Abstract
We previously discovered carpelloid stamens when breeding cytoplasmic male sterile lines in broccoli (Brassica oleracea var. italica). In this study, hybrids and multiple backcrosses were produced from different cytoplasmic male sterile carpelloid stamen sources and maintainer lines. Carpelloid stamens caused dysplasia of the flower structure and led to hooked or coiled siliques with poor seed setting, which were inherited in a maternal fashion. Using four distinct carpelloid stamens and twelve distinct normal stamens from cytoplasmic male sterile sources and one maintainer, we used 21 mitochondrial simple sequence repeat (mtSSR) primers and 32 chloroplast SSR primers to identify a mitochondrial marker, mtSSR2, that can differentiate between the cytoplasm of carpelloid and normal stamens. Thereafter, mtSSR2 was used to identify another 34 broccoli accessions, with an accuracy rate of 100%. Analysis of the polymorphic sequences revealed that the mtSSR2 open reading frame of carpelloid stamen sterile sources had a deletion of 51 bases (encoding 18 amino acids) compared with normal stamen materials. The open reading frame is located in the coding region of orf125 and orf108 of the mitochondrial genomes in Brassica crops and had the highest similarity with Raphanus sativus and Brassica carinata. The current study has not only identified a useful molecular marker to detect the cytoplasm of carpelloid stamens during broccoli breeding, but it also provides evidence that the mitochondrial genome is maternally inherited and provides a basis for studying the effect of the cytoplasm on flower organ development in plants.
Introduction
Broccoli (Brassica oleracea L. var italica), sometimes referred to as Calabrese, is the most important commercial form of Brassica [1]. Driven by its reported abundance of nutrients and health benefits, broccoli has been well received by consumers. Its production and consumption has risen sharply in recent years [2–10], especially in the United Kingdom. Broccoli has an important commercial position, with a planting area of more than 7,000 hectares and a production value of more than £50 million (approximately US$77 million) each year [1, 11].
Broccoli displays obvious heterosis: using a cytoplasmic male sterile (CMS) line and an inbred line to create hybrids can reduce the cost of producing hybrids and improve their purity. However, hybrid production is difficult and produces low yields; therefore, CMS sources are crucial in improving the yield of hybrids. The CMS sources of broccoli have mostly been transferred from cabbage, radish, and other related cruciferous species through backcrossing. However, the floral organs often show extremely complex morphological variations because of the lack of coordination between the nucleus and cytoplasm during the transfer process [12–13]. These variations are different among varying genetic backgrounds in broccoli [14], thereby increasing the difficulty of transferring CMS materials. We previously found that CMS lines of broccoli obtained through some male sterile sources displayed carpelloid stamens. When we used these lines to produce hybrids, the pods were abnormal and seed yields were very low. These consequences dramatically limit the use value of these lines.
Carpelloid stamen phenomena have been studied in Arabidopsis thaliana and are regulated by class B genes of the classic ABC model. If the MADS-box transcription factors APETALA3 (Ap3) or PISTILLATA (PI), which are class B genes involved in conferring identities of the stamen and petal, are mutated or deleted, homeotic conversions of stamens to carpels and petals to sepals can occur [15–17]. Furthermore, the carpelloid stamen phenomenon has been reported in B. juncea var. tumida [18], B. rapa subsp. chinensis [19–20], B. napus [21–23], B. juncea [24], and Daucus carota sativus [25]. Thus far, carpelloid stamens caused by allo-cytoplasmic inheritance have not been documented in CMS lines of broccoli.
Mitochondrial and chloroplast genes are mainly inherited in a maternal pattern, with a slow rate of mutation; therefore, mitochondrial and chloroplast DNA have been widely used in evolutionary and phylogenetic studies [26–31]. The mitochondrial genome has a complicated multipartite structure [32], whereas the structure of the chloroplast genome is more conserved [28, 33–35]. To date, the entire mitochondrial genomes of Arabidopsis thaliana, B. napus (Nap and Pol), B. rapa (Cam), B. oleracea, B. juncea, and B. carinata have been determined [36–39]. The complete chloroplast genomes of Arabidopsis thaliana and B. napus have also been determined [40–41]. Owing to the large number of mitochondrial and chloroplastic sequences available, more and more markers have been developed to analyze genetic diversity [42–48] and variation of cytoplasmic [49–52] and CMS types [53–55]. However, as far as we know, mitochondrial and chloroplastic markers have not been used to distinguish between carpelloid and normal stamens in broccoli. The objectives of this study were: (1) to understand the inheritance pattern and morphological characteristics of carpelloid stamens; (2) to develop chloroplast and mitochondrial simple sequence repeat (SSR) markers that can distinguish between CMS sources of carpelloid and non-carpelloid stamens in broccoli; (3) to confirm the sequence features of the polymorphic bands by cloning and sequencing; and (4) to analyze the similarity of the genes related to carpelloid stamens.
Materials and Methods
Plant material and DNA extraction
The plant materials used in this study are detailed in Table 1. In total, 51 broccoli accessions, including 19 CMS lines and 23 hybrids were studied. Nine high-generation maintainers were included as references, and the backcross generations of CMS lines were continuously backcrossed to 2014.
Table 1. List of 51 broccoli accessions used in this study and their stamen types, identified by a PCR assay.
