest genera of plants with over 1,200 species. Seven primer pairs were evaluated (matK, rbcL1, rbcL2, rbcL3, rpoC, trnL and ITS1) from four plastid regions, matK, rbcL, rpoC, and trnL, and the nuclear ribosomal marker ITS1. The primer pair derived from the ITS1 region was the single most effective region for the identification of the tested species, whereas the rbcL1 primer pair gave the lowest resolution. It was observed that the ITS1 barcode was the most useful DNA barcoding region overall for species discrimination out of all of the regions and primers assessed. Our Bar-HRM results here also provide further support for the hypothesis that both sequence and base composition affect DNA duplex stability.
Introduction
Classification of Croton and uses in ethnomedicine
Croton (Euphorbiaceae) is one of the largest genera of flowering plants, with between 1,200 and 1,300 species. It is widespread in tropical areas, with habits ranging from large woody trees through climbing lianas to simple and prostrate weeds [1,2]. In Southeast Asia and Thailand there are at least 80 species and 30 species of Croton, respectively [3]. The complex subgeneric taxonomy of Croton relies on a provisional revision of the sections of the genus from the early 1990s [1]. Several studies, based on both classical taxonomy and phylogenetic analyses, have demonstrated the complex taxonomy of the Croton genus (e.g. [1,4]). An example of this complexity is highlighted by C. acutifolius Esser in Thailand. This species is superficially similar to C. robustus Kurz, C. laccifer L. and C. caudatus Geiseler which can however be distinguished by their fruit size, and C. argyratus Blume which is characterized by silvery pubescent mature leaves [3]. Additionally, C. griffithii Hook.f. has been confused with C. oblongus Burm.f. by several authors, as the two species show similarity in their indumentum and floral characters [3].
Many species of Croton are used in traditional herbal medicine around the world. In Asia, and Thailand in particular, there has been a resurgence of interest in traditional medicine, aided by government programs to promote research into medicinal plants as a potential source of new remedies. Among the most popular remedies are those based on Croton species. Many Croton species from this region have been used in traditional medicine and have had their biological activities assessed. These include Croton caudatus Geiseler, Croton crassifolius Geiseler, C. kongensis Gagnep., C. oblongifolius Roxb., C. sublyratus Kurz., C. tiglium L., and C. tonkinensis Gagnep. [5–10]. At the same time, it is widely known from scientific and folk literature that many species of Croton are toxic and cause irritation [11], and therefor the correct identification of the species is important to avoid negative health effects.
In Thailand, C. stellatopilosus H.Ohba, known locally as ‘Plao Noi’, is a popular natural remedy used for stomach disorders. Commercial products containing ‘Plao Noi’ are sold as tea bags, capsules, and in powder form in herbal markets and are claimed to have anti-ulcer, anti-cancer, and anti-inflammatory effects. However, confusion has arisen because the same vernacular name is used for a number of species. At least six Croton species, C. columnaris Airy Shaw, C. delpyi Gagnep., C. longissimus Airy Shaw, C. kongensis Gagnep., C. stellatopilosus and C. thorelii Gagnep., share the same Thai common name ‘Plao Noi’ [3]. The different species vary in their secondary metabolite spectra, which results in variation in their potential efficacy and safety.
Identification of plant products in trade through DNA barcoding
Medicinal plant products are commonly sold in processed or modified forms such as powders, dried material, tablets, capsules and tea bags, making it almost impossible to accurately identify the constituent species using morphology [12,13]. Incorrect identification of the constituent plants may lead to the inclusion of undesirable, unrelated species, with potential health risks to end users. Substitution of the product’s ingredients either intentionally or inadvertently can have negative effects on both consumers and producers. Herbal products are often perceived to be safe due to their natural origin. However, counterfeited, substituted and adulterated products can put consumers in danger [14–16]. There is an urgent need to find an approach that could help with the quality control of these herbal products in order to ensure consumer safety and which can also produce reliable species identifications even when morphology-based identification is impossible.
Molecular identification through DNA barcoding is a powerful method for the identification of plant species, including medicinal plants and products. Many studies have shown the potential for DNA barcoding to effectively distinguish among medicinal plants, as well as to identify constituent species in processed herbal medicines [12,17–20]. Several reviews have highlighted the increasing and diverse applications of medicinal plant barcoding [21–23]. However, DNA barcoding in plants does have limitations. These include the inability to amplify marker regions due to degraded DNA in some processed samples [24], limited binding site universality [12,25,26], low rates of marker discrimination [12,27], overlapping intraspecific and infraspecific genetic variation in some groups of plants [28], and low applicability of chloroplast markers for the identification of species of hybrid origin [28]. Another limitation of DNA barcoding is the associated costs, as it requires a molecular laboratory, costly equipment, chemicals and disposables, and DNA sequencing facilities. The lack of access to DNA sequencing facilities and the high costs of sequencing often impedes the wider implementation of DNA barcoding in developing countries [29].
Bar-HRM for species discrimination
Developing and validating sequencing-free methods that are reliable, yet faster and more economical than DNA barcoding is challenging, but will be beneficial for the advancement of herbal product identification routines in developing countries. High resolution melting (HRM) is an emerging method for monitoring DNA dissociation (“melting”) kinetics, and is a powerful technique for the detection of point mutations, indels, and methylated DNA [30,31]. In addition to standard PCR equipment and reagents, HRM requires a generic DNA intercalation fluorescent dye. This dye is added to previously amplified PCR products and as the double-stranded DNA samples dissociate with increasing temperature the dye is progressively released and fluorescence diminishes. These denaturation thermodynamics are based on the binding affinities of individual nucleotide pairs, which vary due to indels, mutations and methylations. These differences are inferred by fluorescent measurements collected at standard temperature increments, which are plotted as a melting curve. The curve’s shape and peak are characteristic for each sample, allowing for comparison and discrimination among samples. By using HRM, single base changes between samples can be readily detected and identified [32,33].
