Abstract
Purpose
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma and includes a PAX3- or PAX7-FOXO1 fusion-positive subtype. Amplification of chromosomal region 12q13-q14, which contains the CDK4 proto-oncogene, was identified in an aggressive subset of fusion-positive RMS. CDK4/6 inhibitors have antiproliferative activity in CDK4-amplified liposarcoma and neuroblastoma, suggesting CDK4/6 inhibition as a potential therapeutic strategy in fusion-positive RMS.
Experimental Procedures
We examined the biological consequences of CDK4 knockdown, CDK4 overexpression, and pharmacologic CDK4/6 inhibition by LEE011 in fusion-positive RMS cell lines and xenografts.
Results
Knockdown of CDK4 abrogated proliferation and transformation of 12q13-14-amplified and non-amplified fusion-positive RMS cells via G1-phase cell cycle arrest. This arrest was mediated by reduced RB phosphorylation and E2F-responsive gene expression. Significant differences in E2F target expression, cell cycle distribution, proliferation, or transformation were not observed in RMS cells overexpressing CDK4. Treatment with LEE011 phenocopied CDK4 knockdown, decreasing viability, RB phosphorylation, and E2F-responsive gene expression and inducing G1-phase cell cycle arrest. Though all fusion-positive cell lines showed sensitivity to CDK4/6 inhibition, there was diminished sensitivity associated with CDK4 amplification and overexpression. This variable responsiveness to LEE011 was recapitulated in xenograft models of CDK4-amplified and non-amplified fusion-positive RMS.
Conclusions
Our data demonstrate that CDK4 is necessary but overexpression is not sufficient for RB-E2F-mediated G1-phase cell cycle progression, proliferation, and transformation in fusion-positive RMS. Our studies indicate that LEE011 is active in the setting of fusion-positive RMS and suggest that low CDK4-expressing fusion-positive tumors may be particularly susceptible to CDK4/6 inhibition.
Keywords: rhabdomyosarcoma, PAX3-FOXO1, CDK4, LEE011, CDK4/6 inhibition
INTRODUCTION
Rhabdomyosarcoma (RMS) is a family of pediatric soft tissue tumors associated with the skeletal muscle lineage (1). As the most common soft tissue sarcoma of children and adolescents (2), RMS comprises two major subtypes: fusion-positive and fusion-negative. Although most fusion-positive RMS tumors are characterized by the PAX3-FOXO1 gene fusion (3), a smaller subset of cases has a PAX7-FOXO1 fusion (4). Fusion-negative RMS tumors do not harbor recurrent gene fusions. These molecular differences correspond to clinically distinct phenotypes, as fusion-positive (~80% of alveolar) RMS is more aggressive and has an unfavorable prognosis compared to fusion-negative (>95% of embryonal) RMS (5–7). This clinical difference is attributable to the propensity of fusion-positive RMS for early dissemination, poor response to therapy, and frequent relapse (6). The estimated 5-year overall survival rate for fusion-positive RMS cases is ~25% compared to ~75% for fusion-negative tumors (8), which underscores the need for more effective therapeutic strategies in fusion-positive RMS.
A particularly aggressive subset of fusion-positive RMS tumors was found to harbor amplification of chromosomal region 12q13-q14 (9, 10). This region, which contains the cyclin-dependent kinase 4 (CDK4) locus, was amplified in ~25% of PAX3-FOXO1-positive tumors and ~4% of PAX7-FOXO1-positive tumors (9). CDK4 amplification or overexpression occurs in numerous adult malignancies, including breast carcinoma, lymphoma, melanoma, and sarcoma (11–13), most notably in >95% of well-differentiated and dedifferentiated liposarcomas (14–16). In addition to RMS, CDK4 is also amplified or overexpressed in other pediatric tumor types, such as neuroblastoma (17, 18).
As one of three interphase CDKs that promote cell cycle progression from G1 to S phase, CDK4 is a well-established proto-oncogene (11, 19). Upon mitogenic stimulation, CDK4 and CDK6 form active complexes with D-type cyclins and initiate inactivation of retinoblastoma (RB) and related proteins via direct phosphorylation (20). Phosphorylation of RB proteins results in their dissociation from transcriptional repressor complexes, thereby activating E2F-dependent expression of genes that promote the G1-S-phase transition of the cell cycle and ultimately drive proliferation (19, 20). The tumor suppressor p16INK4A negatively regulates this signaling cascade by inhibiting assembly and activation of cyclin D-CDK4/6 complexes (12, 13).
Recent development of a new generation of highly selective small molecule inhibitors targeting CDK4/6 has renewed attention to CDK4/6 inhibition as a potential therapeutic strategy in various tumor types. Three orally bioavailable, selective CDK4/6 inhibitors, including PD0332991, LY2835219, and LEE011, have entered clinical trials, with PD0332991 being the most advanced in development (21). LEE011 is currently undergoing evaluation as a single agent or in combination therapy in Phase I/II studies in several tumor settings, such as breast cancer (NCT01872260, NCT02088684, NCT01919229), pediatric malignancies, including malignant rhabdoid tumors and neuroblastoma (NCT01747876), and tumors with CDK4/6 pathway activation (NCT02187783). In accordance with previous studies positively associating CDK4 amplification with vulnerability to CDK4/6 inhibition (17, 22), one criterion for inclusion in the latter trial is amplification of CDK4. This Phase II study underscores the prevailing dogma of CDK4/6-targeted therapies.
Here, we demonstrate that CDK4 is necessary for proliferation and tumor progression but overexpression is not sufficient to increase RB-E2F signaling, cell cycle progression, proliferation, or transformation in fusion-positive RMS. Additionally, we report the first preclinical evaluation of LEE011 in the setting of fusion-positive RMS and propose a novel model of tumor sensitivity to CDK4/6 inhibition in which CDK4 amplification and resultant overexpression confer reduced rather than enhanced susceptibility to LEE011 and PD0332991. Although responses varied, all fusion-positive RMS cell lines evaluated demonstrated sensitivity to CDK4/6 inhibition. Thus, our data support the clinical development of CDK4/6-targeted therapies in this refractory pediatric tumor and suggest that low CDK4-expressing fusion-positive RMS tumors may be especially vulnerable to CDK4/6 inhibition.
MATERIALS & METHODS
Cell culture
Cell lines and their source are as follows: Rh30—American Type Culture Collection; Rh28—Dr. Beverly Emanuel; Rh5, IMR5, and SKNAS—Dr. Javed Khan; CW9019—Dr. Jaclyn Biegel; Rh41 and SMS-CTR—Dr. Corinne Linardic; RD—Dr. Lee Helman; Rh18—Dr. Maria Tsokos; Rh6—Dr. Peter Houghton; OsACL—Dr. David Shapiro; primary human myoblasts—Dr. Grace Pavlath. Verification of cell line identity was performed in July 2014 by short tandem repeat genotyping analysis using the AmpFLSTR profiler plus PCR amplification kit (Applied Biosystems). Genotyping results are consistent with publicly available data and confirm that all cell lines are clonally independent. RMS and neuroblastoma cells were cultured using DMEM or RPMI-1640 media (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals) and antibiotic-antimycotic (Life Technologies) at 37 °C in 5% CO2. Primary human myoblasts were cultured as previously described (23).
Inducible RNAi expression
For isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible RNAi, the pLKO-puro-IPTG-3xLacO vector (Sigma) expressing non-targeting (NT) control shRNA (Sigma) or 3 different shRNAs targeting CDK4 (TRCN0000000362, TRCN0000000363, and TRCN0000010520; Thermo Scientific) were used. Cells were treated with IPTG (Sigma) for 48 h to induce CDK4 knockdown.
Inducible cDNA expression
The pINDUCER10 lentiviral vector (24) was provided by Dr. Ji Luo. Human CDK4 or CDK6 were PCR amplified from the CDK4-MigR1 retroviral vector (25) or the CDK6-pCMV6-XL6 vector (Origene), respectively, subcloned into the AgeI and MluI sites of pINDUCER10, and sequence verified. Cells were incubated with doxycycline (Sigma) for 24 h to induce CDK4 or CDK6 expression.
Western blot analyses
Cells were lysed with RIPA buffer containing Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Proteins were resolved on pre-cast gels using Criterion (Bio-Rad) or Bolt Mini Gel (Life Technologies) systems. Chemiluminescence was measured using a ChemiDoc XRS+ system (Bio-Rad), and images were analyzed with Image Lab software (Bio-Rad). Additional details and antibodies are provided in Supplemental methods.
Co-Immunoprecipitation (Co-IP) analysis
Dynabeads Protein G (Life Technologies) coupled to non-immune mouse serum (Santa Cruz Biotechnology, sc-2025) or anti-CDK4 antibody (Santa Cruz Biotechnology, sc-23896) was incubated with whole cell lysates (100 µg) overnight at 4 °C. After three washes, proteins were eluted in BOLT LDS sample buffer (Life Technologies) and analyzed by Western blot.