| Code | Line name | Type | Backcross generations | Origin | Stamens state |
|---|---|---|---|---|---|
| B1 | 93219 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B2 | CMS0412×93219 | Cytoplasmic male sterile line | 8 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B3 | CMS0413×93219 | Cytoplasmic male sterile line | 8 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B4 | CMS05738×93219 | Cytoplasmic male sterile line | 7 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B5 | CMS04S132×93219 | Cytoplasmic male sterile line | 8 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B6 | CMS10Q688×93219 | Cytoplasmic male sterile line | 2 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B7 | LvFu | Hybrid | - | Variety introduction (Wong Ching Ho Co., Ltd., Hong Kong, China) | Normal |
| B8 | BT-2006 | Hybrid | - | Variety introduction (Beijing Honor Seeds Co., Ltd., Beijing, China) | Normal |
| B9 | BT-2007 | Hybrid | - | Variety introduction (Beijing Honor Seeds Co., Ltd., Beijing, China) | Normal |
| B10 | L×2J | Hybrid | - | Variety introduction (Tianjin Kernel Vegetable Research Institute, Tianjin, China) | Normal |
| B11 | L×FJ | Hybrid | - | Variety introduction (Tianjin Kernel Vegetable Research Institute, Tianjin, China) | Normal |
| B12 | JingYou | Hybrid | - | Variety introduction (Wong Ching Ho Co., Ltd., Hong Kong, China) | Normal |
| B13 | NaiHanYouXiu | Hybrid | - | Variety introduction (Sakata Seed Corporation, Japan) | Normal |
| B14 | Tie mountain | Hybrid | - | Variety introduction (Seminis Seeds Beijing Co., Ltd., Beijing, China) | Normal |
| B15 | HeHuan007 | Hybrid | - | Variety introduction (Ho-Huan Agricultural Product Co., Ltd., Taiwan, China) | Carpellody |
| B16 | SaLi’Ao 55 | Hybrid | - | Variety introduction (Syngenta China Company, Beijing, China) | Normal |
| B17 | NanXiu366 | Hybrid | - | Variety introduction (Seminis Seeds Beijing Co., Ltd., Beijing, China) | Carpellody |
| B18 | 8554 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B19 | 90196 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B20 | 93213 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B21 | 94177 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B22 | YN23 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B23 | YN36 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B24 | 05726 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B25 | 05732 | Inbred line | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B26 | CMS0412×93219 | Cytoplasmic male sterile line | BC9 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B27 | CMS0413×93219 | Cytoplasmic male sterile line | BC9 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B28 | CMS05738×93219 | Cytoplasmic male sterile line | BC8 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Normal |
| B29 | CMS04S132×93219 | Cytoplasmic male sterile line | BC6 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B30 | CMS04S132×93219 | Cytoplasmic male sterile line | BC7 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B31 | CMS04S132×93219 | Cytoplasmic male sterile line | BC9 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B32 | CMS04S132×YN36 | Cytoplasmic male sterile line | BC6 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B33 | CMS10QB688×93219 | Cytoplasmic male sterile line | BC3 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B34 | CMS10QB688×90196 | Cytoplasmic male sterile line | BC2 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B35 | CMS10QB688×90196 | Cytoplasmic male sterile line | BC3 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B36 | CMS10QB688×93213 | Cytoplasmic male sterile line | BC2 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B37 | CMS10QB688×93213 | Cytoplasmic male sterile line | BC3 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B38 | CMS10QB688×94177 | Cytoplasmic male sterile line | BC2 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B39 | CMS10QB688×94177 | Cytoplasmic male sterile line | BC3 | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B40 | (CMS04S132×05732) ×YN23 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B41 | (CMS04S132×YN23) 6 ×93219 | hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B42 | (CMS04S132×YN36) 6 ×93219 | hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B43 | (CMS04S132×93219 5×YN263)×05732 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B44 | (CMS04S132×93219) 5×YN36×05726 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B45 | (CMS04S12×93219) 4 ×YN36 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B46 | HeHuan007×8554 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B47 | HeHuan007×93213 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B48 | HeHuan007×93219 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B49 | NanXiu366×8554 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B50 | NanXiu366×93213 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
| B51 | NanXiu366×93219 | Hybrid | - | Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China) | Carpellody |
Note
‘-’ means no backcross generations.
These accessions were grown in experimental greenhouses under standard field conditions at the Institute of Vegetable and Flowers, Chinese Academy of Agricultural Sciences, Changping, Beijing, China, from 2011 to 2014. When the diameter of the main bouquet on each plant was 8–10 cm, we pruned the bouquet, leaving three lateral balls, and then left the plant to develop branches naturally. Plantlets at the eight to ten leaves stage were randomly chosen from each accession for total genomic DNA isolation, using the modified hexadecyltrimethylammonium bromide method [56].
Flowering and fruiting characteristics observation
When the plants produced flowers, pollination was performed by hand or naturally (free pollination using bees in hives placed along the greenhouse, at positions corresponding to 1/8, 3/8, 5/8, and 7/8 of its length, from early flowering to the end of flowering) in 2011–2014. Flower morphologies, the pod shapes of each material, and the grain number per pod of maintainer 93219 (B1; Table 1) and different generations of CMS line CMS0412×93219, CMS0413×93219, CMS05738×93219, and CMS04S132×93219 (B2–B5, B26–B31; Table 1) were observed.
PCR and sequence analysis
Twenty-one pairs of primers for Brassica napus mitochondrial genome sequences (Gen Bank GI: 112253843) and 32 pairs of primers for Arabidopsis thaliana chloroplast sequences (Gen Bank GI: 7525012) developed by Zhang [57] were used to screen for makers that distinguished carpelloid from normal stamens (Table 2). All primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China).
Table 2. PCR primers used in this study.