The first study reporting the use of Bar-HRM to evaluate herbal medicine substitution came from a recent investigation of species substitution among three medicinal species of Acanthaceae [29]. Bar-HRM has also been used in a number of comparable applications [34], such as for authentication of an EU Protected Designation of Origin product made from Lathyrus clymenum [35], for the identification of olive oil and adulterants [36], for subspecies cultivar identification in eggplants [37], for the identification of closely related species of Sideritis and Helleborus [38,39], for species distinction in Mediterranean pines [40], for the detection of allergenic hazelnut contamination [41], and for the identification of processed bean crops [42–44].
Research questions and hypothesis
The use of Bar-HRM has been reported for species identification and detection in food and herbal and agricultural products. However, all previous studies have looked at species discrimination in complexes of limited species diversity [29,35–37,40–42]. In this study we test the hypothesis that reduced melting discrimination resolution among twelve closely related species in the same genus could be overcome by marker optimization and combination of data from multiple Bar-HRM markers. Here, we use a dataset made up of twelve species in the genus Croton to answer the following research questions relevant to the model group: 1) Can Bar-HRM primers sets that enable universal amplification of selected plastid and nuclear markers be designed?; 2) Can these twelve related Croton species be discriminated by Bar-HRM?; 3) Which single Bar-HRM marker has the highest rate of successful discrimination?; 4) Which minimum number of combined Bar-HRM markers that together give the highest rate of successful identification?
Methods
DNA mining of barcode regions
Sequences of four selected plastid DNA regions (matK, rbcL, rpoC and trnL) and one nuclear regions (ITS1) of Croton species from the family Euphobiaceae were extracted from GenBank (at the end of November 2014) using the key phrases “the name of locus” and “the name of genus” in the annotations. Generally, sequences obtained from public databases, including GenBank, are of low quality with no known associated herbarium vouchers. For this reason, all of the sequences were subjected to critical evaluation and any low-quality sequences were removed. After processing, multiple sequence alignments were made from the selected sequences using MEGA6 [45] and variable characters were calculated in order to design primers to be used for high resolution melting (HRM) analyses. The species names and accession numbers of all analyzed sequences are listed in S1 Table.
Plant material and DNA isolation
Dried plant tissues for DNA extraction were kindly provided by Queen Sirikit Botanic Garden (QSBG) (Table 1). The plant material was ground with liquid nitrogen, and 100 mg of fine powder was then used for DNA extraction with the Nucleospin Plant II kit (Macherey-Nagel, Germany) following the manufacturer’s instructions. DNA concentrations of all samples were adjusted to a final concentration of 20 ng/μL. The DNA was stored at −20°C for further use.
Table 1. Plants included in this study.
| Species | Abbrev. | Sample No. | Thai Vernacular name | Voucher No. |
|---|---|---|---|---|
| Croton cascarilloides Raeusch. 1 | CAS | 1 | Plao ngoen; ‘เปล้าเงิน’ | QBG38236 |
| Croton caudatus Geiseler 2 | CAU1CAU2 | 23 | Plao; ‘เปล้า’ | QBG8395QBG3389 |
| Croton crassifolius Geiseler | CR1CR2 | 45 | Pang khi; ‘ปังคี’ | QBG20547QBG10911 |
| Croton delpyi Gagnep. 3 | CD | 6 | Plao noi; ‘เปล้าน้อย’ | QBG13889 |
| Croton griffithii Hook.f. 2 | CG | 7 | Plao; ‘เปล้า’ | QBG32192 |
| Croton hutchinsonianus Hosseus 4 | CH1CH2 | 89 | Plao lueat; ‘เปล้าเลือด’ | QBG21732QBG2327 |
| Croton kongensis Gagnep. 1 , 3 | CK1CK2CK3 | 101112 | Plao noi; ‘เปล้าน้อย’, Plao ngoen; ‘เปล้าเงิน’ | QBG6677QBG3807QBG33093 |
| Croton poilanei Gagnep. 4 , 5 | CP | 13 | Plao luang; ‘เปล้าหลวง’, Plao yai; ‘เปล้าใหญ่’, Plao lueat; ‘เปล้าเลือด’ | QBG12427 |
| Croton robustus Kurz 5 | CRB1CRB2 | 1415 | Plao yai; ‘เปล้าใหญ่’ | QBG8309QBG5954 |
| Croton persimilis Müll.Arg. 5 , * | CRX1CRX2 | 1617 | Plao luang; ‘เปล้าหลวง’, Plao yai; ‘เปล้าใหญ่’ | QBG984QBG5997 |
| Croton tiglium L. | CB | 19 | Hat sakhuen; ‘หัสคืน’ | QBG10124 |
| Croton thorelii Gagnep. 3 | CT | 20 | Plao noi; ‘เปล้าน้อย’ | QBG29049 |
1 share the vascular name ‘Plao Ngoen’
2 share the vascular name ‘Plao’
3 share the vascular name ‘Plao Noi’
4 share the vascular name ‘Plao Lueat’
5 share the vascular names ‘Plao Luang’ and ‘Plao Yai’
* Synonym Croton roxburghii N.P.Balakr.
Real-time PCR amplification and high resolution melting (HRM) analysis
PCR amplification, DNA melting and fluorescence measurements were performed in a total reaction volume of 20 μL on an Eco Real-Time PCR system (Illumina, San Diego, USA) to determine the characteristic melting temperature (Tm) for each sample that could be used to distinguish among the plants (species?) in genus Croton. Each reaction mixture contained 20 ng of genomic DNA, 10 μL of MeltDoctor HRM Master Mix (Applied Biosystems, California, USA) and 0.2 μL of 10 mM forward and reverse primers. The nucleotide compositions of the five candidate pairs of barcoding primers are shown in Table 2. The real-time PCR reaction conditions were; an initial denaturing step at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 57°C for 30 s and 72°C for 20 s. Subsequently, the PCR amplicons were denatured for HRM at 95°C for 15 s and then annealed at 50°C for 15 s to form random DNA duplexes. Melting curves were generated after the last extension step. The temperature was increased from 60 to 95°C at 0.1°C/s. The melting curves were analyzed with the EcoStudy Software v5.0.
