Abstract
The growing public threat of Alzheimer’s disease (AD) has raised the urgency to discover and validate prognostic biomarkers in order to predicting time to onset of AD. It is anticipated that both whole genome single nucleotide polymorphism (SNP) data and high dimensional whole brain imaging data offer predictive values to identify subjects at risk for progressing to AD. The aim of this paper is to test whether both whole genome SNP data and whole brain imaging data offer predictive values to identify subjects at risk for progressing to AD. In 343 subjects with mild cognitive impairment (MCI) enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI-1), we extracted high dimensional MR imaging (volumetric data on 93 brain regions plus a surface fluid registration based hippocampal subregion and surface data), and whole genome data (504,095 SNPs from GWAS), as well as routine neurocognitive and clinical data at baseline. MCI patients were then followed over 48 months, with 150 participants progressing to AD. Combining information from whole brain MR imaging and whole genome data was substantially superior to the standard model for predicting time to onset of AD in a 48-month national study of subjects at risk. Our findings demonstrate the promise of combined imaging-whole genome prognostic markers in people with mild memory impairment.
Keywords: Alzheimer’s disease, genetics, magnetic resonance imaging, proportional hazards models, risk
INTRODUCTION
The growing public threat of Alzheimer’s disease (AD) has raised the urgency to discover and validate prognostic biomarkers that may identify subjects at greatest risk for future cognitive decline and accelerate the testing of preventive strategies [1, 2]. In this regard, studies of combinatorial biomarkers may have greater ability to capture the heterogeneity and multifactorial complexity of AD, than a traditional single biomarker study [3].
Prior studies of subjects at risk for AD have examined the utility of various individual biomarkers, such as cognitive tests, fluid markers, imaging measures, and some individual genetic markers (e.g., ApoE4) [1]. In particular, imaging markers such as hippocampal volume and shape, cortical regional volumes and thickness, and positron emission tomography (PET) (amyloid imaging, FDG) abnormalities have all been linked in one or more studies to faster progression in at risk subjects [4–16], but are not yet optimally predictive at an individual level.
More recently, genome-wide association study (GWAS) data has been used to characterize several potential genetic risk factors for AD with several cross-sectional studies also correlating these data with imaging and fluid biomarkers [17]. There are also some studies combining imaging and genetics information to predict the conversion of MCI to AD [18, 19], however, they only consider the conversion of MCI to AD as a binary response, and they do not investigate the risk of progression to AD for each specific MCI individual. To our knowledge, no prior study has leveraged both GWAS SNP data, as well as high dimensional whole brain imaging data to examine their combined value in identifying subjects at greatest risk for progressing to AD.
METHODS
Alzheimer’s Disease Neuroimaging Initiative
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://www.loni.usc.edu/ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (aMCI) and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California - San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research— approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with aMCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. For up-to-date information, see http://www.adni-info.org.
Study sample
We considered 343 subjects with mild cognitive impairment (MCI) enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI-1). These MCI patients were then followed over 48 months, with 150 participants progressing to AD (Table 1). MCI converters did not differ from MCI noncoverters in gender, handedness, marital status, retirement percentage, and age (p-values>0.05), but as expected, differed from them in APOE4 status as well as baseline cognition (p<0.05) (Table 1). Mean follow up time was 75 days longer in converters (p=0.06). From them, we extracted high dimensional MR imaging and whole genome data, as well as routine neurocognitive and clinical data at baseline.
Table 1.
Study sample: comparison of converters and nonconverters.