Proliferation and focus formation assays
For proliferation assays, 50,000 cells per well were seeded in a 6-well plate, and cell number was counted using a Cellometer Vision cytometer (Nexcelom Bioscience) on the days indicated by the trypan blue exclusion method. To measure cell viability using CellTiter-Glo (Promega), 5000 cells/well were seeded in 96-well plates. For focus formation assays, 1000 RMS cells were seeded in 10 cm plates in the presence of 1 million NIH 3T3 cells, and medium containing IPTG or doxycycline was replenished every 72 h. Foci were stained 2 weeks later with Giemsa solution (Sigma) and counted using ImageJ software (National Institutes of Health [NIH]).
Quantitative Real-Time-PCR (qPCR)
Total RNA was isolated from cells using the RNeasy kit (Qiagen) and was reverse transcribed with the high-capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was performed on a ViiA7 Real-Time PCR system with ViiA7 software (Applied Biosystems). Taqman gene expression assays (Life Technologies) were used to amplify CDK4 (Hs01565683), CDC25A (Hs00947994), CCNE2 (Hs00180319), and GAPDH (Hs02758991).
Pharmacologic inhibition of CDK4/6
For cell culture studies, PD0332991 (Sigma) and LEE011 (MedChem Express) were reconstituted in water and DMSO, respectively, according to manufacturer’s instructions. Cells were treated with CDK4/6 inhibitors for 72 h. IC50 values were calculated using nonlinear regression on normalized fluorescence measurements and log-transformed inhibitor concentrations (GraphPad Prism). For in vivo studies, LEE011 salt (MedChem Express) was dissolved in 0.5% methylcellulose (Sigma).
Cell cycle analysis
Cells were labeled with bromodeoxyuridine (BrdU), fixed, permeabilized, and stained using an anti-BrdU fluorescent antibody and 7-AAD according to the APC BrdU flow kit (BD Pharmingen) instructions. Flow cytometry was performed on a Becton Dickinson FACS Calibur using Cell Quest software (BD). Data were analyzed using FlowJo software (Tree Star).
Tissue microarray (TMA) of RMS patient tumors
We obtained a RMS TMA from the Children’s Oncology Group (COG) BioPathology Center. The TMA was comprised of tumor cores from control tissues and 70 diagnostic patient tumors, of which 58 tissues (n=29 fusion-positive tumors; n=29 fusion-negative cases) were evaluable.
TMAs of RMS cell line-derived xenograft tumors and Rh30-derived xenograft tumors expressing inducible CDK4-targeting shRNA
Tissues were fixed in formalin, and xenograft TMAs were constructed with 1.00 mm diameter paraffin-embedded tumor cores using a Beecher MTA-1 instrument (26). Selection of tissue for inclusion was based on review of tissues stained with hematoxylin and eosin.
Immunohistochemistry (IHC)
IHC was performed for CDK4 with a mouse monoclonal antibody from Life Technologies (clone DCS-31) at a dilution of 1:1000. Slides were deparaffinized in xylene, rehydrated in graded alcohols, and subjected to antigen retrieval in citrate buffer (pH 9) in a pressure cooker for 20 min. After incubation with primary antibody for 60 min. at room temperature, the antigen-antibody complex was detected with Dako Envision+ detection system and 3,3'-diaminobenzidine. Concurrent positive and negative controls were stained. Tissues were imaged at 100X total magnification. Immunoreactivity was scored on a scale of 0–4 for 1) staining intensity (negative, low, medium, high, very high) and 2) percentage of positive cells (0, <25%, 25–50%, >50%, ~100%). Scores of the two parameters were multiplied for a staining score of 0–16.
Fluorescence in situ hybridization (FISH)
FISH assays were performed on 5 micron formalin-fixed paraffin-embedded tumor sections using a laboratory standardized protocol with slight modifications (27). Slides were analyzed on the BioView Duet-3 fluorescent scanning station using 63X-oil objective and SpectrumOrange/SpectrumGreen/DAPI single band pass filters (Semrock). For detection of 12q13-14 amplification, a FISH probe mix consisting of BAC clones RP11-571M6 and RP11-970A5 labeled with Spectrum Red and a chromosome 12 control probe labeled with Spectrum Green or Aqua were used. The probes were obtained from Empire Genomics. For each specimen, a minimum of 60 qualifying interphase nuclei were analyzed. We calculated the average numbers of 12q and control signals and their ratio. The sample was called positive for amplification if the ratio was ≥ 2 or the average 12q copy number was ≥ 6.0 and the sample was called negative if the ratio was < 2 or the average 12q copy number was < 4.0.
Expression microarray analysis of CDK4 knockdown
Total RNA (100 ng) was run on Affymetrix Human Transcriptome 2.0 GeneChip arrays at Frederick National Laboratory for Cancer Research (FNLCR). Genes downregulated upon CDK4 knockdown were defined as those with expression changes < 0.8. Unsupervised clustering of genes with E2F-binding sites was performed using the Molecular Signatures Database (MSigDB; Broad Institute) v4.0 gene set V$E2F1DP2_01 (C3: TFT collection) (28). Additional methods are described in Supplemental materials.
Microarray analysis of tumor samples with and without 12q13-q14 amplification
Twelve tumors (n=6 with 12q13-q14 amplification; n=6 without 12q13-q14 amplification) were selected from 57 cases reported in a previous study (9). Criteria for selection included PAX3-FOXO1 expression, prior analysis by copy number arrays (9), and availability of RNA. Oligonucleotide microarray expression analysis was performed on the 12 selected cases using the Affymetrix GeneChip Human Gene 1.0 ST Array at the University of Pennsylvania Microarray Facility. We defined a gene as overexpressed in a sample if its expression level was >1 standard deviation (SD) from the mean expression level of that gene across all samples. More stringent analysis was also performed by defining overexpression as >2 SD from the mean. The total number of overexpressed genes in each sample was used as a relative measure of E2F pathway activation. Additional details are described in Supplemental methods.
Xenograft tumor models
Female, 4–6-week-old NOD/SCID mice were purchased from Charles River Laboratories or FNLCR. Two million cells were injected orthotopically into the gastrocnemius muscle of the left hind leg. For IPTG-inducible CDK4 knockdown studies, drinking water containing 10 mM dioxane-free IPTG (Fisher Scientific) was replenished every 72 h. In all xenograft studies, tumor volume was determined according to the formula length × width2 × π/6, where length represents the largest tumor diameter and width represents the perpendicular tumor diameter. All animal studies were conducted in accordance with NIH Animal Care and Use Committee guidelines.
RESULTS
CDK4 is overexpressed in fusion-positive RMS harboring amplification of chromosomal region 12q13-q14
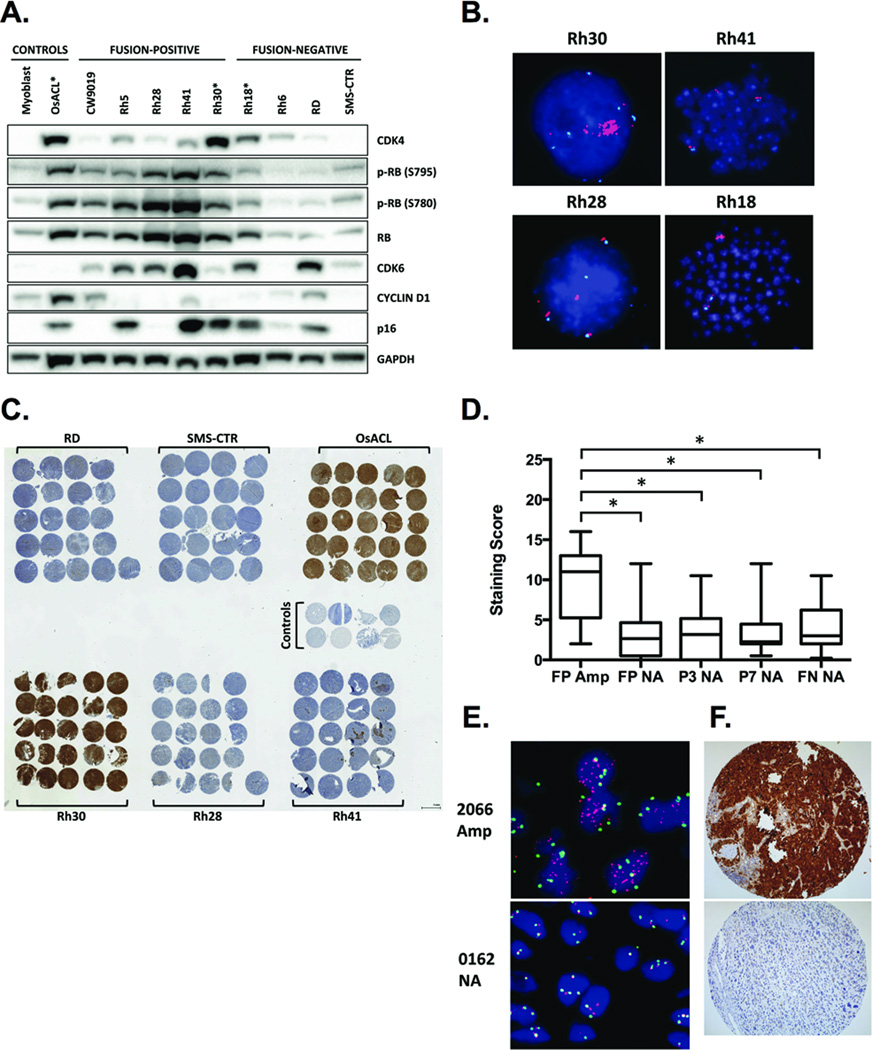
We investigated whether CDK4, a well-characterized proto-oncogene, drives the oncogenic phenotype in fusion-positive RMS with 12q13-q14 amplification. To test this, we first evaluated CDK4 protein expression in RMS cell lines. As predicted, CDK4 levels were highest in Rh30 cells, which harbor the 12q13-q14 amplicon, compared to fusion-positive and fusion-negative cells lacking 12q13-q14 amplification (Fig. 1A and 1B). Low-level amplification of the 12q13-q14 region was also evident in Rh18 cells (Fig. 1B), which expressed elevated CDK4 levels relative to non-amplified, fusion-negative cells but decreased CDK4 compared to Rh30 and the CDK4-amplified (29) osteosarcoma cell line OsACL (Fig. 1A). Xenograft tumors derived from fusion-positive and fusion-negative RMS cell lines confirmed immunoblot data, as only Rh30-derived xenografts demonstrated CDK4 staining intensity comparable to those derived from OsACL (Fig. 1C).