| Name | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| ACP1 | GAACGACGGGAATTGAACC | GGTGGAATTTGCTACCTTTTT |
| ACP2 | GAAAAATGCAAGCACGGTTT | TACGATCCGTAGTGGGTTGC |
| ACP4 | TACCCGTATTAGGCACTA | TTTGTAAGACCACGACTG |
| ACP7 | TGGAGAAGGTTCTTTTTCAAGC | CGAACCCTCGGTACGATTAA |
| ACP9 | AACCATAATCATAGAAATAGAG | GTCGAACAAAGTAATCGG |
| ACP10 | GTATTAAATCCGAAACTC | ACTTGACATAAAACTTGG |
| ACP15 | CGACCAATCCTTCCTAAT | GAATGTTTGCTACCCTGA |
| ACP17 | TGCACTCTTCATTCTCGTTCC | GCGTTCCTTTCATTTAAGACG |
| ACP18 | AATGAAGAGTGCAGTAGC | CTAGGTTTTAGAAAGGAAA |
| ACP20 | CTCAACCGCCATCATACT | CGAAACTTAACCCTCTTT |
| ACP25 | CCCAAACCAAAGAGTGTA | GCTCGCAAGGCTCATAAC |
| ACP26 | AGAGGACCAAGAAACCAA | AACAGGCCATTCAACAAG |
| ACP27 | GAAGAGCAAACAAGGGAT | GAAGGGTTAGTCAATCAAAT |
| ACP29 | GGCCATAGGCTGGAAAGTCT | GTTTATGCATGGCGAAAAGG |
| ACP32 | TTCATAAGCGAAGAACAA | TCAGAGTAAGCAAAACAT |
| ACP33 | AGGGATAGTAATAAGAATAG | CAGATGTAAGAACGAAAA |
| ACP35 | ATTGGCTTACTTCTTGCG | GGTTTCCGGGATGTTATT |
| ACP38 | GGTTCGTTAGCAGGTTTA | GTTCCCTAGCAACACTTT |
| ACP39 | AGACGGGTGAATAGAGTG | GTTATGCTTTTCGACGAT |
| ACP40 | GCAGAATGAGACGGGATA | AGACACTTTGGGATTGCT |
| ACP41 | GGCTCCACAATGGAATTGAC | GCACATTTCAGCGTCACAAA |
| ACP42 | CTTTCTCGATAAAGTCGGTTGA | GGAAGAAGCCCGTTCAGG |
| ACP43 | GCTGTCGTGGATGAGTGG | AAGTGCTTTCTGGGTCGT |
| ACP44 | ATTGTAGATTCTGGGAGG | ATCGATGCTGTATTCATG |
| ACP45 | TTACATTGCTTTTCTTACAG | CTCGTTGGTTTAGGATTA |
| ACP46 | CTACCATTTCACCACCAA | GGACCCTATTCACCTCTT |
| ACP47 | TGTACTTATGGGAAAGCG | CTGGGTTCTTCTACTTCATT |
| ACP48 | AGGCAAGATGATAGGATA | CAAGTCAAGATGATACGG |
| ACP49 | AATAGCTCGACGCCAGGAT | TTCGGATGTGAAAGTGCC |
| ACP50 | TCCGAGTGAATGGAAAAG | GATGGAATTACAAATGGAGTAG |
| ACP51 | TTCTTATTCACTGGTTTG | GTGGAAATCTTTGTTCTA |
| ACP58 | GCTATCCCAAGTTTCTGC | TGGCTTTGATCCGTTATT |
| mtSSR1 | CTCCCGCTTCCTCACATC | GCTCCAATAAGGCGTTCC |
| mtSSR2 | ACCAAGATTGAGCCAGAT | CGTCCACTACCGAAAGAG |
| mtSSR3 | GGCTGCTTTCTCATACCG | TCCTAAATGCTGCCCTTC |
| mtSSR4 | GATTCTAAGGGTACGGGACA | ACCGACGACAAACAATAACA |
| mtSSR5 | TATTGTTTGTCGTCGGTTAT | GGTAGGCAAGTTGGTAGG |
| mtSSR7 | CGTTAGGGGTATTTAGCA | CTCTATTCCGTTTCCACA |
| mtSSR8 | CCCGAGAAGCACTGTTGA | ACGGAGTGACAAAGGAGC |
| mtSSR9 | CGGTGAAAGAGGGCAAAG | AAACAAATACCAGCTAACG |
| mtSSR10 | GCCATTTCATTTCCTTTG | ATCCTCCTTCGCCTTTCT |
| mtSSR22 | TACTAATCGGTGACTTGCT | AGTCTTTATGGATGTGCC |
| mtSSR46 | TACTTGCTGCACTTCCTG | ACAAATGCCACTTCTTCC |
| mtSSR48 | CCTTCTGGGTTGACTTGA | AGTGGTGCCCTCCTCTAA |
| mtSSR61 | GGCGGTGGACAGAAATGG | AGGGAAGCCCAACGAATG |
| mtSSR92 | GCCGCTTTCATTGTTGTA | TTCGGTTTATTAGCTCTTCC |
| mtSSR98 | GTGCCAGATGCAACAAAG | GAGGCCATAGGGAAAGTC |
| mtSSR110 | CGGGTGCTTGCATCATTT | TCTAGCCATTCCAGGTTT |
| mtSSR128 | AATCCTATCCCATCCGAGTC | AGCCTTTCCTTTCCCACC |
| mtSSR129 | CTTCCCTCAGTTGGTTTG | TGCCCTCTGTCCTTTATT |
| mtSSR133 | GCTGCTCATCACTACCTG | CACTACGCTCACTGAAACTA |
| mtSSR152 | AAGAAAGAAGAGCGACAA | GGGTACGGTACTAAAGGT |
| mtSSR156 | TACTCATCAAATGGCACTC | CAAAGGGAAAGAAGAAAG |
PCR amplifications were carried out in a reaction volume of 25 μL, which contained 2 μL (30 ng μL−1) genomic DNA, 12.5 μL Dream Taq™ Green PCR Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA), and 1 μL of each primer (10 pmol μL−1). The following amplification protocol was carried out in an ABI Veriti 96-well PCR thermal cycler (Applied Biosystems, Foster City, CA, USA). Initial denaturation was performed at 94°C for 4 min; followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and a final extension at 72°C for 10 min. PCR products were analyzed on 2.0% (w/v) agarose gels in 0.5×TBE buffer and visualized after staining with Goldview (Solarbio Technology Co., Ltd, Beijing, China). The bands were photographed under ultraviolet light under a Universal Hood II (Bio-Rad, Hercules, CA, USA).
For each single polymorphic locus, the cloned products were sequenced by Bo Maide Biotech Co., Ltd, Beijing, China. Alignment and similarity analyses of the obtained sequences were performed using the Align and BLAST tools, respectively, in the Universal Protein Resource (UniProt) database.
Results
Flowering and fruiting characteristics of carpelloid stamen plants
To determine the mechanism of inheritance of the carpelloid stamen phenomenon, we carried out crosses or successively backcrosses between maintainer lines with normal stamens (B1, B18, B19, B20, B21; Table 1) and CMS lines with carpelloid stamens (B6, B15, B17, B29; Table 1) materials in 2011–2014. The results showed that the offspring obtained by crossing or backcrossing all had carpelloid stamens. Therefore, the phenomenon of CMS carpelloid stamens was caused by allo-cytoplasmic inheritance in broccoli; i.e., it was mainly inherited in a maternal pattern and the heritability rate was 100%.