| Sample Size | MCI converters n=150 |
MCI nonconverters n=193 |
Test statistics |
|---|---|---|---|
| Mean Age (in years) | 74.7 (7.0) | 75.1 (7.5) | T test: p=0.69 |
| Gender (Male percentage) | 60.0% | 66.8% | Chi-square test: p=0.23 |
| Handedness (Right hand percentage) | 92.0% | 90.7% | Chi-square test: p=0.81 |
| Marital Status (Widowed percentage) | 10.7% | 11.9% | Chi-square test: p=0.95 |
| Marital Status (Divorced percentage) | 4.7% | 6.2% | |
| Marital Status (Never Married percentage) | 1.3% | 1.0% | |
| Mean Education Length (in years) | 15.6 (2.9) | 15.8 (3.0) | T test: p=0.52 |
| Retirement percentage | 80.0% | 79.3% | Chi-square test: p=0.98 |
| APOE 4 carriers (%): APOE4 first allele (genotype 3 %) | 78.7% | 81.4% | Chi-square test: p=0.02 |
| APOE4 first allele (genotype 4 %) | 17.3% | 9.3% | |
| APOE4 second allele (genotype 4 %) | 30.0% | 56.0% | Chi-square test: p<0.0001 |
| Mean ADAS-Cog 11 Score | 13.2 (4.0) | 10.2 (4.3) | T test: p<0.0001 |
| Mean Duration of Follow up (in days) | 1009.9 | 934.4 | T test: p=0.06 |
MCI, mild cognitive impairment; ADAS-Cog, Alzheimer’s Disease Assessment Scale-cognitive subscale
MRI imaging
These scans on 343 subjects were performed on a 1.5 T MRI scanners by using a sagittal MPRAGE sequence with the following parameters: repetition time (TR) = 2400 ms, inversion time (TI) = 1000 ms, flip angle = 8°, and field of view (FOV) = 24 cm with a 256 x 256 x 170 acquisition matrix in the x-, y-, and z-dimensions, which yields a voxel size of 1.25 x 1.261 x 2.
We processed the MRI data by using standard steps including anterior commissure and posterior commissure correction, skull-stripping, cerebellum removing, intensity inhomogeneity correction, segmentation, and registration [20]. After segmentation, we segmented the brain data into four different tissues: grey matter, white matter, and cerebrospinal fluid. Moreover, we automatically labeled 93 regions of interest (ROIs) on the Jacob atlas [21], and transferred the labels following the deformable registration of subject images [22]. In addition, we chose 23 ROIs, which may significantly influence MCI progression from the existing literature [10, 23, 24]. The 23 ROIs were bilateral entorhinal cortices, bilateral hippocampal formation, bilateral amygdala, bilateral caudate nuclei, bilateral putamen, bilateral posterior limb of internal capsule including cerebral peduncle, bilateral nucleus accumbens, bilateral lateral ventricles, bilateral thalamus, bilateral fornix, bilateral cingulate, and the corpus callosum.
Hippocampus image preprocessing
We adopted a surface fluid registration based hippocampal subregional analysis package [25], which uses isothermal coordinates and fluid registration to generate one-to-one hippocampal surface registration for following surface statistics computation. This software package has been adopted by various studies [22, 26–30].
Given the 3D MRI scans, hippocampal substructures were segmented with FIRST [31] and hippocampal surfaces were automatically reconstructed with the marching cube method [32]. We applied an automatic algorithm, topology optimization, to introduce two cuts on a hippocampal surface to convert it into a genus zero surface with two open boundaries. The locations of the two cuts were at the front and back of the hippocampal surface, representing its anterior junction with the amygdala, and its posterior limit as it turns into the white matter of the fornix. Then holomorphic 1-form basis functions were computed [33]. These induced conformal grids the hippocampal surfaces, which were consistent across subjects. With this conformal grid, we computed the conformal representation of the surface [25], i.e., the conformal factor and mean curvature, which represent the intrinsic and extrinsic features of the surface, respectively. The “feature image” of a surface was computed by combining the conformal factor and mean curvature and linearly scaling the dynamic range into [0, 255]. Next, we registered the feature image of each surface in the dataset to a common template with an inverse consistent fluid registration algorithm [26]. With conformal parameterization, we essentially converted a 3D surface registration problem into a 2D image registration problem. The flow induced in the parameter domain establishes high-order correspondences between 3D surfaces. Finally, various surface statistics were computed on the registered surface, such as multivariate tensor-based morphometry statistics [33], which retain the full tensor information of the deformation Jacobian matrix, together with the radial distance [34], which retains information on the deformation along the surface normal direction.
SNP data
The subjects’ genotype variables were acquired based on the Human 610-Quad Bead-Chip (Illumina, Inc., San Diego, CA) in the ADNI database, which resulted in 620,901 SNPs. To reduce the population stratification effect, we used 749 Caucasians from all 818 subjects with complete imaging measurements at baseline. Quality control procedures included (i) call rate check per subject and per SNP marker, (ii) gender check, (iii) sibling pair identification, (iv) the Hardy-Weinberg equilibrium test, (v) marker removal by the minor allele frequency, and (vi) population stratification. The second line preprocessing steps include removal of SNPs with (a) more than 5% missing values, (b) minor allele frequency smaller than 5%, and (c) Hardy-Weinberg equilibrium p-value < 1e–6. Remaining missing genotype variables were imputed as the modal value. 747 subjects and 504,095 SNPs remained.