Figure 1. CDK4 is overexpressed in fusion-positive RMS harboring 12q13-q14 amplification.
A, Western blot analysis of a panel of fusion-positive and fusion-negative RMS cell lines. Primary human myoblasts are normal cells included as a negative control, and OsACL is a CDK4-amplified osteosarcoma cell line included as a positive control. Asterisks denote cell lines with 12q13-q14 amplification. B, Copy number analysis of the 12q13-q14 region by FISH in one fusion-negative and three fusion-positive RMS cell lines. Red, 12q13-14 probe; green or aqua, chromosome 12 centromere probe; blue, DAPI-stained nuclei. C, IHC staining for CDK4 in intramuscular xenograft tumors derived from fusion-positive and fusion-negative RMS cell lines. OsACL is a 12q13-q14-amplified osteosarcoma cell line included as a positive control. CDK4 positivity in non-amplified cell lines, such as Rh41, is associated with necrosis and thus is artifactual. D, CDK4 protein expression analyzed by IHC in a TMA comprising fusion-positive (FP), including PAX3-FOXO1 (P3) and PAX7-FOXO1 (P7) cases, and fusion-negative (FN) tumors harboring 12q13-14 amplification (Amp) or no amplification (NA). * p < 0.05 by Student’s t-test. E, Representative images of chromosomal region 12q13-q14 copy number analysis by FISH (red, 12q13-14 probe; green, chromosome 12 centromere probe; blue, DAPI-stained nuclei) and (F) CDK4 protein expression analysis by IHC in tumors from patients with (2066) and without (0162) 12q13-q14 amplification.
We further examined CDK4 protein levels in RMS patient samples to verify that CDK4 overexpression occurs in RMS tumors with 12q13-q14 amplification and is not simply a cell culture phenomenon. We used FISH to analyze copy number of the 12q13-q14 region in a TMA including 29 fusion-positive RMS tumors and 29 fusion-negative RMS tumors from the COG (Fig. 1D and 1E). Of the 29 fusion-positive cases, 10 tumors harbored amplification of 12q13-q14 (9 of 21 PAX3-FOXO1-positive; 1 of 8 PAX7-FOXO1-positive). Two of 29 fusion-negative tumors also contained the 12q13-14 amplicon (data not shown). These findings are consistent with previous data indicating preferential occurrence of 12q13-q14 amplification in fusion-positive, particularly PAX3-FOXO1-positive, RMS tumors (9). Immunohistochemical analyses of the TMA revealed significantly higher CDK4 protein expression in tumors with 12q13-q14 amplification compared to tumors lacking amplification (Fig. 1D and 1F). Taken together, these data demonstrate that CDK4 is overexpressed in fusion-positive RMS cell lines and tumors harboring the 12q13-q14 amplicon.
Depletion of CDK4 inhibits fusion-positive RMS cell proliferation and transformation via RB-E2F-mediated G1-phase cell cycle arrest
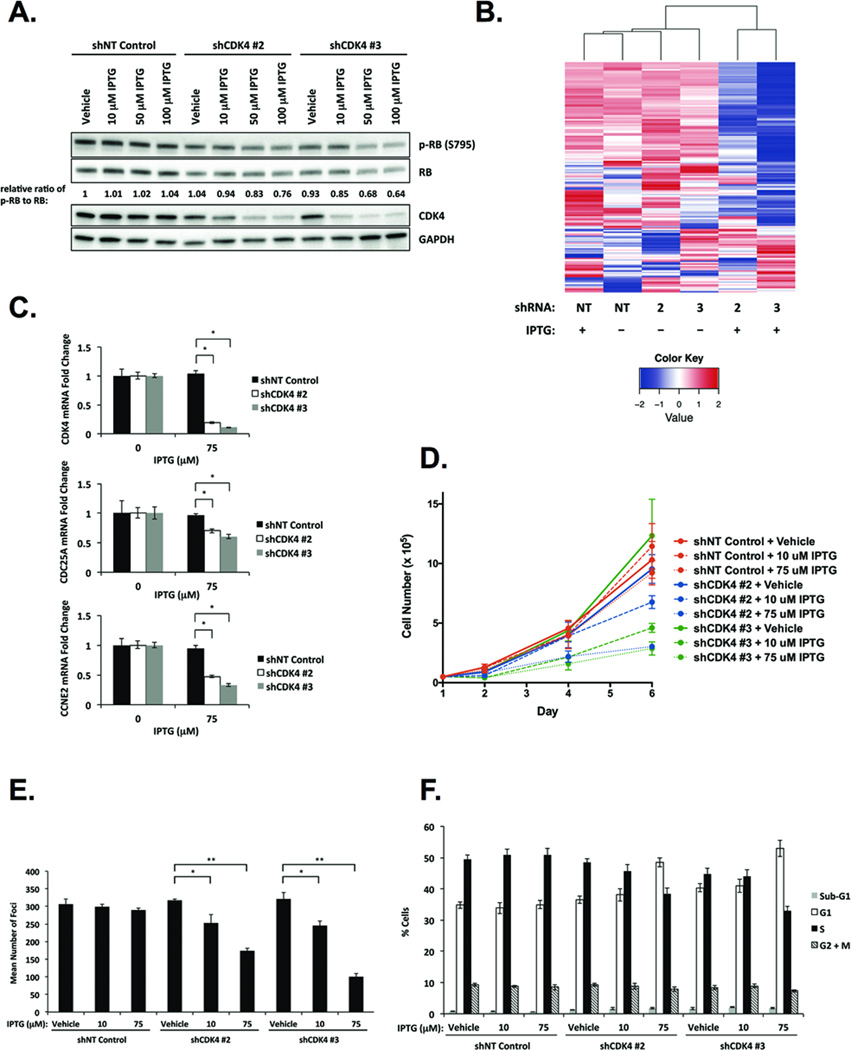
We next investigated whether high-level expression of the CDK4 oncoprotein is required for proliferation and transformation in fusion-positive RMS with amplification of 12q13-q14. To test this, we generated Rh30 cells stably expressing IPTG-inducible NT control shRNA or three different IPTG-inducible shRNAs targeting CDK4 (shCDK4). CDK4-targeting shRNAs #2 and #3 were employed, as they yielded the greatest reduction in CDK4 protein (Supplemental Fig. 1A). Treatment of shCDK4 #2 or #3-expressing cells with IPTG resulted in CDK4 mRNA and protein depletion in a dose-dependent manner, with 75–100 µM IPTG inducing maximal knockdown of CDK4 (Supplemental Fig. 1B and 1C). Levels of CDK4 knockdown achieved by 75 µM IPTG were generally comparable to endogenous CDK4 protein expression in non-amplified RMS cell lines (Supplemental Fig. 1A). Consistent with canonical cyclin D1/CDK4-RB signaling (20, 30) and previous studies of CDK4 knockdown in other tumor categories (22), phosphorylation of RB at Ser795–a CDK4-specific phosphorylation residue (31)–was diminished in Rh30 cells depleted of CDK4 (Fig. 2A). CDK4 knockdown also resulted in reduced total RB, though RB hypophosphorylation was the predominant effect, as evidenced by decreased phospho-RB to RB ratios compared to NT control or vehicle-treated shCDK4-expressing cells (Fig. 2A). Reduction in both RB phosphorylation and total RB was observed upon CDK4 knockdown in previous studies (17, 22), suggesting that CDK4 may regulate RB via phosphorylation as well as stability/degradation mechanisms (32).