The growth characteristics of maintainer 93219 (B1; Table 1) and the sixth generation to ninth generation of CMS04S132×93219 (B5, B29, B30, B31; Table 1) plants were observed from 2011 to 2014. The growth and morphological characteristics of the two lines were similar from seed germination to plant bolting, but differences appeared from the big bud stage to before early flowering. In maintainer 93219 plants, the buds were plump with smooth surfaces, and the floral organs developed normally, with six stamens and opened petals (Fig 1A). In addition, the pistils and seed pods were all erect (Fig 1D) with 12.72 ± 0.86 grains per silique. In the different generations of CMS04S132×93219, the buds were soft with slightly wrinkled surfaces, the flowers showed developmental abnormalities, the petals were smaller and deformed, and three to six stamens appeared that strongly varied (Fig 1B and 1C). Interestingly, the stamens mutated into carpelloid structures and pseudo pistils, which were entangled with the pistil; some small green beads, resembling ovules, were observed inside the stamens (Fig 1C). The carpelloid stamens adhered to the pistil, causing the pistil to bend before flowering. The stamens unfolded gradually with flowering; however, most could not completely separate from the pistil. Moreover, the plants could fruit via artificial pollination during the bud and flowering stages. The small green beads in the stamens stopped growing and died away as the siliques developed. The siliques were hooked or coiled (Fig 1E–1G) and were set at a rate of 80.90 ± 3.30% per plant and 6.10 ± 0.53 grains per silique. The number of siliques per plant was much lower than the other CMS lines (B2, B3, and B4; Table 1) that had undergone free pollination.
Fig 1. Morphological characteristics of flowers and siliques of maintainer 93219 and CMS04S132×93219.
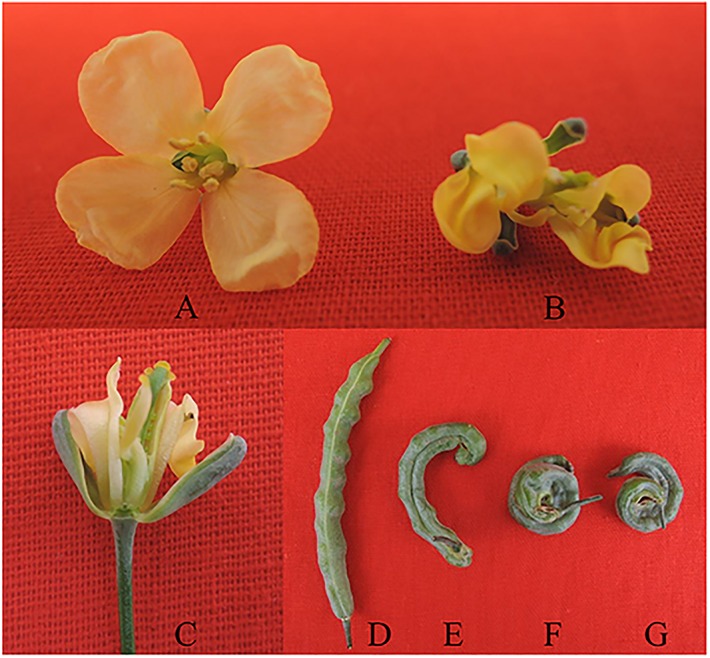
A: flower of maintainer 93219, B and C: flowers of CMS04S132×93219, D: siliques of maintainer 93219, E–G: siliques of CMS04S132×93219.
Identification of carpelloid stamens of broccoli CMS materials using cpSSR and mtSSR molecular markers
We first used 32 pairs of chloroplastic SSR (cpSSR) primers and 21 pairs of mitochondrial SSR (mtSSR) primers developed by Zhang [57] (Table 2) to amplify the total genomic DNA of five CMS lines (B2–B6; Table 1), maintainer 93219 (B1; Table 1), and 11 unequal hybrids of CMS lines (B7–B17; Table 1), which were collected from seven countries. The results demonstrated that all the cpSSR primers and mtSSR primers could successfully amplify a product. However, only one mtSSR primer (mtSSR2) showed obvious polymorphisms between normal and carpelloid stamen materials (Fig 2). We then used the primers of mtSSR2 to screen the other 34 broccoli accessions (B17–B51; Table 1), including eight inbred lines, 14 CMS lines, and 12 CMS hybrids. The results showed that mtSSR2 could distinguish between the normal and carpelloid stamen materials with 100% accuracy.
Fig 2. PCR amplification profiles of 17 broccoli accessions.
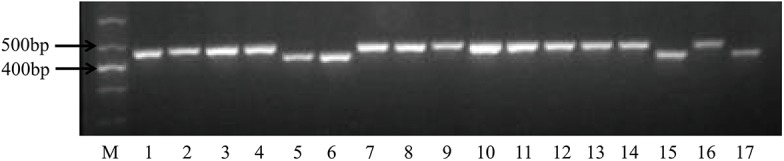
M: marker, 1: maintainer 93219, 2: CMS0412×932198, 3: CMS0413×932198, 4: CMS05738×932197, 5: CMS04S132×932198, 6: CMS10Q688×932192, 7: LvFu, 8: BT-2006, 9: BT-2007, 10: L×2J, 11: L×FJ, 12: JingYou, 13: NaiHanYouXiu, 14: Tie mountain, 15: HeHuan007, 16: SaLi’Ao 55, 17: NanXiu366.
Sequence features of polymorphic bands
The polymorphic bands obtained after PCR using mtSSR2 were sequenced. The amplicons were identical in all maintainer lines (471 bp), whereas they were 420 bp in all carpelloid stamen materials. Analyses after aligning the polymorphic sequences from normal and carpelloid materials indicated that the similarity of the sequences were 88.96%, with one single nucleotide polymorphism at 100 bp (C/T), and the sequences from carpelloid stamen materials had 51 nucleotides deleted between 309 and 359 bp compared with the normal sequences (Fig 3). Open reading frame (ORF) analysis showed that the deleted nucleotides were located between position 102 and 152 bp in the ORF coding region of the normal sequences (Fig 4), and encoded 18 amino acids (Fig 5). These results indicated that the polymorphism of mtSSR2 could be attributed to the fragment insertion or deletion near the SSR loci.