We included information from all the 22 chromosomes. Since each chromosome contains a number of SNPs, we used principal component analysis for each chromosome and picked the first two principal components for each chromosome. We then used the PLINK package (https://pngu.mgh.harvard.edu/~purcell/plink/data.shtml#plink) to perform quality control for the genomic data. The principal component analysis for each chromosome was conducted using ‘svd’ function in R software.
Statistical analyses
A popular model used in literature is the Cox proportional hazards model, which accounts for other covariates that are associated with the timing of the events. Covariates of interest include demographic information (8 covariates), the APOE4 genotype (3 covariates), the Alzheimer’s Disease Assessment Scale-cognitive subscale (ADAS-Cog) score (1 covariate), the hippocampus surface data (7 covariates for each curve, total 14 covariates), the ROI volume data (23 covariates), the chromosomewise information (2 covariates for each chromosome, total 44 covariates), and the significant SNPs information (5 covariates). We used the R function “coxph” to implement the fitting of the Cox proportional hazards model. We fitted a Cox regression model with demographic, clinical, and cognitive (ADAS-Cog score) predictors as well as APOE, referred to as the Clinical-Cognitive model (Model 1), and obtained its estimation and testing results. This model did not include any other imaging and genetic data. We fitted a second Cox regression model with demographic, imaging, and chromosome-wise predictors, but without the ADAS-Cog score and significant SNPs information, referred to as the Imaging-Genetics model (Model 2), and obtained its estimation and testing results.
As a comparison, we also used the results obtained from GWAS to incorporate the genetic information. Specifically, we selected the top 101 significant SNPs by using a kernel machine method [35], and then calculated their top 5 PCs and used them as predictors (significant SNP information). We fitted a third Cox regression model with demographic, imaging, and significant SNP information, but without the ADAS-Cog score and chromosome-wise information, and then obtained parameter estimation and testing results. We referred this model to as the Traditional Imaging-Genetics Model (Model 3).
When we fitted Cox regression models, we treated the left and right hippocampus surface data as functional predictors. For each subject, the radial distance was obtained from baseline hippocampal surfaces data, which yields two 15,000 dimensional vectors denoting data from left hippocampus and right hippocampus, respectively. We applied functional principal component analysis (FPCA) [36, 37]. We selected 7 functional principal component (FPC) scores for each functional predictor, which explain approximately 70% of the variance. For implementation of FPCA, we use the ‘svd’ function in R software.
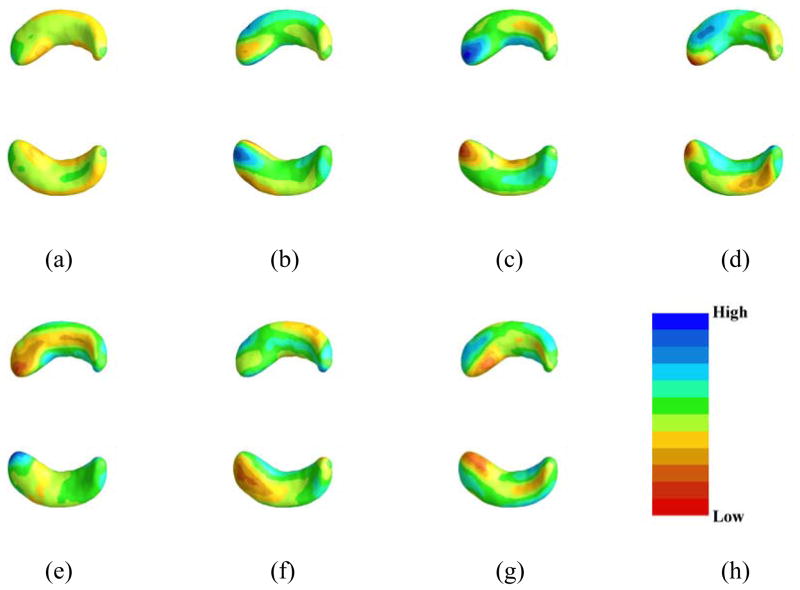
We used FPCA to extract the first 7 FPCs of each of the left and right hippocampus surface data and the first 2 FPCs of the SNP data along each chromosome, then used these as basis functions to represent the regression coefficient functions associated with the hippocampus surface and SNP on all chromosomes in the Cox regression. As an illustration, we plotted the first FPCs for both left and right hippocampi in Figure 1.