Table 2. Oligonucleotide sequences of primers used for HRM analyses.
| Primer HRM | 5' → 3' | Ta (°C) | Expected size (bp) |
|---|---|---|---|
| HRM_rpoCF | CCSATTGTATGGGAAATACTT | 57 | 170 |
| HRM_rpoCR | CTTACAAACTAATGGATGTAA | ||
| HRM_matKF | CTTCTTATTTACGATTAACATCTTCT | 57 | 170 |
| HRM_matKR | TTTCTTTGATATCGAACATAATG | ||
| HRM_trnLF | TGGGCAATCCTGAGCCAAATC | 57 | 120 |
| HRM_trnLR | AACAGCTTCCATTGAGTCTCTGCACCT | ||
| HRM_rbcL1F | GCAGCATTCCGAGTAACTCCTCA | 57 | 100 |
| HRM_rbcL1R | TCCACACAGTTGTCCATGTACC | ||
| HRM_rbcL2F | GGTACATGGACAACTGTGTGGA | 57 | 150 |
| HRM_rbcL2R | ACAGAACCTTCTTCAAAAAGGTCTA | ||
| HRM_rbcL3F | TAGACCTTTTTGAAGAAGGTTCTGT | 57 | 145 |
| HRM_rbcL3R | TGAGGCGGRCCTTGGAAAGTT | ||
| HRM_ITS1FHRM_ITS1R | GGTGAACCTGCGGAAGGATCATTGCCGAGATATCCATTGCCGAGAGTC | 57 | 300 |
Results and Discussion
Data mining
Sequences from all five selected barcode regions were extracted from GenBank for species in the genus Croton, and the variable characters and average %GC content were calculated for all samples using MEGA6. Sequence data were present for most markers, except for rpoC. In total, 62 sequences of matK, 148 of rbcL, 240 of trnL and 770 of ITS1 were retrieved, of which 19, 39, 136 and 293 sequences of each respective region were deemed useful for further analysis (Table 3). The rpoC data were excluded from this study due to the absence of a sufficient number of rpoC reference sequences for the target species (only 1). A single set of primers for each of matK, trnL, and ITS1, along with three sets of primers for rbcL (rbcL1-rbcL3), were evaluated. These various primer combinations yielded amplicons ranging from 100 to 320 bp (Table 3). Reed and Wittwer [31] found that amplicons suitable for HRM analysis should be 300 bp or less for optimal results. The trnL and ITS1 primer sets yielded amplicons of variable length, whereas the rbcL and matK primers set yielded amplicons of consistent sizes (Table 3).
Table 3. Characteristics of sequences from selected plastid regions (matK, rbcL, rpoC and trnL) and nuclear genome region (ITS1) retrieved from GenBank (NCBI) from species of Croton.
| Markers | matK | rbcL1 | rbcL2 | rbcL3 | trnL | ITS1 | rpoC |
|---|---|---|---|---|---|---|---|
| Search result from GenBank | 62 | 148 | 148 | 148 | 240 | 770 | 0 |
| Sequences included in dataset | 19 | 39 | 39 | 39 | 136 | 293 | - |
| Expected product size | 125 | 100 | 149 | 145 | 101–115 | 290–320 | - |
| Characters (bp) | 125 | 100 | 149 | 145 | 124 | 353 | - |
| Variable characters (%) | 19 (15.2) | 14 (14) | 13 (8.72) | 12 (8.27) | 29 (23.38) | 239 (67.7) | - |
| Conserved F primer sites/total (%) | 8/26 (30.77) | 20/22 (90.9) | 22/22 (100) | 24/25 (96) | 20/21 (95.24) | 23/24 (95.83) | - |
| Conserved R primer sites/total (%) | 25/27 (92.59) | 24/25 (96) | 24/25 (96) | 20/21 (95.24) | 26/27 (96.29) | 20/24 (83.33) | - |
| Average %GC content (SD) | 32.26 (1.02) | 53.54 (0.59) | 45.1 (0.67) | 43.56 (0.72) | 35.74 (1.1) | 52.17 (2.82) | - |
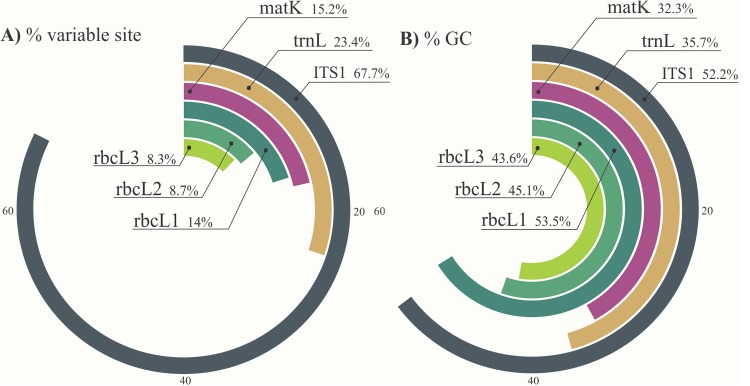
Both the sequence length and the nucleotide variation within sequences influence the dissociation energy of the base pairs and result in different Tm values. Two hundred and thirty nine variable sites (67.7%) were observed within the analyzed fragment of the ITS1 region (353 characters) (Table 3). The ITS1 amplicon sequences were observed to have the highest nucleotide variation, and the relative nucleotide variation within amplicons was found to be: ITS1> trnL > matK > rbcL1 > rbcL2 > rbcL3 (Fig 1A).
Fig 1. Comparison of variable characters (1A) and average %GC content (1B) of Croton matK, rbcL1, rbcL2, trnL and ITS1 sequences retrieved from GenBank.
The regions correspond to the same fragments that were amplified using the primers described in the present study.