Fig 3. Sequence alignment of mtSSR2 amplicons with the B. napus mitochondrial genome.

Fig 4. ORF sequence alignment of mtSSR2 amplicons with the B. napus mitochondrial genome.

Fig 5. Amino acid sequences of ORFs of mtSSR2 amplicons and the B. napus mitochondrial genome.

Analyzing similarity of genes related to carpelloid stamens
Analysis of the amino acid sequences in the ORF region demonstrated that the polymorphic region amplified with mtSSR2 is located in the orf125 coding region of B. napus, B. juncea, B. rapa subsp. oleifera, Eruca sativa, B. oleracea, B. oleracea var. botrytis, and B. juncea var. tumida. It is also located in the coding region of orf108c and orf108 in B. carinata and Raphanus sativus, respectively. However, the proteins were annotated as being hypothetical and as having unknown functions. Similarity analysis of the amino acid sequences revealed that the protein sequences related to the carpelloid stamen have the highest similarity with orf108c and orf108 in B. carinata and Raphanus sativus, respectively (Table 3).
Table 3. Sequence identity between amplified products of mtSSR2 in broccoli CMS normal and carpelloid stamen materials and the corresponding sequences of B. juncea, B. rapa, Eruca sativa, B. oleracea, B. oleracea var. botrytis, B. juncea var. tumida, B. napus, B. carinata, and Raphanus sativus cytoplasmic genomes.
| Entry | Organism | Protein name | Sequence identity (normal stamen) | Sequence identity (carpelloid stamen) |
|---|---|---|---|---|
| G4XYV8 | B. juncea | Orf125 | 100% | 79.5% |
| G4XYB0 | B. rapa subsp. oleifera | Orf125 | 100% | 79.5% |
| A0A088BGJ2 | Eruca sativa | Orf125 | 100% | 79.5% |
| G4XYC4 | B. oleracea | Orf125 | 100% | 79.5% |
| A0A068BCT7 | B. oleracea var. botrytis | Orf125 | 100% | 79.5% |
| A0A023VX75 | B. juncea var. tumida | Orf125 | 100% | 79.5% |
| Q6YSR6 | B. napus | ORF125 | 100% | 79.5% |
| G4XYQ4 | B. carinata | Orf108c | 80.7% | 98.6% |
| R4I1C8 | Raphanus sativus | Orf108 | 80.7% | 98.6% |
Discussion
Carpelloid stamen phenomenon and its inheritance pattern
In higher plants, the process of flowering is complex, involving many interactions between genes or between genes and the environment. The carpelloid stamen phenomenon refers to stamen structures of the flower that are converted to carpels. It not only causes alteration of the flower structure, but also brings about male sterility. This phenomenon is caused by floral homeotic mutations and is controlled by nuclear genes. The B class or C class genes, such as AP3, PI, AGL8, SHP1, SHP2, and NAPc, are involved in the regulation of this phenomenon [15–16, 20, 23, 58–62]. The effect of the plasmon or interactions between cytoplasmic and nuclear genes on the development of floral organs has been reported [14, 21, 25, 63]; however, the molecular mechanisms are not clear. In this study, we confirmed that the carpelloid stamens were caused by allo-cytoplasmic inheritance in CMS lines of broccoli, using hybrid and backcross methods combined with field observation for several years. Stamen carpellody was passed on in a maternal inheritance fashion. Furthermore, the phenotypic features of the carpelloid stamen materials were similar to the carpellody of stamens in B. napus and B. rapa obtained from the mutation of nuclear genes: the flowers showed dysplasia, the petals were smaller and deformed, the stamens were seriously deformed, the pistil was deviated, and the stamens were entangled with the pistil [20, 23]. The siliques were erect and the seed setting and combining ability performed well in CMS carpelloid stamen materials of B. napus and B. rapa. In contrast, the siliques were hooked or coiled, with few grains per silique observed in this study, which would reduce the utility of CMS lines in hybrid seed production practice in broccoli. Therefore, the occurrence of carpelloid stamens should be avoided in broccoli breeding.
Maternal inheritance and application of the organelle SSR markers
Chloroplasts and mitochondria are mainly inherited uniparentally in seeded plants [64]. The chloroplast genome is usually maternally inherited in most angiosperm species, including cruciferous crops, such as cabbage, broccoli, and cauliflower [47, 65], although it can also be paternally [66–67] or biparentally inherited [68–69]. Mitochondrial DNA is maternally inherited or passed via paternal transmission [67]. However, mitochondrial inheritance in broccoli has not been documented so far, perhaps because no polymorphisms have been found. In our study, mtSSR2 could be detected in carpelloid stamen materials for four successive backcross generations and there was no sequence variation in the mitochondrial PCR products studied. Our results provide evidence not only that the mitochondrial genome is maternally and stably inherited in broccoli, but also that broccoli mtDNA has advantages for evolutionary studies.
The main advantage of organelle genomes is that no recombination can occur between two alleles, because there is only one allele per cell and per organism [69]. Furthermore, organelle DNA can be easily obtained because it exists in many copies in each cell. Based on organelle genomes’ inheritance patterns and characteristics, organelle SSR markers have been used widely with total genomic DNA. Kaundun and Matsumoto [70] used cpSSR markers to analyze the variation of heterologous nuclear and chloroplast DNA in tea (Camellia sinensis). Zhang et al. [47] studied the inheritance of the cabbage chloroplast and assessed subspecies diversity of B. oleracea using cpSSR primers. Cheng et al. [71] distinguished somatic hybrids in citrus using cpSSR primers. Moreover, cpSSR markers have also been used in neotropical orchids [72] and cruciferous crops [42, 44, 48, 55]. For mitochondrial SSR markers, Wang et al. [55] used mtSSR primers to distinguish allo-cytoplasmic inheritance in cabbage. To the best of our knowledge, the identification of carpelloid and normal stamens using cpSSR or mtSSR in broccoli, as well as other species, has not been reported. In this study, mtSSR2 could accurately distinguish carpelloid from normal stamen materials with the same polymorphic products as reported in cabbage, which can distinguish OguCMSR1-2, OguCMSHY, and NigCMS (420 bp) from pol CMS and OguCMSR3 (471 bp) [55]. The polymorphic band amplified in CMS carpelloid stamen materials was 420 bp, which suggests that the original CMS source came from OguCMSHY, OguCMSR1-2, or NigCMS and was transferred to broccoli. Alignment analysis of the sequences showed that the deleted sequences in broccoli were not at the same location as in cabbage. This will benefit the selection of CMS types and backcross breeding in broccoli; moreover, it will also provide a basis for evolutionary studies in crucifers.