Figure 1.
Estimated coefficient functions associated with the hippocampus surface data. Panel (a)-(g) are the estimates of the first seven functional principal components corresponding to the sorted seven eigenvalues, in which (a) corresponds the largest eigenvalue and (h) is the color bar with 11 lines representing 11 equally spaced points between [−0.0462, 0.0421].
Since we do not have the validation data set, we investigated the predictive performance of the candidate models using the receiving operating characteristic (ROC) curve. In particular, we calculated the area under the curve (AUC), which is often used to measure the prediction of survival models. In particular, we first randomly picked 200 subjects for the training data and fit all the candidate models. After that, we used the remaining 143 individuals for the testing and calculated the AUC [38]. The method can be implemented by using the R function “AUC.cd”. We repeated the above steps for 100 times, i.e., randomly separated the data for 100 times and obtained 100 AUCs. The mean and standard deviations can be obtained using these 100 AUCs.
RESULTS
We compared the predictive value of standard of care (clinical demographic variables, APOE4, the ADAS-Cog score, Model 1) versus imaging-genetic markers (MRI volumes and surface data plus GWAS SNP and demographics, Model 2). We have obtained the first five principal components of the 101 top SNPs obtained from GWAS and added it into our Model 2 (call it Model 3). We have compared our Model 2 versus Model 3 as well.
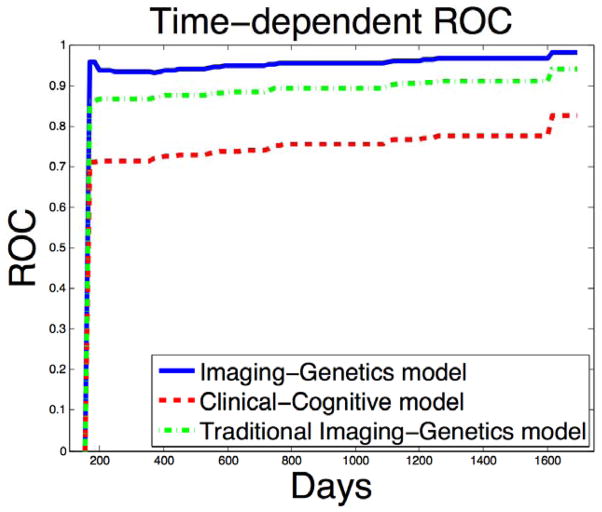
ROC analysis revealed that Model 1 (combining just routine clinical demographics and cognitive data with a single genotype APOE4) had a low predictive value at 48 months (AUC 0.75) (Figure 2, Supplementary Table 1, Supplementary Figure 3). In this model, APOE4 and ADAS-Cog were the significant predictors. In contrast, Model 2 (combining full genetic SNP and high dimension imaging data with demographics but without any cognitive data) had a much higher predictive value (AUC 0.95) (Figure 2, Supplementary Table 2, Supplementary Figure 3). SNPS on chromosome 2, 10, 11, 15, 17, and 18 (Supplementary Figure 2), APOE4 status, surface morphology data of both hippocampi (especially anterior regions, Figure 1, Supplementary Figure 1) and volumes of hippocampus, amygdala, and thalamus contributed significantly. The 100-fold cross validation using a test and training data set revealed AUC of 0.95 (±0.014) for the imaging-genetic model and 0.75 (±0.024) for the clinical-cognitive model. For the Traditional Imaging-Genetics model, we can only obtain a predictive value (AUC 0.90) (Figure 2, Supplementary Table 3, Supplementary Figure 3), which indicates the advantages of using the chromosome-wise information instead of the traditional significant SNPs information. The reason may be due to the pitfalls of prediction using significant SNPs [40]. Meanwhile, we have found that combining all variables (cognitive data plus imaging-genetic data) showed high predictive accuracy (AUC 0.96) but was not different from the value provided by imaging-genetics data alone. Besides, combining the significant SNPs information with all the predictors in our Imaging Genetics Model achieved slightly higher predictive accuracy (AUC 0.97), but was not different from the value provided by Imaging-Genetic Model alone.