Figure 2. Depletion of CDK4 represses fusion-positive RMS cell proliferation and transformation via RB-E2F-mediated cell cycle arrest.
A, Western blot analysis of CDK4 and RB upon IPTG-inducible CDK4 knockdown in Rh30 cells. Intensity ratios of phospho-RB to RB are normalized to the ratio in vehicle-treated cells expressing NT control shRNA. B, Unsupervised clustering of genes containing the 8-nucleotide consensus E2F-binding site categorizes Rh30 cells expressing NT control shRNA or vehicle-treated (−) cells expressing IPTG-inducible CDK4 shRNAs #2 and #3 versus IPTG-treated (+) cells expressing IPTG-inducible CDK4 shRNAs #2 and #3 into two distinct clusters. IPTG-inducible depletion of CDK4 (C) diminishes CDC25A and CCNE2 mRNA expression, (D) abrogates cell proliferation, (E) inhibits focus formation, and (F) arrests cells in G1-phase of the cell cycle. Data represent the mean ± SD of three independent experiments performed in triplicate. * p < 0.005, ** p < 0.00005 by Student’s t-test.
Given that RB phosphorylation leads to transcriptional derepression of E2F-regulated gene expression (20), we predicted that CDK4 knockdown would result in reduced expression of E2F target genes. To test this, Rh30 cells stably expressing IPTG-inducible NT control shRNA or shCDK4s #2 and #3 were treated with vehicle or 75 µM IPTG (Fig. 2B) and analyzed by expression microarray. We interrogated whether cells expressing CDK4 relative to cells with CDK4 knockdown could be categorized according to differential expression of E2F-responsive genes. Indeed, unsupervised analysis of genes containing the 8-nucleotide core E2F-binding site (33–35) classified CDK4-expressing compared to CDK4-depleted Rh30 cells into two separate clusters (Fig. 2B). To validate E2F-responsive gene expression changes identified by microarray analysis, we selected two E2F targets, CDC25A and CCNE2 (36–40), and analyzed their expression by qPCR in Rh30 cells with CDK4 knockdown. Consistent with microarray data, mRNA levels of CDC25A and CCNE2 were reduced in cells depleted of CDK4 compared to vehicle-treated cells (Fig. 2C). These results confirm the role of CDK4 as an upstream component in the RB-E2F signaling axis (20) and collectively indicate that, in CDK4-amplified Rh30 cells, high CDK4 expression is required for E2F target gene expression.
To investigate the functional role of CDK4 in the 12q13-q14 amplicon, we evaluated the consequences of CDK4 knockdown on cell proliferation and oncogenic transformation in Rh30 cells. In cells depleted of CDK4, we observed decreased proliferation rates relative to NT control-expressing cells (Fig. 2D). Suppression of proliferation correlated with the degree of CDK4 reduction, as CDK4 knockdown induced by 75 µM IPTG attenuated proliferation more dramatically than that induced by 10 µM IPTG. Similarly, focus formation was reduced upon CDK4 knockdown in an IPTG dose-dependent manner, suggesting that CDK4 is necessary for the transformed phenotype of 12q13-q14 amplified, fusion-positive RMS (Fig. 2E).
Given that CDK4 promotes G1 to S-phase progression in the cell cycle (11), we hypothesized that the diminished proliferative and transformative capacity of cells depleted of CDK4 is mediated by G1-phase cell cycle arrest. Indeed, flow cytometry analyses revealed accumulation of cells in G1 upon CDK4 knockdown (Fig. 2F). The percentage of cells in the sub-G1 phase was unchanged by CDK4 depletion, suggesting that apoptosis is not a major mechanism underlying repression of proliferation and transformation in 12q13-q14 amplified, fusion-positive RMS. Moreover, we observed no morphological differences (data not shown), and levels of myogenin and myosin heavy chain–markers of early and late myogenic differentiation, respectively–remained constant in cells depleted of CDK4 compared to control cells, indicating that reduced proliferation and focus formation are not attributable to cells undergoing differentiation (Supplemental Fig. 1D). Collectively, these data demonstrate that CDK4 is necessary for RB-E2F-mediated cell cycle progression from G1 to S phase, thereby promoting proliferation and transformation in fusion-positive RMS cells with 12q13-q14 amplification.
Additionally, we examined the functional consequences of IPTG-inducible CDK4 depletion in fusion-positive RMS cells that lack 12q13-q14 amplification (Fig. 1B). IPTG treatment of Rh41 cells stably expressing shCDK4s #2 and #3 induced CDK4 depletion in a dose-dependent manner at the protein (Supplemental Fig. 2A) and mRNA levels (Supplemental Fig. 2B). As endogenous CDK4 expression is lower in Rh41 cells compared to Rh30 cells (Fig. 1A), CDK4 knockdown in this non-amplified cell line diminished CDK4 protein to nearly undetectable levels (Supplemental Fig. 2A). Knockdown of CDK4 inhibited proliferation (Supplemental Fig. 2C) and focus formation (Supplemental Fig. 2D) in Rh41 cells as robustly as that in Rh30 cells (Fig. 2D and 2E), suggesting that CDK4 is necessary for proliferation and transformation in fusion-positive RMS independent of 12q13-q14 amplification status.
CDK4 knockdown suppresses tumor growth of fusion-positive RMS harboring 12q13-q14 amplification in vivo
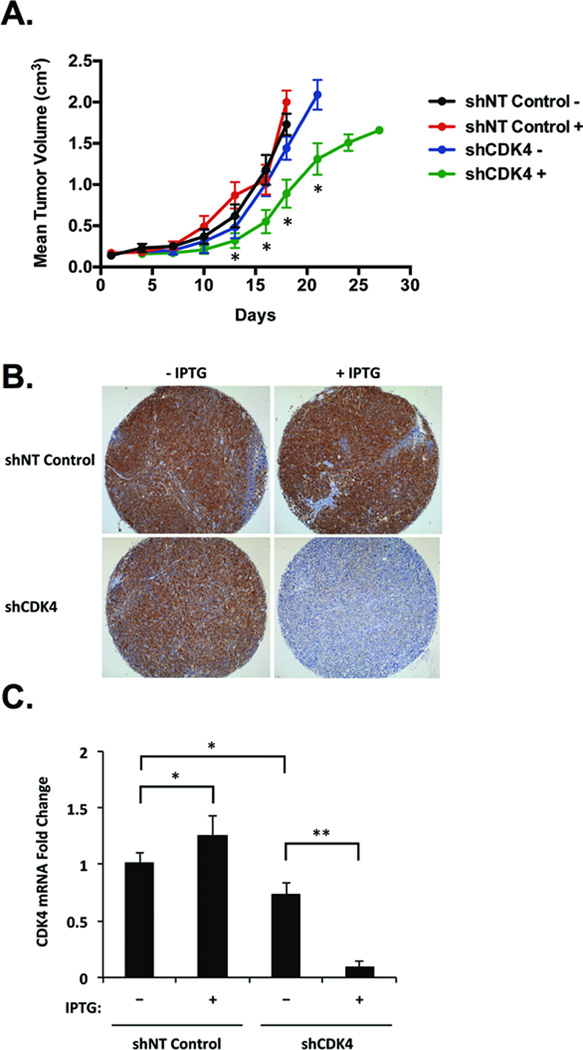
We next investigated whether depletion of CDK4 could repress 12q13-q14-amplified, fusion-positive RMS tumor growth in vivo. Intramuscular xenograft tumors were derived from Rh30 cells stably expressing IPTG-inducible NT control shRNA or shCDK4 #3, and mice received either no IPTG or 10 mM IPTG in the drinking water after formation of a palpable tumor. In mice with shCDK4 that received IPTG, tumor progression was significantly retarded compared to NT control or vehicle-treated shCDK4 xenograft tumors (Fig. 3A). CDK4 knockdown was apparent at the protein (Fig. 3B) and mRNA levels (Fig. 3C) in xenograft tumors induced to express shCDK4. Reduced CDK4 mRNA levels were evident in vehicle-treated shCDK4 tumors relative to NT control tumors (Fig. 3C), though tumor growth rates in these cohorts were comparable (Fig. 3A). Thus, leaky expression of CDK4-targeting shRNA appears to be inconsequential, as it failed to manifest phenotypically in our xenograft models. Consistent with previous reports, we observed no deleterious effects of IPTG (41). These data indicate that CDK4 is required for optimal tumor progression in 12q13-q14-amplified, fusion-positive RMS.