The forward matK primer matched the consensus sequence of the target species at the binding sites in only 8 out of 26 sites (30.77%) (Table 3). High universality at the initial bases of the primer site is crucial for primer annealing and subsequent elongation initiation by the DNA polymerase. The matK locus is one of the most variable plastid coding regions and has high interspecific divergence and good discriminatory power. However, it can be difficult to amplify with the standard barcoding primers due to high substitution rates at the primer sites [46,47]. The average %GC content of amplicons was calculated in order to predict variation in melting curves for the different markers. The matK region had the lowest average %GC content, with 32.26%, followed by trnL, rbcL3, rbcL2, ITS1, and rbcL1 with 35.74, 43.56, 45.1, 52.17 and 53.54% respectively (Fig 1B).
Evaluation of primers and amplicons for discrimination among four Croton species
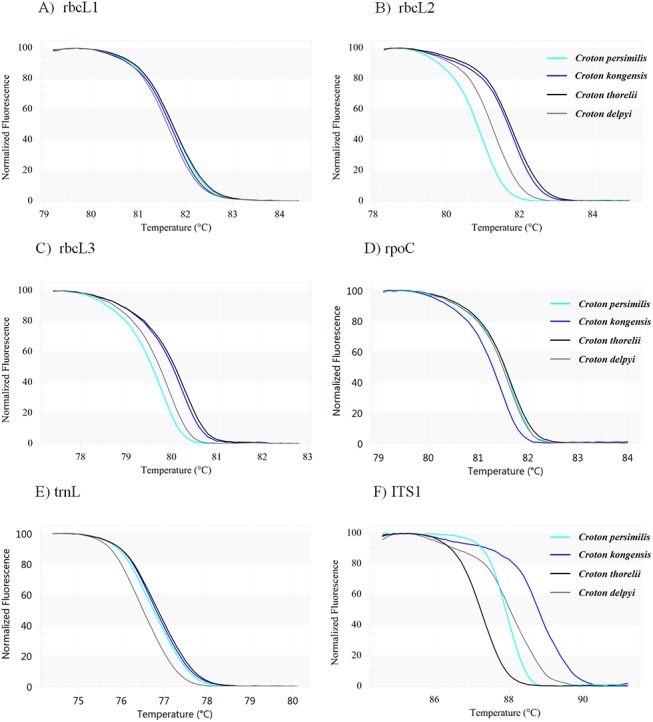
The seven HRM primer sets were used for the amplification of DNA-fragments from four Croton species, C. delpyi, C. kongensis, C. persimilis and C. thorelii, and the resulting amplicons were analyzed using HRM to define the melting temperature (Tm). These seven primer sets amplified fragments from matK, rbcL1, rbcL2, rbcL3, rpoC, trnL and ITS1 and yielded amplicons of 125 bp, 100 bp, 150 bp, 150 bp, 170 bp, 120 bp, and 300 bp, respectively. HRM analysis was performed in triplicate on each of the four taxa to establish the Tm for each primer set. The shapes of the melting curves were analyzed using EcoStudy Software v 5.0 to distinguish among the different plant species. The melting profiles of all amplicons are illustrated in Fig 2A–2F. The mean of the melting temperatures obtained from each primer pair, along with the melting curve shape, was used to measure the discriminatory power of each region regarding the species tested. The discrimination power of each region ranged from 0% (rbcL1) to 100% (ITS1). The primer pairs could be divided into four classes as follows: 1) none of the tested species could be distinguished from each other (rbcL1) (Fig 2A), 2) two out of the four species could be distinguished (rpoC and trnL) (Fig 2D and 2E), 3) three out of the four species could be distinguished (rbcL2 and rbcL3) (Fig 2B and 2C), and 4) all four species could be distinguished (ITS1) (Fig 2F). Although matK has been proposed as one of the best plant barcodes in terms of species discrimination [46,48], we found that the amplification of the fragment of matK that we targeted had such a low success rate that it could not be used for further analysis. Based on these results, it was predicted that the ITS1 primer pair would perform well in HRM analyses with an expanded sampling of Croton species. Several other studies have also shown the accuracy and universality of ITS1 for DNA barcoding in plants [12,17,19,48–51].
Fig 2. Melting curves of four Croton species generated by HRM using the following primer pairs: 2A) rbcL1; 2B) rbcL2; 2C) rbcL3; 2D) rpoC; 2E) trnL and 2F) ITS1.
Bar-HRM using ITS1 primers and an expanded Croton sampling
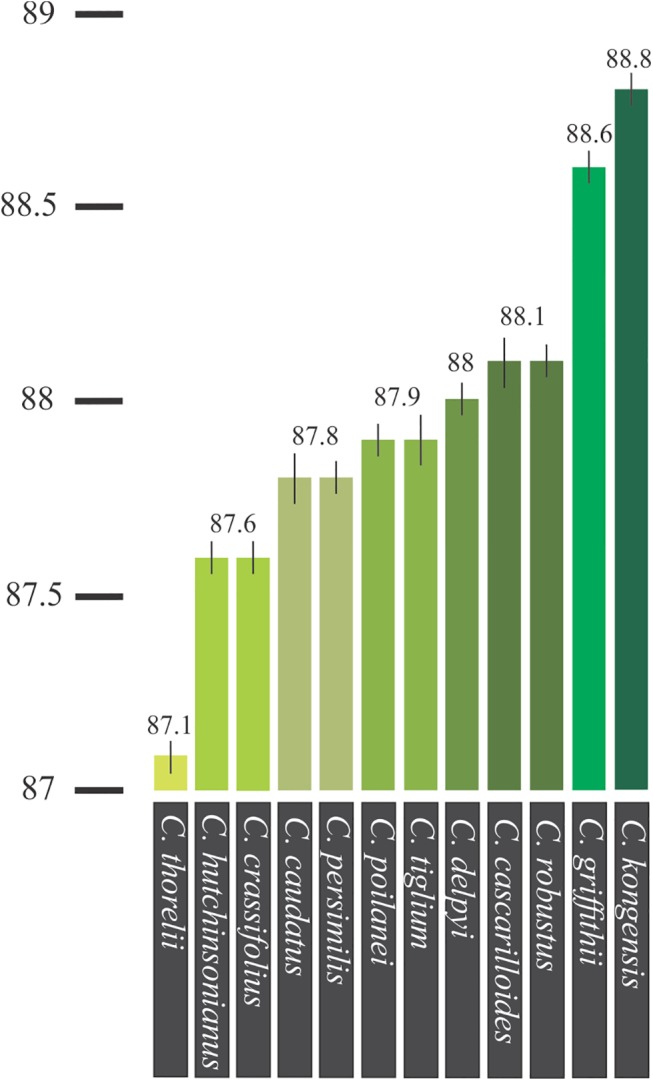
The ITS1 primer set yielded amplicons of the expected size, approximately 300 base-pairs long, and the amplicons were analyzed using HRM to determine the Tm of all 12 tested Croton species (Table 4). Fig 3 depicts the analysis by means of conventional derivative plots, which show the Tm value for the ITS1 fragment from each species. However, not all amplicons from the different species yielded distinctive HRM profiles. The melting curves were reproducibly achieved from each of the tested species in triplicate analyses.