Figure 2.
The ROC curves comparison for the imaging plus genetics model versus the cognitive model plotted using one pair of training and testing data set. The blue solid line denotes the ROC curve for the image-genetics model and the red dash line denotes the ROC curve for clinical-cognitive model.
From the estimation results of our imaging-genetics model, we have found that divorced individuals, older people, individuals with 2nd allele of APOE4 genotype 3 may have smaller hazard function. The individuals with larger ROI volumes in hippocampal left, amygdala right, and thalamus left may have smaller hazard function, while the individuals with larger ROI volumes in hippocampal right, amygdala left, posterior limb of internal capsule, and thalamus right may have larger hazard function.
DISCUSSION
These findings are the first demonstration, to our knowledge, of the value of combined whole brain MR and whole genome SNP data in the 48-month prognosis of subjects at risk for AD. Our finding support prior MRI studies of volumetric hippocampal changes in prodromal AD [8, 39] and extend them by finding that the possible prognostic value of combining information from high dimensional imaging and genetics may be superior to that provided by routine clinical-cognitive testing data.
Our findings also confirm the association between APOE4 status and AD, and identify additional new markers on chromosomes 2, 10, 11, 15, 17, and 18 as having significant effects on conversion. A variety of genes have been identified in prior GWAS studies as potential risk factors for AD such as clusterin (chromosome 8), complement receptor 1 (chromosome 1), phosphatidylinositol binding clathrin assembly protein (chromosome 11), sortilin-related receptor (chromosome 11), triggering receptor expressed on myeloid cells 2 (chromosome 6), and cluster of differentiation 33 (chromosome 19) as well as TOMM40 (chromosome 19) [41]. Our study did not examine any of these newer gene markers specifically but provides support to the notion that there is additional genetic heritability in late-onset AD beyond that accounted for by APOE.
There are some strengths and limitations to our analyses. ADNI is a national biomarker study that utilized rigorous standardized data collection procedures, and well established criteria to select MCI subjects. Rather than using the individual data on all SNPs, we used the more conservative statistical method of doing principal component analysis for each chromosome and picking the first two principal components for each chromosome. Our findings survived internal cross validation but need replication in an independent community based sample. We did not include measures of pathology (e.g., amyloid-β) in our models since cerebrospinal fluid and amyloid-PET were available only in a small subset of individuals in ADNI-1. However, a study of ADNI-2 subjects has shown a robust correlation between the APOE ε4 allele and cortical amyloid burden [42], suggesting that APOE ε4 may have served as a surrogate for cortical amyloid plaque load in our analysis. It is important to confirm the above findings obtained from ADNI-1 in other independent data sets [40].
Prior investigations of prediction of MCI-AD to AD have utilized feature selection to assess the most important biomarkers of prediction of conversion. Here, we have demonstrated the utility of Cox hazard models as a valuable method for identifying the “optimal combination” of early markers of conversion to AD in patients with MCI. If replicated in independent cohorts, such combinatorial markers of early AD could be useful for selecting at risk individuals for prevention trials and for discovering novel targets for discovering disease modifying therapies.
Supplementary Material
Acknowledgments
This work was supported in part by NlH grants RR025747-01, 1UL1TR001111, P01CA142538-01, MH086633, and AG033387 to Dr. Zhu. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
PMD has received research grants and/or advisory fees from several government agencies, advocacy groups and pharmaceutical/imaging companies. PMD received a grant from ADNI to support data collection for this study and he owns stock in Sonexa, Maxwell, Adverse Events and Clarimedix, whose products are not discussed here.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/15-0164r1).