Furthermore, the sequences of the products were located in orf108 or orf125 coding regions of mitochondrial genomes in many Brassica species. Compared with the maintainers, the carpelloid stamen materials showed a deletion of 51 nucleotides, suggesting that the deletion of mitochondrial nucleotides could explain the carpelloid stamen phenotype. Thus, it is likely that orf108 or orf125 encodes genes involved in the development of floral organs. Our findings provide an important reference for further study on the molecular mechanism of the interaction between the nucleus and cytoplasm in floral organ development.
In conclusion, we have identified a useful molecular marker to detect the cytoplasm of carpelloid stamens in broccoli. The results of our study are valuable for improving the efficiency of broccoli breeding and also for forming a solid basis for further study of the molecular mechanisms underlying the CMS carpelloid stamen phenomenon.
Acknowledgments
This work was supported by the China Agriculture Research System (Grant No. CARS-25-A), National Natural Science Foundation of China (Grant No. 31372067), National High Technology Research and Development Program (863 Program) of China (Grant No. 2012AA100105), Key Projects in the National Science and Technology Pillar Program of China (Grant No. 2013BAD01B04), Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China, and Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. CAAS-ASTIP-IVFCAAS).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by China Agriculture Research System (Grant No. CARS-25-A), National Natural Science Foundation of China (Grant No. 31372067), the National High Technology Research and Development Program (863 Program) of China (Grant No. 2012AA100105), the Key Projects in the National Science and Technology Pillar Program of China (Grant No. 2013BAD01B04), the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China, and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. CAAS-ASTIP-IVFCAAS). YML received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Walley PG, Carder J, Skipper E, Mathas E, Lynn J, Pink D, et al. A new broccoli×broccoli immortal mapping population and framework genetic map: tools for breeders and complex trait analysis. Theor Appl Genet 2012; 124: 467–484. 10.1007/s00122-011-1721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lampe JW, Peterson S. Brassica, biotransformation and cancer risk: genetic polymorphisms alter the preventive effects of cruciferous vegetables. J Nutr 2002; 132: 2991–2994. [DOI] [PubMed] [Google Scholar]
- 3. Finley JW. The antioxidant responsive element (ARE) may explain the protective effects of cruciferous vegetables on cancer. Nutr Rev 2003; 61(7): 250–254. [DOI] [PubMed] [Google Scholar]
- 4. Keck AS, Qiao Q, Jeffery EH. Food matrix effects on bioactivity of broccoli-derived sulforaphane in liver and colon of F344 rats. J Agric Food Chem 2003; 51: 3320–3327. 10.1021/jf026189a [DOI] [PubMed] [Google Scholar]
- 5. Murashima M, Watanabe S, Zhou XG, Uehara M, Kurashige A. Phase 1 study of multiple biomarkers for metabolism and oxidative stress after one-week intake of broccoli sprouts. Biofactors 2004; 22: 271–275. [DOI] [PubMed] [Google Scholar]
- 6. Kensler TW, Chen JG, Egner PA, Fahey JW, Jacobson LP, Stephenson KK, et al. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People’s Republic of China. Cancer Epidemiol Biomarkers Prev 2005; 14: 2605–2613. 10.1158/1055-9965.EPI-05-0368 [DOI] [PubMed] [Google Scholar]
- 7. Canene-Adams K, Lindshield BL, Wang S, Jeffery EH, Clinton SK, Erdman JW Jr. Combinations of tomato and broccoli enhance antitumor activity in dunning r3327-h prostate adenocarcinomas. Cancer Res 2007; 67: 836–843. 10.1158/0008-5472.CAN-06-346 [DOI] [PubMed] [Google Scholar]
- 8. Dinkova-Kostova AT, Fahey JW, Wade KL, Jenkins SN, Shapiro TA, Fuchs EJ, et al. Induction of the phase 2 response in mouse and human skin by sulforaphane-containing broccoli sprout extracts. Cancer Epidemiol Biomarkers Prev 2007; 16: 847–851. EPI-06-0934 [DOI] [PubMed] [Google Scholar]
- 9. Jeffery EH, Araya M. Physiological effects of broccoli consumption. Phytochem Rev 2009; 8(1): 283–298. [Google Scholar]
- 10. Sun B, Xu YJ, Xu TF, Yuan GF, Guo RF, Wang BL, et al. Variation of bioactive compounds and nutrients among different organs of broccoli (Brassica oleracea var. italica Planck). Acta Hortic Sin 2010; 37(1): 59–64. [Google Scholar]
- 11. Department for Environment, Food and Rural Affairs (DEFRA). Basic horticultural statistics for the United Kingdom DEFRA, 2010; UK: http://www.defra.gov.uk/. [Google Scholar]
- 12. Fang ZY, Sun PT, Liu YM, Yang LM, Wang XW, Zhuang M. Investigation of different type of male sterility and application of dominant male sterility in cabbage. China Veg 2001; (1): 6–10. [Google Scholar]
- 13. Fang ZY, Liu YM, Yang LM, Wang XW, Zhuang M, Zhang YY, et al. Breeding and seed production technique of dominant genic male sterile line and cytoplasmic male sterile line in cabbage. Sci Agri Sin 2004; 37 (5): 717–723. [Google Scholar]
- 14. Zhu YY, Yao WY, Zhang SQ, Ling C, Shen FY, Gong J, et al. Breeding and utilization of male sterile lines with Ogura cytoplasm in cabbage. Acta Agric Shanghai 1998; 14(2): 19–24. [Google Scholar]
- 15. Jack T, Brockman LL, Meyerowitz EM. The homeotic gene apetala 3 of Arabidopsis thaliana encodes a Mads Box and is expressed in petals and stamens. Cell 1992; 68(4): 683–697. [DOI] [PubMed] [Google Scholar]