Table 4. Average Tm with standard deviation (SD) of the ITS1 amplicon for all twelve Croton species.
| Species | SampleNo. | Tm (°C) | SD | Average Tm(°C) | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| Croton cascarilloides | 1 | 88.1 | 88.2 | 88.1 | 0.06 | 88.1 |
| Croton caudatus | 3 | 87.8 | 87.8 | 87.7 | 0.06 | 87.8 |
| Croton crassifolius | 4 | 87.6 | 87.6 | 87.5 | 0.06 | 87.6 |
| Croton delpyi | 6 | 88 | 87.9 | 88.1 | 0.10 | 88.0 |
| Croton griffithii | 7 | 88.6 | 88.6 | 88.5 | 0.06 | 88.6 |
| Croton hutchinsonianus | 8 | 87.6 | 87.6 | 87.5 | 0.06 | 87.6 |
| Croton kongensis | 11 | 88.8 | 88.7 | 88.9 | 0.10 | 88.8 |
| Croton poilanei | 13 | 87.9 | 87.9 | 88 | 0.06 | 87.9 |
| Croton robustus | 14 | 88.1 | 88.2 | 88 | 0.10 | 88.1 |
| Croton persimilis | 16 | 87.7 | 87.8 | 87.8 | 0.06 | 87.8 |
| Croton tiglium | 19 | 87.8 | 87.8 | 87.9 | 0.06 | 87.8 |
| Croton thorelii | 20 | 87.1 | 87.1 | 87.2 | 0.06 | 87.1 |
Fig 3. The normalized plot of each amplified fragment derived from ITS1 region shows the differentiation of melting temperature (Tm) of each ITS1 amplicon from each species, generated by high resolution melting (HRM) analysis.
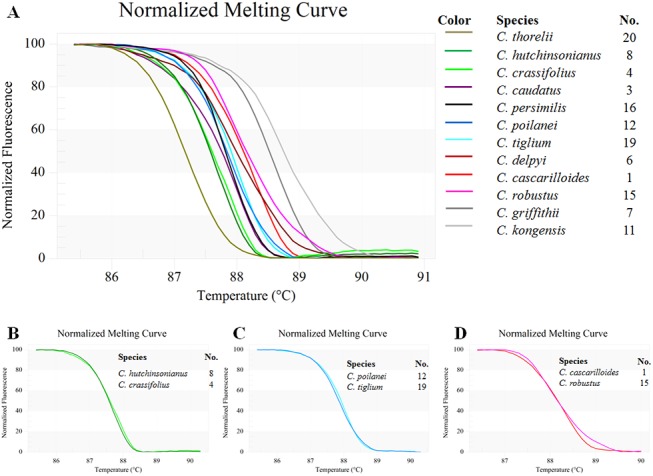
3A) Twelve Croton species, 3B) C. hutchinsonianus and C. crassifolius, 3C) C. poilanei and C. tiglium, 3D) C. cascarilloides and C. robustus.
The tested species formed nine clusters, denoted as A-I, when both Tm and the curve shape were taken into account. These were: A) C. thorelii (Sample No. 20, cf. Table 1), B) C. hutchinsonianus (8) and C. crassifolius (4) (Fig 3B), C) C. caudatus (3), D) C. persimilis (16), E) C. poilanei (12) and C. tiglium (19) (Fig 3C), F) C. delpyi (6), G) C. cascarilloides (1) and C. robustus (15) (Fig 3D), H) C. griffithii (7), and I) C. kongensis (11) (Fig 4).
Fig 4. Average Tm of the ITS1 amplicon for all twelve Croton species.
To corroborate the results of the HRM analysis, the ITS1 sequences of the 12 tested species were used for further analysis. Sequence data from six species, C. cascarilloides (CAS), C. caudatus (CAU), C. crassifolius (CRA), C. hutchinsonianus (HUT), C. kongensis (KON) and C. persimilis (PER), were obtained. Pairwise comparison of the species’ respective ITS1 sequences showed the number of variable sites ranged from 4 sites for PER-HUT to 21 for CAS-CRA (Table 5). HRM can detect differences among samples of as little as one base pair, but in order to allow for intra-specific and geographical variations within species, an arbitrary confidence interval cut-off of 90% was chosen based on previous molecular studies. The sequence variation in ITS1 among thirty samples of Croton stellatopilosus collected from different parts of Thailand shows that these can be divided into three groups based on the occurrence of indels [52].