Figure 3. CDK4 knockdown retards fusion-positive RMS tumor growth in vivo.
Mice were injected into the gastrocnemius muscle with Rh30 cells stably expressing NT control shRNA or shRNA #3 targeting CDK4 and were randomized to receive no IPTG (n=10) or 10 mM IPTG in the drinking water (n=10) upon formation of a palpable tumor. Mice bearing Rh30-derived intramuscular xenograft tumors expressing shRNA targeting CDK4 and receiving IPTG (+) compared to shCDK4-expressing tumors receiving no IPTG (−) or to tumors expressing NT control shRNA treated with (+) or without (−) IPTG show (A) delayed tumor growth (* p < 0.005 by 2-tailed t-test for shCDK4 + versus each other group), (B) reduced CDK4 protein levels by IHC analysis, and (C) decreased CDK4 mRNA expression by qPCR analysis. qPCR data represent the mean ± SD of quadruplicate samples from at least five mice per treatment group. * p < 0.05, ** p < 0.0005 by Student’s t-test.
CDK4 overexpression fails to increase RB-E2F signaling, cell cycle progression, proliferation, or transformation in fusion-positive RMS lacking 12q13-q14 amplification
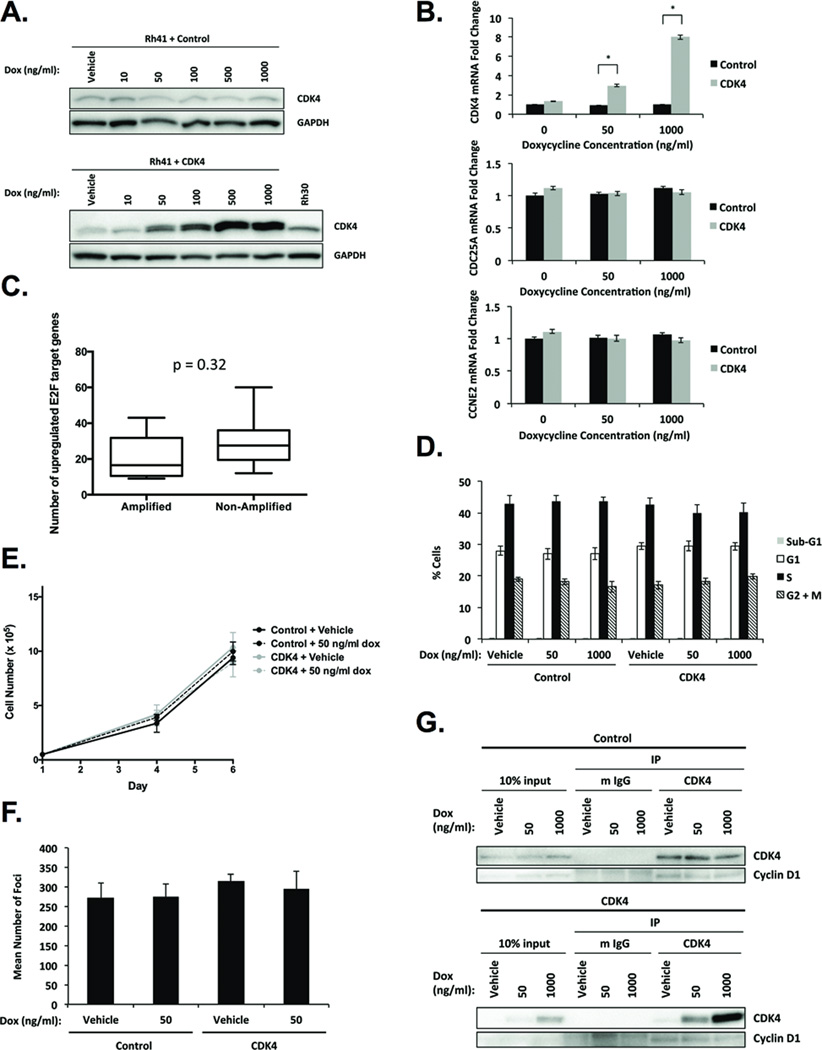
Given that CDK4 knockdown resulted in diminished RB-E2F signaling (Fig. 2A, 2B, and 2C), we investigated whether E2F-regulated transcription would increase in response to CDK4 overexpression. To test this, we generated Rh41 cells stably expressing doxycycline-inducible empty vector control or CDK4. CDK4 protein levels in CDK4-transduced cells treated with 50 ng/ml doxycycline were comparable to endogenous CDK4 protein expression in 12q13-q14-amplified Rh30 cells, and 1 µg/ml doxycycline resulted in maximal CDK4 expression (Fig. 4A). CDC25A and CCNE2 mRNA levels were unaffected by CDK4 overexpression induced by either doxycycline concentration (Fig. 4B), suggesting that CDK4 overexpression is not sufficient to enhance E2F-responsive gene expression in Rh41 cells.
Figure 4. CDK4 overexpression fails to alter RB-E2F signaling, cell cycle progression, proliferation, or transformation in fusion-positive RMS.
A, Western blot analysis of Rh41 cells stably expressing doxycycline-inducible empty vector control or CDK4. B, qPCR analysis indicates that CDC25A and CCNE2 mRNA expression is unaffected by doxycycline-inducible CDK4 overexpression. Data represent the mean ± SD of quadruplicate samples from three independent experiments. * p < 0.0005 by Student’s t-test. C, The number of upregulated genes harboring E2F-binding motifs per sample is comparable between fusion-positive RMS tumors with and without 12q13-14 amplification. p = 0.32 by Student’s t-test. Doxycycline-inducible CDK4 overexpression has no significant effect on (D) cell cycle distribution, (E) cell proliferation, or (F) focus formation. G, Co-IP analysis of CDK4 and cyclin D1 in Rh41 cells stably expressing doxycycline-inducible empty vector control or CDK4.
To further examine this issue in human tumor samples, we compared genome-wide expression in fusion-positive RMS tumors with and without 12q13-q14 amplification. Although CDK4 as well as other genes in the amplicon were significantly overexpressed in tumors harboring 12q13-q14 amplification versus those lacking amplification, no significant difference in expression of RB-E2F pathway genes was observed between cohorts (data not shown). As an alternative strategy, we analyzed expression of genes containing the consensus E2F-binding site (33) in these 12q13-q14-amplified and non-amplified fusion-positive RMS tumors but did not detect any significant difference between groups in the number of overexpressed E2F-responsive genes per sample (Fig. 4C; p=0.32).
Next, we investigated the functional consequences of overexpressing CDK4 in Rh41 cells. Consistent with unaltered expression of downstream RB targets, CDK4 overexpression failed to significantly enhance cell cycle progression (Fig. 4D), proliferation (Fig. 4E), or focus formation (Fig. 4F). Co-IP experiments demonstrate that levels of CDK4-associated cyclin D1 remain constant as CDK4 expression increases (Fig. 4G), suggesting that CDK4 overexpression has no significant functional effects because cyclin D1 levels are limiting. Importantly, overexpression of CDK4 in the absence of cyclin D1 overexpression recapitulates the CDK4 and cyclin D1 expression profiles identified in our analyses of fusion-positive RMS tumors with and without 12q13-q14 amplification. Taken together, these data indicate that CDK4 overexpression is not sufficient to increase E2F-regulated transcription, G1-S-phase cell cycle transition, proliferation, or transformation in fusion-positive RMS.
Fusion-positive RMS cell lines are sensitive to pharmacologic inhibition of CDK4/6
Given the marked attenuation of proliferation and transformation observed in 12q13-q14-amplified and non-amplified fusion-positive RMS cells depleted of CDK4, we hypothesized that CDK4/6 inhibition may be a promising therapeutic strategy in fusion-positive RMS. Comparable RB-E2F signaling between 12q13-14-amplified and non-amplified RMS tumors, however, implied that CDK4 overexpression might be less exploitable than in other tumor contexts for CDK4-targeted therapeutic response. We first interrogated the antiproliferative effects of pharmacologic CDK4/6 inhibition on fusion-positive RMS in vitro by treating five fusion-positive RMS cell lines with a five-log dose range (0.01–100 µM) of LEE011. RMS cell lines demonstrated differential sensitivity to CDK4/6 inhibition, with Rh28 cells being the most sensitive and Rh30 cells being the least sensitive (Fig. 5A). Sensitivity to LEE011 suggested an inverse correlation with CDK4 protein expression, though this association is based on a small number of cell lines (Fig. 1A). Relative sensitivities of fusion-positive cell lines to the CDK4/6 inhibitor PD0332991 also corresponded to CDK4 protein levels and were consistent with the range of sensitivities observed upon LEE011 treatment (Supplemental Fig. 3A). Although we focused evaluation of CDK4/6 inhibition on fusion-positive RMS cell lines, preliminary studies of fusion-negative RMS cell lines generally revealed attenuated sensitivity to LEE011 compared to fusion-positive cells (Supplemental Fig. 3B). In accordance with their relative insensitivity to CDK4/6 inhibition, fusion-negative RMS cell lines displayed lower levels of RB than fusion-positive cell lines (Fig. 1A).