References
- 1.Weiner MW, Veitcha DP, Aisen PS, Beckett LA, Cairnsh NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykino AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ Alzheimer’s Disease Neuroimaging Initiative . The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 2012;8:S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doraiswamy PM, Sperling RA, Johnson K, Reiman EM, Wong TZ, Sabbagh MN, Sadowsky CH, Fleisher AS, Carpenter A, Joshi AD, Lu M, Grundman M, Mintun MA, Skovronsky DM, Pontecorvo MJ. AV45-A11 Study Group (2014) Florbetapir F 18 amyloid PET and 36-month cognitive decline: a prospective multicenter study. Mol Psychiatry. 19:1044–1051. doi: 10.1038/mp.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atluri G, Padmanabhan K, Fang G, Steinbach M, Petrella JR, Lim K, Macdonald A, 3rd, Samatova NF, Doraiswamy PM, Kumar V. Complex biomarker discovery in neuroimaging data: Finding a needle in a haystack. Neuroimage Clin. 2013;3:123–131. doi: 10.1016/j.nicl.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu CY, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Wahlund LO, Spenger C, Toga AW, Lovestone S, Simmons A. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage. 2011;56:212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin SE. Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32:2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickerson BC, Wolk DA. Alzheimer’s Disease Neuroimaging Initiative. Biomarker-based prediction of progression in MCI: comparison of AD signature and hippocampal volume with spinal fluid amyloid-β and tau. Front Aging Neurosci. 2013;5:1–9. doi: 10.3389/fnagi.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckerström C, Olsson E, Bjerke M, Malmgren H, Edman A, Wallin A, Nordlund A. A combination of neuropsychological, neuroimaging, and cerebrospinal fluid markers predicts conversion from mild cognitive impairment to dementia. J Alzheimers Dis. 2013;36:421–431. doi: 10.3233/JAD-122440. [DOI] [PubMed] [Google Scholar]
- 10.Jack CR, Jr, Petersen RC, Xu YC, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu A, Fennema-Notestine C, Dale AM, Miller MI. Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P. Early diagnosis of Alzheimer’s disease using cortical thickness: Impact of cognitive reserve. Brain. 2009;132:2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaffe JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Coleman RE, Doraiswamy PM Alzheimer’s Disease Neuroimaging Initiative. Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology. 2013;266:583–591. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toledo JB, Weiner MW, Wolk DA, Da X, Chen K, Arnold SE, Jagust W, Jack C, Reiman EM, Davatzikos C, Shaw LM, Trojanowski JQ Alzheimer’s Disease Neuroimaging Initiative. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta Neuropathol Commun. 2014;2:26. doi: 10.1186/2051-5960-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohman TJ, Koran ME, Thornton-Wells TA Alzheimer’s Disease Neuroimaging Initiative. Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimers Dement. 2014;10:637–645. doi: 10.1016/j.jalz.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh N, Wang AY, Sankaranarayanan P, Fletcher PT, Joshi S. Genetic, structural and functional imaging biomarkers for early detection of conversion from MCI to AD. Med Image Comput Comput Assist Interv. 2012;15:132–140. doi: 10.1007/978-3-642-33415-3_17. [DOI] [PubMed] [Google Scholar]
- 19.Lemoine B, Rayburn S, Benton R. Data Fusion and Feature Selection for Alzheimer’s Diagnosis. In: Yao Y, Sun R, Poggio T, Liu J, Zhong N, Huang J, editors. BI 2010. LNAI 6334. Springer-Verlag; Berlin Heidelberg: 2010. pp. 320–327. [Google Scholar]
- 20.Shen DG, Davatzikos C. Measuring temporal morphological changes robustly in brain MR images via 4-dimensional template warping. Neuroimage. 2004;21:1508–1517. doi: 10.1016/j.neuroimage.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Bryant C, Giovanello KS, Ibrahim JG, Chang J, Shen D, Peterson BS, Zhu HT Alzheimer’s Disease Neuroimaging Initiative . Mapping the genetic variation of regional brain volumes as explained by all common SNPs from the ADNI study. PloS One. 2013;8:e71723. doi: 10.1371/journal.pone.0071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Song Y, Rajagopalan P, An T, Liu K, Chou Y-Y, Gutman B, Toga AW, Thompson PM. Surface-based TBM boosts power to detect disease effects on the brain: an N= 804 ADNI study. Neuroimage. 2011;56:1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fennema-Notestine C, Hagler DJ, McEvoy LK, Fleisher AS, Wu EH, Karow DS, Dale AM. Structural MRI biomarkers for preclinical and mild Alzheimer’s disease. Hum Brain Mapp. 2009;30:3238–3253. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan NS, Keihaninejad S, Shakespeare TJ, Lehmann M, Crutch SJ, Malone IB, Thornton JS, Mancini L, Hyare H, Yousry T, Ridgway GR, Zhang H, Modat M, Alexander DC, Rossor MN, Ourselin S, Fox NC. Magnetic resonance imaging evidence for presymptomatic change in thalamus and caudate in familial Alzheimer’s disease. Brain. 2013;136:1399–1414. doi: 10.1093/brain/awt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Wang Y, Ceschin R, An X, Lao Y, Vanderbilt D, Nelson MD, Thompson PM, Panigrahy A, Lepore N. A multivariate surface- based analysis of the putamen in premature newborns: Regional differences within the ventral striatum. PloS One. 2013;8:e66736. doi: 10.1371/journal.pone.0066736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Thompson PM, Gutman B, Wang Y. Surface fluid registration of conformal representation: Application to detect disease burden and genetic influence on hippocampus. NeuroImage. 2013;78:111–134. doi: 10.1016/j.neuroimage.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, Lepor N, Gutman BA, Thompson PM, Baxter LC, Caselli RL, Wang Y Alzheimer’s Disease Neuroimaging Initiative . Genetic influence of apolipoprotein E4 genotype on hippocampal morphometry: An N = 725 surface-based Alzheimer’s disease neuroimaging initiative study. Hum Brain Mapp. 2014;35:3903–3018. doi: 10.1002/hbm.22447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monje M, Thomason ME, Rigolo L, Wang Y, Waber DP, Sal-lan SE, Golby AJ. Functional and structural differences in the hippocampus associated with memory deficits in adult survivors of acute lymphoblastic leukemia. Pediatr Blood Cancer. 2013;60:293–300. doi: 10.1002/pbc.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colom R, Stein JL, Rajagopalan P, Martınez K, Hermel D, Wang Y, Alvarez-Linera J, Burgaleta M, Quiroga M, Shih PC, Thompson PM. Hippocampal structure and human cognition: Key role of spatial processing and evidence supporting the efficiency hypothesis in females. Intelligence. 2013;41:129–140. doi: 10.1016/j.intell.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luders E, Thompson PM, Kurth F, Hong JY, Phillips OR, Wang Y, Gutman BA, Chou YY, Narr KL, Toga AW. Global and regional alterations of hippocampal anatomy in long-term meditation practitioners. Hum Brain Mapp. 2013;34:3369–3375. doi: 10.1002/hbm.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorensen WE, Cline HE. Marching cubes: A high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput Graph. 1987;21:163–169. [Google Scholar]
- 33.Wang Y, Zhang J, Gutman B, Chan TF, Becker JT, Aizenstein HJ, Lopez OL, Tamburo RJ, Toga AW, Thompson PM. Multivariate tensor-based morphometry on surfaces: application to mapping ventricular abnormalities in HIV/AIDS. Neuroimage. 2010;49:2141–2157. doi: 10.1016/j.neuroimage.2009.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizer SM, Fritsch DS, Yushkevich PA, Johnson VE, Chaney EL. Segmentation, registration, and measurement of shape variation via image object shape. IEEE Trans Med Imaging. 1999;18:851–865. doi: 10.1109/42.811263. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Cai T, Wu MC, Zhou Q, Liu G, Christiani DC, Lin X. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genet Epidemiol. 2011;35:620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice JA, Silverman BW. Estimating the mean and covariance structure nonparametrically when the data are curves. J R Stat Soc Series B Stat Methodol. 1991;53:233–243. [Google Scholar]
- 37.Ramsay JO, Silverman BW. Springer Series in Statistics. 2. Springer; New York: 2005. Functional Data Analysis. [Google Scholar]
- 38.Champless LE, Diao G. Estimation of time-dependent area under the curve for long-term risk prediction. Stat Med. 2006;25:3474–3486. doi: 10.1002/sim.2299. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Liu Y, Gong P, Zhang C, Ye J Alzheimer’s Disease Neuroimaging Initiative. Hierarchical interactions model for predicting mild cognitive impairment (MCI) to Alzheimer’s disease (AD) conversion. PLoS One. 2014;9:e82450. doi: 10.1371/journal.pone.0082450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagyinszky E, Youn YC, An SS, Kim S. The genetics of Alzheimer’s disease. Clin Interv Aging. 2014;9:535–51. doi: 10.2147/CIA.S51571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, Petrella JR, Wong TZ, Doraiswamy PM Alzheimer’s Disease Neuroimaging Initiative. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. Neuroimage. 2013;78:474–480. doi: 10.1016/j.neuroimage.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.