Table 5. Variable sites found between sequence pairs of two species (total length 320 bp).
| Species | PER | CAS | CAU | CRA | HUT | KON |
|---|---|---|---|---|---|---|
| PER | ||||||
| CAS | 15 | |||||
| CAU | 15 | 20 | ||||
| CRA | 16 | 21 | 6 | |||
| HUT | 4 | 18 | 18 | 19 | ||
| KON | 15 | 16 | 14 | 16 | 18 |
PER = Croton persimilis, CAS = C. cascarilloides, CAU = C. caudatus, CRA = C. crassifolius, HUT = C. hutchinsonianus, KON = C. kongensis
The melting profiles of the six Croton species (Fig 5) were compared to the pairwise sequence analyses (Table 5). As expected from the sequence analyses, melting curves of C. caudatus and C. crassifolius were nearly the same (6 variable sites in 320 bp fragment) (Fig 6A). The melting curves of C. hutchinsonianus and C. persimilis (4 variable sites) were also expected to be similar to each other but the result showed that they are not (Fig 6B). The most surprising result emerging from the HRM analysis is that the melting curves of C. crassifolius and C. hutchinsonianus are nearly identical to each other (Fig 6C), despite the presence of 19 variable sites among the two species’ ITS1 sequences. Comparison of the base composition of the amplicons for each of the nucleotides (Fig 6A–6F) yielded a possible explanation for these unexpected results. A small number of nucleotide variations were found between C. hutchinsonianus and C. persimilis (4 variable sites; Fig 6B), but differences in their base composition are as high as other pairs with different melting curves, e.g. C. cascarilloides and C. hutchinsonianus (18 variable sites; Fig 6D), C. caudatus and C. kongensis (14 variable sites; Fig 6E), and C. cascarilloides and C. crassifolius (21 variable sites; Fig 6F). It can thus be suggested that in the C. crassifolius and C. hutchinsonianus pair, where a high number of variable sites was found between the two sequences, low differences in the overall inter-sequence base composition resulted in similar melting curve shapes. This combination of findings provides some support for the conceptual premise that it is not only the nucleotide variation but also the nucleotide composition of each sample that contributes to the melting curve shape. Our results seem to be consistent with other research which found that DNA duplex stability can be determined by both sequence and base composition as in the nearest-neighbor (NN) model [53–55]. The NN model for nucleic acids assumes that the stability of a given base pair depends on the identity and orientation of neighboring base pairs. The NN model has long been engaged in several works on DNA melting analysis (e.g. [56–58]), even before the introduction of HRM technology. Several computer programs such as OligoCalc [59], uMELT [60] and MELTING [61] that can be used to predict melting temperatures and/or generate melting curves for DNA duplexes of interest are built on the NN model, and it is one of the most frequent approaches used for predicting melting temperature.
Fig 5. Melting profiles of ITS1 amplicons from six Croton species generated from HRM analysis using ITS1 primers.
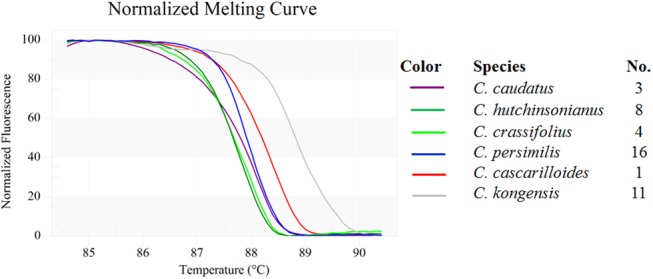
Fig 6. Normalized curves of pairwise comparisons between Croton ITS1 sequences, with a pairwise comparison of nucleotide composition and number of variable sites.
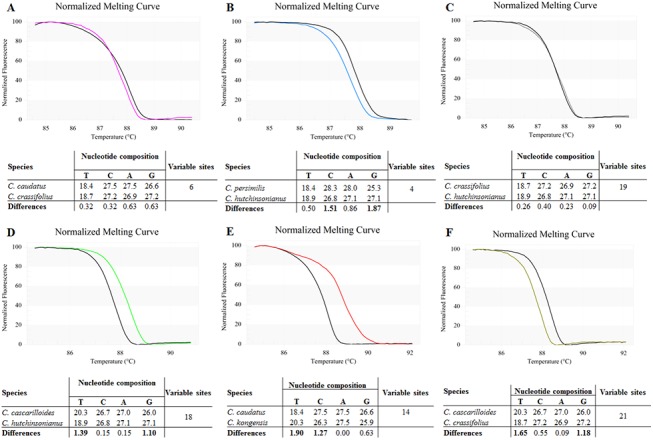
6A) C. caudatus and C. crassifolius; 6B) C. hutchinsonianus and C. persimils; 6C) C. crassifolius and C. hutchinsonianus; 6D) C. cascarilloides and C. hutchinsonianus; 6E) C. caudatus and C. kongensis; 6F) C. cascarilloides and C. crassifolius.
Although the ITS1 was found to be optimal primer pair in this study, it is likely that other markers and/or combinations thereof might perform better in other plant groups as the results shown in this study are limited to the examined genus (Croton).
Conclusions
Bar-HRM has proven to be a cost-effective and reliable method for the identification of the twelve Croton species tested in this study. The hybrid method of DNA barcoding and High Resolution Melting is dependable, fast, and sensitive enough to distinguish between species. The fragment amplified using the ITS1 primer set had the highest single marker species discrimination rate out of the seven regions coming from the tested plastid and nuclear primer pairs, and it is likely that other ITS1 based primer sets will perform well in other plant groups. The Bar-HRM primer sets developed in this study are not only useful for identification of Croton plant vouchers, but can also be used for species discrimination, authentication, and detection of adulteration in samples lacking diagnostic morphological characters, such as herbal products.