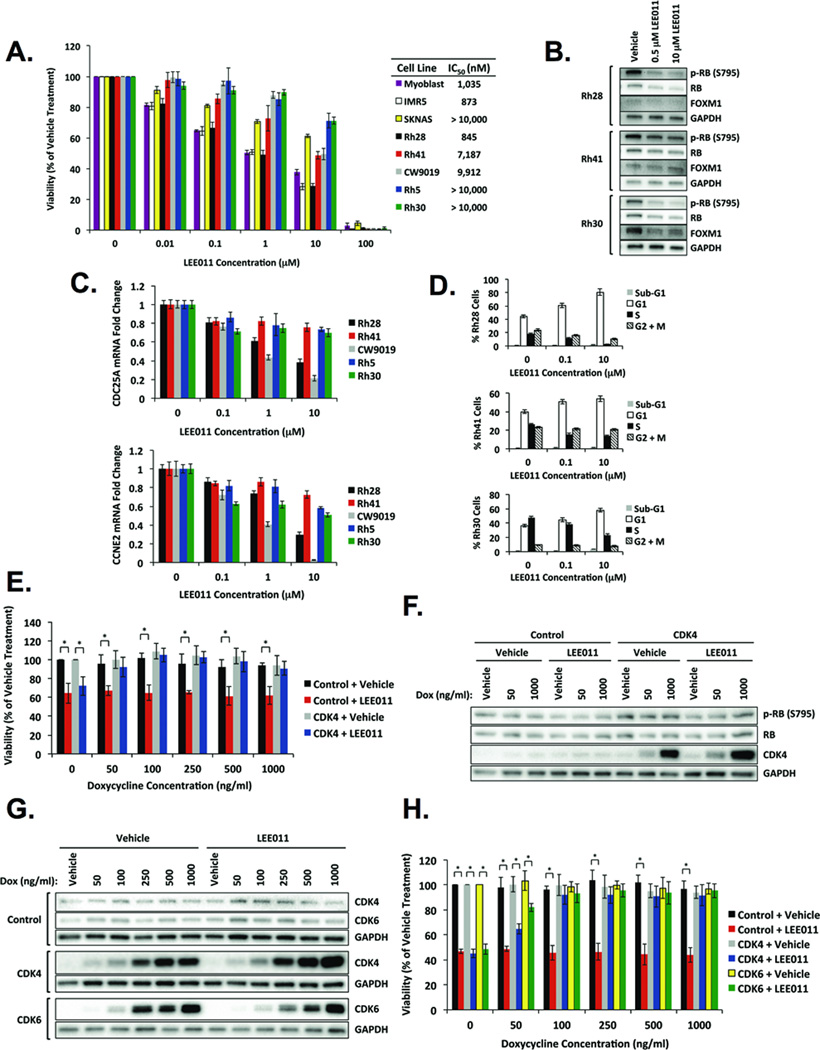
Figure 5. Fusion-positive RMS cell lines exhibit sensitivity to pharmacologic inhibition of CDK4/6.
A, LEE011 decreases viability of fusion-positive RMS cells. Primary human myoblasts represent normal cells, and IMR5 and SKNAS are neuroblastoma cell lines indicative of high and low sensitivity to LEE011, respectively. Fusion-positive cells treated with LEE011 demonstrate (B) diminished levels of FOXM1 and phospho-RB by Western blot analysis, (C) reduced CDC25A and CCNE2 mRNA expression by qPCR analysis, and (D) accumulation in G1-phase of the cell cycle by flow cytometry analysis. E, Doxycycline-inducible overexpression of CDK4 enhances viability of Rh41 cells treated with LEE011. Cells were treated with a range of doxycycline concentrations in the presence of vehicle or 10 µM LEE011. Data represent the mean ± SD of five replicates from three independent experiments. * p < 0.005 by Student’s t-test. F, Western blot analysis of Rh41 cells stably expressing doxycycline-inducible empty vector control or CDK4 treated with two different doxycycline concentrations in the presence of vehicle or 500 nM LEE011. G, Western blot analysis of Rh28 cells stably expressing doxycycline-inducible empty vector control, CDK4, or CDK6 treated with the indicated doxycycline concentrations in the presence of vehicle or 1 µM LEE011. H, Doxycycline-inducible overexpression of CDK4 or CDK6 increases viability of Rh28 cells treated with LEE011. Cells were treated with a range of doxycycline concentrations in the presence of vehicle or 1 µM LEE011. Data represent the mean ± SD of five replicates from three independent experiments. * p < 0.005 by Student’s t-test.
If LEE011 specifically inhibits CDK4/6, then we predicted that LEE011 treatment should phenocopy CDK4 depletion. Indeed, RB phosphorylation and FOXM1, another CDK4 target (42), were reduced in cells treated with LEE011 (Fig. 5B). Moreover, mRNA levels of E2F targets, including CDC25A and CCNE2, were diminished in all fusion-positive RMS cell lines evaluated, though decreases in their expression did not strictly correlate with relative sensitivities to LEE011 (Fig. 5C). As observed upon CDK4 knockdown (Fig. 2F) and consistent with previous studies of CDK4/6 inhibition (17, 22, 43), treatment with LEE011 resulted in G1-phase cell cycle arrest (Fig. 5D). Rh28 cells, which exhibited the highest sensitivity to CDK4/6 inhibition (Fig. 5A), also demonstrated the greatest accumulation of cells in G1, implying that the antiproliferative effects of LEE011 on fusion-positive RMS cell lines are mediated by G1-phase cell cycle arrest.
Based on our findings that low CDK4-expressing Rh28 cells were most sensitive to LEE011 while high CDK4-expressing Rh30 cells were least sensitive, we investigated whether overexpression of CDK4 attenuates sensitivity to CDK4/6 inhibition in fusion-positive RMS. To test this in an isogenic system, we treated Rh41 cells stably expressing doxycycline-inducible empty vector control or CDK4 with 10 µM LEE011 and a range of doxycycline concentrations (50–1000 ng/ml). Viability of control cells and CDK4-transduced cells not receiving doxycycline was reduced by ~30% upon LEE011 treatment (Fig. 5E). In contrast, viability of CDK4-transduced cells treated with 50–1000 ng/ml doxycycline was not significantly decreased by LEE011 relative to vehicle-treated cells (Fig. 5E). Similar results were found using PD0332991 (Supplemental Fig. 3C). Consistent with enhanced viability (Fig. 5E and Supplemental Fig. 3C), rescue of phospho-RB and total RB was evident in LEE011-treated cells overexpressing CDK4 (Fig. 5F).
Given that LEE011 is a dual CDK4/6 inhibitor, we reasoned that CDK6 expression might also influence responsiveness to LEE011 in fusion-positive RMS. As Rh41 cells endogenously express the highest levels of CDK6 (Fig. 1A), we employed another isogenic system in which Rh28 cells stably expressing doxycycline-inducible control, CDK4, or CDK6 constructs were treated with 1 µM LEE011 and 50–1000 ng/ml doxycycline (Fig. 5G). LEE011 decreased viability of control cells and non-doxycycline-treated CDK4- or CDK6-transduced cells by ~50%, and viability of CDK4- and CDK6-transduced cells treated with 100–1000 ng/ml doxycycline was rescued (Fig. 5H). These data demonstrate that fusion-positive RMS cell lines are sensitive to LEE011, and overexpression of CDK4 or CDK6 renders cells less sensitive to CDK4/6 inhibition.