- 16. Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene pistillata. Genes & Development 1994; 8(13): 1548–1560. [DOI] [PubMed] [Google Scholar]
- 17. Lamb RS, Irish VF. Functional divergence within the AP-ETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci USA 2003; 100(11): 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen ZJ, Gao QK, Wu GL, Wang BL, Sun WG, Gong L. Morphology and genetic variation of flower of cytoplasmic male-sterile in tuber mustard. Acta Agric Zhejiangensis 1993; 5(3): 172–176. [Google Scholar]
- 19. Zhang YF, Wang XF, Zhang X, Li DR, Liang ZS. The discovery and study on Homeotic Mutant Male Sterile Line HGMS of non-heading Chinese Cabbage. Acta Agric Bor-occid Sin 2005; 14(6): 164–168. [Google Scholar]
- 20. Zhang YF, Wang XF, Zhang WX, Yu F, Tian JH, Li DR, et al. Functional analysis of the two Brassica AP3 genes involved in apetalous and carpelloid stamen phenotypes. Plos one 2011; 6(6): e20930 10.1371/journal.pone.0020930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leino M, Teixeira R, Landgren M. Brassica napus lines with rearranged Arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor Appl Genet 2003; 106: 1156–1163. [DOI] [PubMed] [Google Scholar]
- 22. Lu GY, Wu XM, Chen BY, Xu K, Gao GZ, Li XZ. Discovery and characteristics of homeotically mutated male sterility in rapeseed. Chin J of Oil Crop Sci 2005; 27(2): 28–31. [Google Scholar]
- 23. Wang DJ, Yang CL, Liu Z, Dong L. Cytological characterization and gene expression of carpelloid stamen of apetalous flower in Brassica napus L.. Plant Physiology Journal 2014; 50 (3): 290–296. [Google Scholar]
- 24. Malik M, Vyas P, Rangaswamy NS. Development of two new cytoplasmic male-sterile lines in Brassica juncea through wide hybridization. Plant Breeding 1999; 118: 75–78. [Google Scholar]
- 25. Linke B, Nothnagel T, Börner T. Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS . The Plant Journal 2003; 34: 27–37. [DOI] [PubMed] [Google Scholar]
- 26. Palmer JD, Shields CR, Cohen DB, Orton TJ. An unusual mitochondrial DNA plasmid in the genus Brassica. Nature 1983a; 301: 725–728. [Google Scholar]
- 27. Palmer JD, Shields CR, Cohen DB, Orton TJ. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor Appl Genet 1983b; 65: 181–189. 10.1007/BF00308062 [DOI] [PubMed] [Google Scholar]
- 28. Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNAs. Proc Natl Acad Sci USA 1987; 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soltis DE, Soltis PS. Contributions of plant molecular systematic to studies of molecular evolution. Plant Mol Biol 2000; 42: 45–75. [PubMed] [Google Scholar]
- 30. Nishiyama T, Wolf PG, Kugita M, Sinclair RB, Sugita M, Sugiura C, et al. Chloroplast phylogeny indicates that Bryophytes are monophyletic. Mol Biol Evol 2004; 21: 1813–1819. [DOI] [PubMed] [Google Scholar]
- 31. Lehnebach CA, Cano A, Monsalve C, McLenachan P, Hörandl E, Lockhart P. Phylogenetic relationships of the monotypic Peruvian genus Laccopetalum (Ranunculaceae). Plant Syst Evol 2007; 46: 109–116. [Google Scholar]
- 32. Avise JC. Molecular markers, natural history and evolution New York: Chapman and Hall; 1994. [Google Scholar]
- 33. Palmer JD. Plastid chromosomes: structure and evolution In: Vasil IK, Bogorad L, editors. Cell culture and somatic cell genetics in plants, the molecular biology of plastids. San Diego: Academic Press; 1991. pp. 5–53. [Google Scholar]
- 34. Raubeson LA, Jansen RK. Chloroplast genomes of plants In: Henry R, editor. Diversity and evolution of plants-genotypic variation in higher plants. London: CABI Publishing; 2005. pp. 45–68. [Google Scholar]
- 35. Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science 2006; 311: 1727–1730. [DOI] [PubMed] [Google Scholar]
- 36. Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 1997; 15(1): 57–61. [DOI] [PubMed] [Google Scholar]
- 37. Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana . Nucleic Acids Res 2003; 20: 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen J, Guan R, Chang S, Du T, Zhang H. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS ONE 2011; 6:e17662 10.1371/journal.pone.0017662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang SX, Yang TT, Du TQ, Huang YJ, Chen JM, Yan JY, et al. Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica . BMC Genomics 2011; 12: 497 10.1186/1471-2164-12-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana . DNA Res 1999; 6(5): 283–90. [DOI] [PubMed] [Google Scholar]
- 41. Hu ZY, Hua W, Huang SM, Wang HZ. Complete chloroplast genome sequence of rapeseed (Brassica napus L.) and its evolutionary implications. Genet Resour Crop Evol 2010; 58: 875–887. [Google Scholar]
- 42. Flannery ML, Mitchell FJG, Coyne S, Kavanagh TA, Burke JI, Salamin N, et al. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor Appl Genet 2006; 113: 1221–1231. [DOI] [PubMed] [Google Scholar]
- 43. Handa H. Investigation of the origin and transmission of linear mitochondrial plasmid based on phylogenetic analysis in Japanese rapeseed varieties. Genome 2007; 50: 234–240. [DOI] [PubMed] [Google Scholar]
- 44. Allender CJ, Allainguillaume J, Lynn J, King GJ. Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n = 9) wild relatives. Theor Appl Genet. 2007; 114: 609–618. [DOI] [PubMed] [Google Scholar]
- 45. Allender CJ, King GJ. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 2010; 10: 54 10.1186/1471-2229-10-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao HX, Li ZJ, Hu SW, Sun GL, Chang JJ, Zhang ZH. Identification of cytoplasm types in rapeseed (Brassica napus L.) accessions by a multiplex PCR assay. Theor Appl Genet 2010; 121: 643–650. 10.1007/s00122-010-1336-3 [DOI] [PubMed] [Google Scholar]