Supporting Information
(DOCX)
Acknowledgments
We thank the late Mr. James F. Maxwell (Curator of CMUB herbarium, Chiang Mai University) and Mr. Kittiphong Kertsawang (The Botanical Garden Organization, Ministry of Natural Resources and Environment, Thailand) for providing the samples.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work received funding from the National Research Council of Thailand. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Webster GL. A provisional synopsis of the sections of the genus Croton (Euphorbiaceae). Taxon. 1993; 793–823. [Google Scholar]
- 2.Govaerts R, Frodin DG, Radcliffe-Smith A, Carter S. World checklist and bibliography of Euphorbiaceae (with Pandaceae). 2000; Available: http://agris.fao.org/agris-search/search.do?recordID=US201300050879
- 3. Chayamarit K, van Welzen PC. Flora of Thailand, Volume 8, Part 1: Euphorbiaceae (Genera A-F). Bangkok, Thailand: Bangkok Forest Herbarium; 2005. [Google Scholar]
- 4. Berry PE, Hipp AL, Wurdack KJ, Van Ee B, Riina R. Molecular phylogenetics of the giant genus Croton and tribe Crotoneae (Euphorbiaceae sensu stricto) using ITS and trnL-trnF DNA sequence data. Am J Bot. 2005;92: 1520–1534. 10.3732/ajb.92.9.1520 [DOI] [PubMed] [Google Scholar]
- 5. Thongtan J, Kittakoop P, Ruangrungsi N, Saenboonrueng J, Thebtaranonth Y. New Antimycobacterial and Antimalarial 8, 9-Secokaurane Diterpenes from Croton kongensis . J Nat Prod. 2003;66: 868–870. [DOI] [PubMed] [Google Scholar]
- 6. Dey S, Mukherjee D, Chakraborty S, Mallick S, Dutta A, Ghosh J, et al. Protective effect of Croton caudatus Geisel leaf extract against experimental visceral leishmaniasis induces proinflammatory cytokines in vitro and in vivo. Exp Parasitol. 2015;151: 84–95. 10.1016/j.exppara.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 7. Sun L, Meng Z, Li Z, Yang B, Wang Z, Ding G, et al. Two new natural products from Croton kongensis Gagnep. Nat Prod Res. 2014;28: 563–567. 10.1080/14786419.2013.867856 [DOI] [PubMed] [Google Scholar]
- 8. Ahmed B, Alam T, Varshney M, Khan SA. Hepatoprotective activity of two plants belonging to the Apiaceae and the Euphorbiaceae family. J Ethnopharmacol. 2002;79: 313–316. [DOI] [PubMed] [Google Scholar]
- 9. El-Mekkawy S, Meselhy MR, Nakamura N, Hattori M, Kawahata T, Otake T. Anti-HIV-1 phorbol esters from the seeds of Croton tiglium . Phytochemistry. 2000;53: 457–464. [DOI] [PubMed] [Google Scholar]
- 10. Minh PTH, Ngoc PH, Taylor WC, Cuong NM. A new ent-kaurane diterpenoid from Croton tonkinensis leaves. Fitoterapia. 2004;75: 552–556. [DOI] [PubMed] [Google Scholar]
- 11. Rates SMK. Plants as source of drugs. Toxicon. 2001;39: 603–613. [DOI] [PubMed] [Google Scholar]
- 12. Kool A, de Boer HJ, Krüger Å, Rydberg A, Abbad A, Björk L, et al. Molecular identification of commercialized medicinal plants in Southern Morocco. PLOS ONE. 2012;7: e39459 10.1371/journal.pone.0039459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veldman S, Otieno J, Gravendeel B, Andel T van, de Boer HJ. Conservation of Endangered Wild Harvested Medicinal Plants: Use of DNA Barcoding. Gurib-Fakim A, editor. Nov Plant Bioresour Appl Food Med Cosmet. 2014; 81–88.
- 14. Skalli S, Alaoui I, Pineau A, Zaid A, Soulaymani R. L’intoxication par le chardon à glu (Atractylis gummifera L.): à propos d’un cas clinique. Bull Soc Pathol Exot. 2002;95: 284–286. [PubMed] [Google Scholar]
- 15. Ize-Ludlow D, Ragone S, Bruck IS, Bernstein JN, Duchowny M, Pena BMG. Neurotoxicities in infants seen with the consumption of star anise tea. Pediatrics. 2004;114: e653 10.1542/peds.2004-0058 [DOI] [PubMed] [Google Scholar]
- 16. Ouarghidi A, Powell B, Martin GJ, de Boer HJ, Abbad A. Species substitution in medicinal roots and possible implications for toxicity in Morocco. Econ Bot. 2012;66: 370–382. [Google Scholar]
- 17. Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5: 1–8. 10.1371/journal.pone.0008613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coghlan M, Haile J, Houston J, Murray D, White N, Moolhuijzen P, et al. Deep sequencing of plant and animal DNA contained within traditional chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012;8: e1002657 10.1371/journal.pgen.1002657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Boer HJ, Ouarghidi A, Martin G, Abbad A, Kool A. DNA Barcoding Reveals Limited Accuracy of Identifications Based on Folk Taxonomy. PLOS ONE. 2014;9: e84291 10.1371/journal.pone.0084291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newmaster SG, Grguric M, Shanmughanandhan D, Ramalingam S, Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11: 222 10.1186/1741-7015-11-222 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Li M, Cao H, But PP-H, Shaw P-C. Identification of herbal medicinal materials using DNA barcodes. J Syst Evol. 2011;49: 271–283. [Google Scholar]
- 22. Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25: 103–110. 10.1016/j.copbio.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 23. De Boer HJ, Ichim MC, Newmaster SG. DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf. [In Press]; [DOI] [PubMed] [Google Scholar]
- 24. Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT. How to Open the treasure chest? Optimising DNA extraction from herbarium specimens. PLOS ONE. 2012;7: e43808 10.1371/journal.pone.0043808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Piredda R, Simeone MC, Attimonelli M, Bellarosa R, Schirone B. Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. Mol Ecol Resour. 2011;11: 72–83. 10.1111/j.1755-0998.2010.02900.x [DOI] [PubMed] [Google Scholar]
- 26. Sass C, Little DP, Stevenson DW, Specht CD. DNA barcoding in the cycadales: testing the potential of proposed barcoding markers for species identification of cycads. PLOS ONE. 2007;2: e1154 10.1371/journal.pone.0001154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S, Little DP. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep. 2011;1: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, et al. Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol Ecol Resour. 2009;9: 130–139. 10.1111/j.1755-0998.2009.02652.x [DOI] [PubMed] [Google Scholar]