CDK4/6 inhibition abrogates fusion-positive RMS tumor growth in vivo
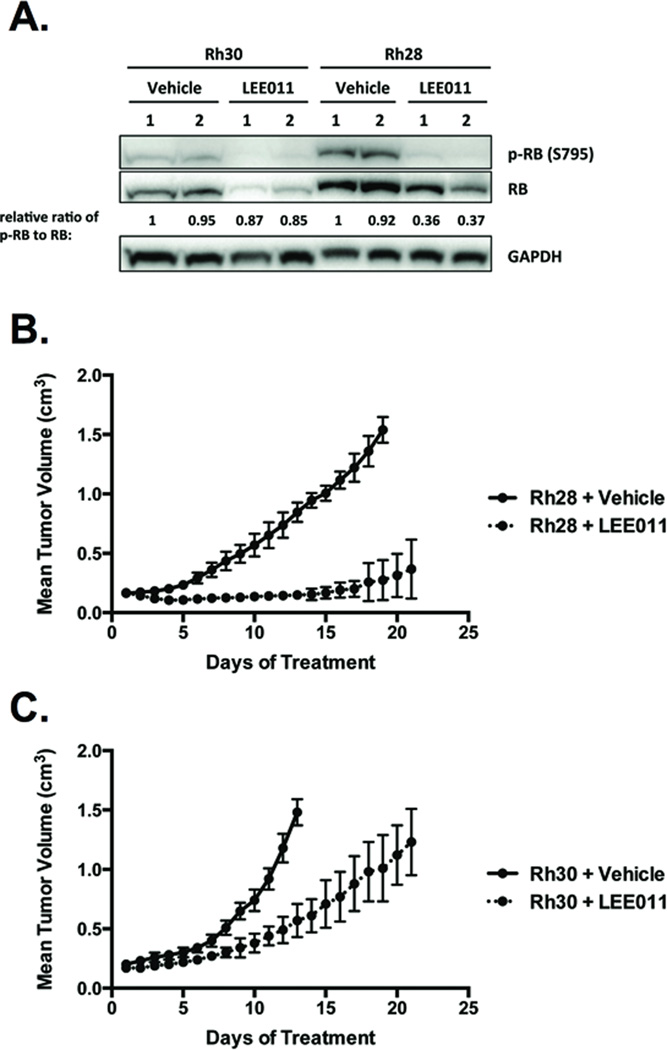
We interrogated whether the relative in vitro sensitivities of fusion-positive RMS cells to CDK4/6 inhibition could be recapitulated in xenograft tumors derived from Rh30 and Rh28 cells, which represented the extremes of response measured in cell culture. As observed in vitro, LEE011 reduced RB phosphorylation (Fig. 6A). Consistent with CDK4 knockdown experiments (Fig. 2A) and prior investigation of LEE011 in vivo (22), total RB levels also decreased in response to LEE011 (Fig. 6A), further supporting CDK4-mediated regulation of RB at the levels of phosphorylation and stability/degradation (32). LEE011 significantly delayed tumor progression in both models, but activity was more robust in Rh28-derived (Fig. 6B) compared to Rh30-derived (Fig. 6C) xenograft tumors. Similar to previous studies (17, 22), we dosed mice with 200 mg/kg daily; however, Phase I studies of LEE011 defined the recommended Phase II dose at 600 mg/day (44). Despite the high dosage administered relative to that achievable in patients, LEE011 was well tolerated, as no significant weight loss or adverse events were observed in mice (data not shown). These data demonstrate in vivo antitumor activity of LEE011 as a single agent in fusion-positive RMS and are consistent with the proposed inverse relationship between CDK4 expression and CDK4/6 inhibitor sensitivity in this pediatric tumor setting.
Figure 6. CDK4/6 inhibition antagonizes fusion-positive RMS tumor growth in vivo.
Mice were injected intramuscularly with Rh30 or Rh28 cells. After formation of a palpable tumor, mice were randomized to receive vehicle (n=10) or 200 mg/kg LEE011 (n=10) daily by oral gavage for 21 days. A, LEE011 diminishes phosphorylation of RB in xenograft tumors derived from Rh30 and Rh28 cells. Intensity ratios of phospho-RB to RB are normalized to the ratio in vehicle-treated mouse #1. LEE011 abrogates tumor progression in (B) Rh28-derived and (C) Rh30-derived xenograft models.
DISCUSSION
In the context of fusion-positive RMS, we introduce a novel model of CDK4-amplified tumor sensitivity in which CDK4 amplification and resultant overexpression reduce rather than enhance sensitivity to CDK4/6 inhibition. Analysis of the antiproliferative effects of pharmacologic CDK4/6 inhibitors provided initial evidence of this notion, as lowest CDK4-expressing Rh28 cells showed the highest sensitivity to both LEE011 and PD0332991, while highest CDK4-expressing Rh30 cells exhibited the lowest sensitivity. Moreover, rescue of cell viability upon overexpression of CDK4 in Rh41 and Rh28 cells treated with LEE011 indicated that elevated CDK4 expression confers decreased sensitivity to CDK4/6 inhibition. Our in vivo results of robust LEE011 activity in Rh28-derived xenograft tumors compared to more modest, albeit statistically significant, antitumor effects in Rh30-derived xenografts further support this finding. Thus, our studies oppose the predominant paradigm in which target overexpression renders tumors more vulnerable to target-directed inhibition—the premise of molecularly targeted therapy and personalized medicine. The potential inverse relationship between CDK4 expression and inhibitor sensitivity in fusion-positive RMS is not unprecedented, as BCR-ABL amplification and overexpression is a well-characterized mechanism of imatinib resistance in chronic myeloid leukemia (45–47). As our data demonstrate no significant functional consequences of CDK4 overexpression, stoichiometric competition is the likely resistance mechanism underlying the relationship between CDK4 expression and LEE011 sensitivity in fusion-positive RMS.
Although sensitivities of Rh30 and Rh28 cells, which represented the lower and upper limits of responses, respectively, to LEE011 suggested an inverse correlation between CDK4 expression and inhibitor sensitivity, neither the relative expression of CDK4 nor CDK6 exactly correlated with the antiproliferative effects of LEE011 in fusion-positive RMS cell lines. Similarly, previous studies of PD0332991 in RMS (43) and LEE011 in non-CDK4-amplified neuroblastoma (17) found no strict association between CDK4 or CDK6 levels and response to CDK4/6 inhibition. It is clear, however, that sensitivity to CDK4/6 inhibitors depends on the presence of functional RB. Indeed, RB status has been proposed as a selective biomarker of CDK4/6 inhibitor utility (22), though additional work is needed to refine such predictive biomarkers of response. Investigations in ovarian cancer revealed that high RB and low p16 levels conferred greatest sensitivity to PD0332991 (48). Interestingly, in our studies, Rh28 cells, which exemplified this model of high RB and low p16 expression, were the most responsive to LEE011 and PD0332991, warranting further examination of this expression signature as a potentially useful biomarker in predicting response to CDK4/6 inhibition in the context of fusion-positive RMS.
Our preliminary evaluation of fusion-negative RMS cell lines showed lower RB protein levels and relative resistance to LEE011 compared to fusion-positive cell lines. In one study of RMS tumors, RB protein expression was generally comparable between fusion-negative and fusion-positive cases (49); however, protein expression of cyclin E and E2F1—both E2F targets (37)—was significantly elevated in fusion-positive relative to fusion-negative tumors (49). This finding suggests that fusion-positive tumors exhibit enhanced activation of the RB-E2F pathway compared to fusion-negative tumors, a molecular observation that may contribute both to the more aggressive clinical behavior of the fusion-positive category and its higher sensitivity to CDK4/6 inhibitors. Further characterization of RB in the setting of fusion-negative RMS will provide insight into the utility of CDK4/6 inhibition as a viable therapeutic strategy for RMS tumors lacking PAX-FOXO1 expression.
In addition to RB status, MYCN amplification has been proposed as a predictive biomarker of response to CDK4/6 inhibition, as MYCN-amplified neuroblastoma cell lines showed higher sensitivity to LEE011 than non-amplified cells (17). Several studies have identified MYCN amplification in RMS (1), with predominant occurrence in fusion-positive tumors (50). Although none of the fusion-positive RMS cell lines evaluated in the present studies harbor amplification of MYCN, additional investigation of the correlation between MYCN amplification status and CDK4/6 inhibitor sensitivity is warranted in fusion-positive RMS.
Consistent with findings in well-differentiated and dedifferentiated liposarcoma (22), CDK4 knockdown studies revealed that CDK4 drives RB-E2F pathway-mediated cell cycle progression from G1 to S phase and is necessary for proliferation and transformation in fusion-positive RMS regardless of 12q13-q14 amplification status. Although CDK4 knockdown significantly suppressed xenograft tumor growth, compensation by CDK6 may have partially attenuated the observed antitumor effects. In contrast, CDK4 overexpression experiments and analysis of patient tumors indicated that CDK4 overexpression does not enhance activation of E2F-responsive gene expression in 12q13-q14-amplified relative to non-amplified fusion-positive RMS tumors. Moreover, CDK4 overexpression studies in non-amplified fusion-positive RMS cells demonstrated that CDK4 does not increase cell cycle progression, proliferation, or transformation. Co-IP experiments suggested that cyclin D1 binding is saturated with endogenous levels of CDK4, and thus cyclin D1 expression may limit the biological consequences of CDK4 overexpression in fusion-positive RMS. Analysis of fusion-positive RMS tumors with and without 12q13-q14 amplification revealed comparable cyclin D1 expression between cohorts, implying that CDK4 overexpression may not functionally contribute to the oncogenic phenotype of 12q13-q14-amplified tumors.