- 47. Zhang YY, Fang ZY, Wang QB, Liu YM, Yang LM, Zhuang M, et al. Chloroplast subspecies-specific SNP detection and its maternal inheritance in Brassica oleracea L. by using a dCAPS marker. Journal of Heredity 2012; 103(4): 606–611. 10.1093/jhered/ess006 [DOI] [PubMed] [Google Scholar]
- 48. Zhang RJ, Hu SW, Yan JQ, Sun GL. Cytoplasmic diversity in Brassica rapa L. investigated by mitochondrial markers. Genet Resour Crop Evol 2013; 60: 967–974. [Google Scholar]
- 49. Lannér C. Relationships of wild Brassica species with chromosome number 2n = 18, based on the comparison of the DNA sequence of the chloroplast intergenic region between trnL (UAA) and trnF (GAA). Can J Bot 1998; 76: 228–237. [Google Scholar]
- 50. Soranzo N, Provan J, Powell W. An example of microsatellite length variation in the mitochondrial genome of conifers. Genome 1999; 42: 158–161. [PubMed] [Google Scholar]
- 51. Panda S, Martín JP, Aguinagalde I. Chloroplast and nuclear DNA studies in a few members of the Brassica oleracea L. group using PCR-RFLP and ISSR-PCR markers: a population genetic analysis. Theor Appl Genet 2003; 106: 1122–1128. [DOI] [PubMed] [Google Scholar]
- 52. Hu JB, Li JW, Zhou XY. Analysis of cytoplasmic variation in a cucumber germplasm collection using chloroplast microsatellite markers. Acta Physiol Plant 2009; 31: 1085–1089. [Google Scholar]
- 53. Ashutosh, Dwivedi KK, Kumar VD, Prakash S, Bhat SR. rep-PCR helps to distinguish different alloplasmic cytoplasmic male sterile lines of Brassica juncea . Plant Sci 2005; 168: 1083–1087. [Google Scholar]
- 54. Zhang YY, Fang ZY, Wang QB, Liu YM, Yang LM, Zhuang M, et al. Molecular distinction of two Ogura CMS sources in Brassica oleracea var. capitata L. Sci Agric Sin 2011; 44(14): 2959–2965. [Google Scholar]
- 55. Wang QB, Zhang YY, Fang ZY, Liu YM, Yang LM, Zhuang M. Chloroplast and mitochondrial SSR help to distinguish allo-cytoplasmic male sterile types in cabbage (Brassica oleracea L. var. capitata). Mol Breeding 2012; 30: 709–716. [Google Scholar]
- 56. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 1980; 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang YY. Research on the cytoplasmic DNA diversity in Brassica oleracea vegetables. Ph. D dissertations, Chinese Academy of Agricultural Sciences. 2011. Available:http://cnki.caas.cn/kcms/detail/detail.aspx?filename=1011159218.nh&dbname=CDFD2011&dbcode=CDFD&uid=WEFkWGd4ckdUQ3R6.
- 58. Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature 1991; 353: 31–37. [DOI] [PubMed] [Google Scholar]
- 59. Weigel D, Meyerowitz E. The ABCs of floral homeotic genes. Cell 1994; 78: 203–209. [DOI] [PubMed] [Google Scholar]
- 60. Sablowski RW, Meyerowitz EM, Yanofsky MF. A homolog of NO APICAL ERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA . Cell 1998; 92: 93–103. [DOI] [PubMed] [Google Scholar]
- 61. Liljegren SJ, Ditta GS, Eshed HY, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis . Nature 2000; 404: 766–770. [DOI] [PubMed] [Google Scholar]
- 62. Pinyopich A, Ditta GS, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003; 424: 85–88. [DOI] [PubMed] [Google Scholar]
- 63. Zhang Y, Wang XJ, Li CQ, Song HY, Ren XS, Si J. Molecular identification of Brassica oleracea CMS and the morphology response of flower to nuclear background. Acta Hortic Sin 2010; 37(6): 915–922. [Google Scholar]
- 64. Birky CW Jr. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA 1995; 92: 11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reboud X, Zeyl C. Organelle inheritance in plants. Heredity 1994; 72:132–140. [Google Scholar]
- 66. Testolin R, Cipriani G. Paternal inheritance of chloroplast DNA and maternal inheritance of mitochondrial DNA in the genus Actinidia. Theor Appl Genet 1997; 94: 897–903. [Google Scholar]
- 67. Worth JRP, Yokogawa M, Isagi Y. Outcrossing rates and organelle inheritance estimated from two natural populations of the Japanese endemic conifer Sciadopitys verticillata . J Plant Res 2014; 127: 617–626. 10.1007/s10265-014-0646-y [DOI] [PubMed] [Google Scholar]
- 68. Hansen AK, Escobar LK, Gilbert LE, Jansen RK. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am J Bot 2007; 94: 42–46. 10.3732/ajb.94.1.42 [DOI] [PubMed] [Google Scholar]
- 69. Skuza L, Filip E, Szucko I. Use of organelle markers to study genetic diversity in soybean, a comprehensive survey of international soybean research-genetics, physiology, agronomy and nitrogen relationships In: Board J (Ed), ISBN: 978-953-51-0876-4, 2013. InTech, 10.5772/52028 http://www.intechopen.com/books/acomprehensive-survey-of-international-soybean-research-genetics-physiology-agronomy-and-nitrogen-relationships/use-of-organelle-markers-to-study-genetic-diversity-in-soybean. [DOI] [Google Scholar]
- 70. Kaundun SS, Matsumoto S. Heterologous nuclear and chloroplast microsatellite amplification and variation in tea, Camellia sinensis . Genome 2002; 45: 1041–1048. [DOI] [PubMed] [Google Scholar]
- 71. Cheng YJ, Guo WW, Deng XX. cpSSR: a new tool to analyze chloroplast genome of citrus somatic hybrids. Acta Bot Sin 2003; 45: 906–909. [Google Scholar]
- 72. Pinheiro F, Palma-Silva C, de Barros F, Félix LP, Lexer C, Cozzolino S, et al. Chloroplast microsatellite markers for the Neotropical orchid genus Epidendrum, and cross-amplification in other Laeliinae species (Orchidaceae). Conserv Genet Resour 2009; 1: 505–511. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