- 29. Osathanunkul M, Madesis P, de Boer HJ. Bar-HRM for authentication of plant-based medicines: evaluation of three medicinal products derived from Acanthaceae species. PLoS ONE. 2015; 10 (5): e0128476 10.1371/journal.pone.0128476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reja V, Kwok A, Stone G, Yang L, Missel A, Menzel C, et al. ScreenClust: Advanced statistical software for supervised and unsupervised high resolution melting (HRM) analysis. Methods. 2010;50: S10–S14. 10.1016/j.ymeth.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 31. Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50: 1748–1754. [DOI] [PubMed] [Google Scholar]
- 32. Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245: 154–160. [DOI] [PubMed] [Google Scholar]
- 33. Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49: 853–860. [DOI] [PubMed] [Google Scholar]
- 34. Madesis P, Ganopoulos I, Sakaridis I, Argiriou A, Tsaftaris A. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res Int. 2014;60: 163–172. [Google Scholar]
- 35. Ganopoulos I, Madesis P, Darzentas N, Argiriou A, Tsaftaris A. Barcode High Resolution Melting (Bar-HRM) analysis for detection and quantification of PDO “Fava Santorinis” (Lathyrus clymenum) adulterants. Food Chem. 2012;133: 505–512. 10.1016/j.foodchem.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 36. Ganopoulos I, Bazakos C, Madesis P, Kalaitzis P, Tsaftaris A. Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J Sci Food Agric. 2013;93: 2281–2286. 10.1002/jsfa.6040 [DOI] [PubMed] [Google Scholar]
- 37. Ganopoulos I, Xanthopoulou A, Mastrogianni A, Drouzas A, Kalivas A, Bletsos F, et al. High Resolution Melting (HRM) analysis in eggplant (Solanum melongena L.): A tool for microsatellite genotyping and molecular characterization of a Greek Genebank collection. Biochem Syst Ecol. 2015;58: 64–71. [Google Scholar]
- 38. Kalivas A, Ganopoulos I, Xanthopoulou A, Chatzopoulou P, Tsaftaris A, Madesis P. DNA barcode ITS2 coupled with high resolution melting (HRM) analysis for taxonomic identification of Sideritis species growing in Greece. Mol Biol Rep. 2014;41: 5147–5155. 10.1007/s11033-014-3381-5 [DOI] [PubMed] [Google Scholar]
- 39. Schmiderer C, Mader E, Novak J. DNA-based identification of Helleborus niger by high-resolution melting analysis. Planta Med. 2010;76: 1934–1937. 10.1055/s-0030-1249908 [DOI] [PubMed] [Google Scholar]
- 40. Ganopoulos I, Aravanopoulos F, Madesis P, Pasentsis K, Bosmali I, Ouzounis C, et al. Taxonomic identification of Mediterranean pines and their hybrids based on the high resolution melting (HRM) and trnL approaches: from cytoplasmic inheritance to timber tracing. PLOS ONE. 2013;8: e60945 10.1371/journal.pone.0060945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madesis P, Ganopoulos I, Bosmali I, Tsaftaris A. Barcode High Resolution Melting analysis for forensic uses in nuts: A case study on allergenic hazelnuts (Corylus avellana). Food Res Int. 2013;50: 351–360. [Google Scholar]
- 42. Madesis P, Ganopoulos I, Anagnostis A, Tsaftaris A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control. 2012;25: 576–582. [Google Scholar]
- 43. Bosmali I, Ganopoulos I, Madesis P, Tsaftaris A. Microsatellite and DNA-barcode regions typing combined with High Resolution Melting (HRM) analysis for food forensic uses: A case study on lentils (Lens culinaris). Food Res Int. 2012;46: 141–147. [Google Scholar]
- 44. Ganopoulos I, Madesis P, Tsaftaris A. Universal ITS2 barcoding DNA region coupled with high-resolution melting (HRM) analysis for seed authentication and adulteration testing in leguminous forage and pasture species. Plant Mol Biol Report. 2012;30: 1322–1328. [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci. 2009;106: 12794–12797. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci U S A. 2011;108: 19451–19452. 10.1073/pnas.1116812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLOS ONE. 2008;3: e2802 10.1371/journal.pone.0002802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci. 2005;102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci. 2011;108: 19641–19646. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao T, Yao H, Song J, Liu C, Zhu Y, Ma X, et al. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol. 2010;130: 116–121. 10.1016/j.jep.2010.04.026 [DOI] [PubMed] [Google Scholar]
- 52. Rinthong P, Zhu S, Komatsu K, Chanama S, De-Eknamkul W. Genetic variation of Croton stellatopilosus Ohba based on non-coding DNA sequences of ITS, trnK and trnL-F regions. J Nat Med. 2011;65: 641–645. 10.1007/s11418-011-0536-8 [DOI] [PubMed] [Google Scholar]
- 53. Sugimoto N, Katoh M, Nakano S, Ohmichi T, Sasaki M. RNA/DNA hybrid duplexes with identical nearest-neighbor base-pairs have identical stability. FEBS Lett. 1994;354: 74–78. [DOI] [PubMed] [Google Scholar]
- 54. SantaLucia J, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry (Mosc). 1996;35: 3555–3562. [DOI] [PubMed] [Google Scholar]
- 55. Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2009; gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeVoe H, Tinoco I. The stability of helical polynucleotides: base contributions. J Mol Biol. 1962;4: 500–517. [DOI] [PubMed] [Google Scholar]
- 57. Gray DM, Tinoco I. A new approach to the study of sequence-dependent properties of polynucleotides. Biopolymers. 1970;9: 223–244. [Google Scholar]
- 58. Gotoh O, Tagashira Y. Stabilities of nearest-neighbor doublets in double-helical DNA determined by fitting calculated melting profiles to observed profiles. Biopolymers. 1981;20: 1033–1042. [Google Scholar]
- 59. Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35: W43–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dwight Z, Palais R, Wittwer CT. uMELT: prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics. 2011;27: 1019–1020. 10.1093/bioinformatics/btr065 [DOI] [PubMed] [Google Scholar]
- 61. Dumousseau M, Rodriguez N, Juty N, Novère NL. MELTING, a flexible platform to predict the melting temperatures of nucleic acids. BMC Bioinformatics. 2012;13: 101 10.1186/1471-2105-13-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.