Based on our studies, CDK4 overexpression alone is not sufficient to recapitulate the 12q13-q14 amplification event in fusion-positive RMS. Our data suggest that other genes in the amplified region drive the aggressive phenotype in fusion-positive RMS tumors that harbor this genomic feature, encouraging further investigation of other components of the 12q13-q14 amplicon, including both coding and noncoding regions. Moreover, future studies of other overexpressed genes from this amplicon may identify additional useful targets for therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Fusion-positive rhabdomyosarcoma (RMS) represents the more aggressive, refractory subtype of this pediatric cancer. A subset of fusion-positive RMS tumors harbors amplification of the CDK4-containing chromosomal region 12q13-q14. Other tumor types with CDK4 amplification or overexpression, including liposarcoma and neuroblastoma, are sensitive to CDK4/6 inhibition, suggesting that CDK4/6-targeted therapies may provide a new treatment strategy in fusion-positive RMS. To evaluate the potential clinical utility of CDK4/6 inhibition in this disease setting, we examined the activity of LEE011, a highly selective, orally available small molecule inhibitor targeting CDK4/6, in fusion-positive RMS in vitro and in vivo. We demonstrate overall sensitivity to CDK4/6 inhibition in all fusion-positive RMS models tested, with evidence of differential antitumor activity resulting from an inverse relationship between CDK4 expression and inhibitor vulnerability. Collectively, our data provide preclinical evidence supporting further investigation of CDK4/6-targeted therapies in treatment regimens for fusion-positive RMS.
ACKNOWLEDGEMENTS
The authors thank Gail McMullen and Kris Ylaya for technical assistance with animal studies and IHC, respectively. The authors also thank Dr. Ji Luo for supplying the pINDUCER10 plasmid and Dr. Grace Pavlath for providing primary human myoblasts.
Financial Support: This research was supported by the Intramural Research Program of the National Cancer Institute and by the Joanna McAfee Childhood Cancer Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest: F.G. Barr had ownership interest in Novartis prior to January 2014. The other authors have no potential conflicts of interest to disclose.
REFERENCES
- 1.Olanich ME, Barr FG. A call to ARMS: targeting the PAX3-FOXO1 gene in alveolar rhabdomyosarcoma. Expert opinion on therapeutic targets. 2013;17:607–623. doi: 10.1517/14728222.2013.772136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Giovanni C, Landuzzi L, Nicoletti G, Lollini PL, Nanni P. Molecular and cellular biology of rhabdomyosarcoma. Future Oncol. 2009;5:1449–1475. doi: 10.2217/fon.09.97. [DOI] [PubMed] [Google Scholar]
- 3.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nature genetics. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 4.Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer research. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 5.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer. 2009;115:4218–4226. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wachtel M, Runge T, Leuschner I, Stegmaier S, Koscielniak E, Treuner J, et al. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J Clin Oncol. 2006;24:816–822. doi: 10.1200/JCO.2005.03.4934. [DOI] [PubMed] [Google Scholar]
- 7.Mercado GE, Barr FG. Fusions involving PAX and FOX genes in the molecular pathogenesis of alveolar rhabdomyosarcoma: recent advances. Curr Mol Med. 2007;7:47–61. doi: 10.2174/156652407779940440. [DOI] [PubMed] [Google Scholar]
- 8.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 9.Barr FG, Duan F, Smith LM, Gustafson D, Pitts M, Hammond S, et al. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. Genes Chromosomes Cancer. 2009;48:661–672. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park S, Lee J, Do IG, Jang J, Rho K, Ahn S, et al. Aberrant CDK4 amplification in refractory rhabdomyosarcoma as identified by genomic profiling. Scientific reports. 2014;4:3623. doi: 10.1038/srep03623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 12.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res. 2013;19:5320–5328. doi: 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 14.Binh MB, Sastre-Garau X, Guillou L, de Pinieux G, Terrier P, Lagace R, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. The American journal of surgical pathology. 2005;29:1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 15.Sirvent N, Coindre JM, Maire G, Hostein I, Keslair F, Guillou L, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. The American journal of surgical pathology. 2007;31:1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 16.Italiano A, Bianchini L, Gjernes E, Keslair F, Ranchere-Vince D, Dumollard JM, et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res. 2009;15:5696–5703. doi: 10.1158/1078-0432.CCR-08-3185. [DOI] [PubMed] [Google Scholar]
- 17.Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, et al. Dual CDK4/CDK6 Inhibition Induces Cell-Cycle Arrest and Senescence in Neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faussillon M, Monnier L, Junien C, Jeanpierre C. Frequent overexpression of cyclin D2/cyclin-dependent kinase 4 in Wilms' tumor. Cancer letters. 2005;221:67–75. doi: 10.1016/j.canlet.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 20.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33:1890–1903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 21.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res. 2014;20:3379–3383. doi: 10.1158/1078-0432.CCR-13-1551. [DOI] [PubMed] [Google Scholar]
- 22.Zhang YX, Sicinska E, Czaplinski JT, Remillard SP, Moss S, Wang Y, et al. Antiproliferative Effects of CDK4/6 Inhibition in CDK4-Amplified Human Liposarcoma In Vitro and In Vivo. Mol Cancer Ther. 2014;13:2184–2193. doi: 10.1158/1535-7163.MCT-14-0387. [DOI] [PubMed] [Google Scholar]
- 23.Pavlath GK, Gussoni E. Human myoblasts and muscle-derived SP cells. Methods in molecular medicine. 2005;107:97–110. doi: 10.1385/1-59259-861-7:097. [DOI] [PubMed] [Google Scholar]
- 24.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia SJ, Rajput P, Strzelecki DM, Barr FG. Analysis of genetic events that modulate the oncogenic and growth suppressive activities of the PAX3-FKHR fusion oncoprotein. Lab Invest. 2007;87:318–325. doi: 10.1038/labinvest.3700521. [DOI] [PubMed] [Google Scholar]
- 26.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 27.Pack SD, Zhuang Z. Fluorescence in situ hybridization : application in cancer research and clinical diagnostics. Methods in molecular medicine. 2001;50:35–50. doi: 10.1385/1-59259-084-5:35. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khatib ZA, Matsushime H, Valentine M, Shapiro DN, Sherr CJ, Look AT. Coamplification of the CDK4 gene with MDM2 and GLI in human sarcomas. Cancer research. 1993;53:5535–5541. [PubMed] [Google Scholar]
- 30.Baker SJ, Reddy EP. CDK4: A Key Player in the Cell Cycle, Development, and Cancer. Genes & cancer. 2012;3:658–669. doi: 10.1177/1947601913478972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grafstrom RH, Pan W, Hoess RH. Defining the substrate specificity of cdk4 kinase-cyclin D1 complex. Carcinogenesis. 1999;20:193–198. doi: 10.1093/carcin/20.2.193. [DOI] [PubMed] [Google Scholar]
- 32.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nat Rev Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 33.Tao Y, Kassatly RF, Cress WD, Horowitz JM. Subunit composition determines E2F DNA-binding site specificity. Molecular and cellular biology. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovesdi I, Reichel R, Nevins JR. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 35.Reichel R, Kovesdi I, Nevins JR. Activation of a Preexisting Cellular Factor as a Basis for Adenovirus E1a-Mediated Transcription Control. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:387–390. doi: 10.1073/pnas.85.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends in biochemical sciences. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. The Journal of biological chemistry. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- 38.Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. The EMBO journal. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, et al. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Molecular and cellular biology. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & development. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes & development. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saab R, Bills JL, Miceli AP, Anderson CM, Khoury JD, Fry DW, et al. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol Cancer Ther. 2006;5:1299–1308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 44.Lee B, Sandhu S, McArthur G. Cell cycle control as a promising target in melanoma. Current opinion in oncology. 2015;27:141–150. doi: 10.1097/CCO.0000000000000159. [DOI] [PubMed] [Google Scholar]
- 45.Weisberg E, Griffin JD. Mechanism of resistance to the ABL tyrosine kinase inhibitor STI571 in BCR/ABL-transformed hematopoietic cell lines. Blood. 2000;95:3498–3505. [PubMed] [Google Scholar]
- 46.Mahon FX, Deininger MW, Schultheis B, Chabrol J, Reiffers J, Goldman JM, et al. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: diverse mechanisms of resistance. Blood. 2000;96:1070–1079. [PubMed] [Google Scholar]
- 47.le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, et al. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–1766. [PubMed] [Google Scholar]
- 48.Konecny GE, Winterhoff B, Kolarova T, Qi J, Manivong K, Dering J, et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res. 2011;17:1591–1602. doi: 10.1158/1078-0432.CCR-10-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi Y, Oda Y, Kawaguchi K, Tamiya S, Yamamoto H, Suita S, et al. Altered expression and molecular abnormalities of cell-cycle-regulatory proteins in rhabdomyosarcoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2004;17:660–669. doi: 10.1038/modpathol.3800101. [DOI] [PubMed] [Google Scholar]
- 50.Tonelli R, McIntyre A, Camerin C, Walters ZS, Di Leo K, Selfe J, et al. Antitumor activity of sustained N-myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin Cancer Res. 2012;18:796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.