Abstract
Pancreatic cancer is one of the leading causes of cancer related death. Increasing incidence and mortality indicates a lack of detection and post diagnostic management of this disease. Recent evidences suggest that, miRNAs are very attractive target molecules that can serve as biomarkers for predicting development and progression of pancreatic cancer. Furthermore, miRNAs are also promising therapeutic targets for pancreatic cancer. The objective of the present review is to discuss the significance of miRNA in pancreatic cancer development, diagnosis, therapy and prognosis. We extracted and compiled the useful information from PubMed database, which satisfied our criteria for analysis of miRNAs in pancreatic cancer diagnosis, therapy and prognosis. A summary of the most important miRNAs known to regulate pancreatic tumorigenesis is provided. The review also provides a collection of evidence that show miRNA profiles of biofluids hold much promise for use as biomarkers to predict and detect development of pancreatic cancer in its early stages. Identification of key miRNA networks in pancreatic cancer will provide long-awaited diagnostic/therapeutic/prognostic tools for early detection, better treatment options, and extended life expectancy and quality of life in PDAC patients.
Keywords: biomarker, prognosis, pancreatic ductal adenocarcinoma, microRNA, diagnosis, therapeutics
1. Introduction
Pancreatic cancer is the tenth most common malignancy in the world (1). In particular, pancreatic ductal adenocarcinoma (PDAC) is the most common type (90%) of pancreatic cancer diagnosed in the exocrine component of the pancreas (2). It is frequently diagnosed in developed countries and is more common in men than in women (3). Patients with pancreatic cancer have a very low survival rate of ~1% at 5 years, which is mainly due to difficulty in detecting pancreatic cancer at early stages (4). Approximately 280,000 new pancreatic cancer cases are diagnosed globally each year. The incidence of pancreatic cancer is expected to increase by 55% between 2010 and 2030. The most common risk factors for developing pancreatic cancer are smoking, diabetes, hereditary pancreatitis, multiple endocrine neoplasia type 1 syndrome, hereditary nonpolyposis colon cancer, von Hippel-Lindau syndrome, ataxia telangiectasia, and the familial atypical multiple mole melanoma syndrome (FAMMM) (3).
Like several other cancers, early detection of pancreatic cancer increases the probability of a better prognosis. However, pancreatic cancer is difficult to detect and diagnose because it does not exhibit any specific, detectable symptoms and is hidden behind other large abdominal organs (5). Moreover, pancreatic cancer causes symptoms that are characteristic of other common illnesses, such as jaundice, back/abdominal pain, loss of appetite, weight loss and fatigue (6). In addition to the lack of unique symptoms, pancreatic cancer is highly invasive and metastatic and highly resistant to chemotherapy (7,8). The standard treatment options available for patients with pancreatic cancer are surgery, radiation, chemotherapy [5-FU, gemcitabine and cetuximab) (9)], chemoradiation, and some marginally effective targeted therapies [inhibitors for VEGF, EGFR (9,10) mTOR (11)]. Despite remarkable global research efforts by clinicians and cancer scientists, pancreatic cancer remains fatal. Even though the incidence of pancreatic cancer is lower relative to other cancers, it is the fourth leading cause of cancer-related death in the U.S. According to the Pancreatic Cancer Action Network pancreatic cancer is expected to become the 2nd leading cause of cancer-related death by the year 2020. Because there is not an effective treatment for this deadly cancer (12), there is an immediate need to improve our understanding of pancreatic cancer. The better understanding of mechanism of disease pathogenesis will facilitate early detection and development of effective treatments. The focus of the present review is to summarize current knowledge regarding the roles of miRNAs in pancreatic cancer diagnosis, treatment and prognosis.
2. microRNA (miRNA or miR)
The importance of small non-coding microRNA (miRNA) molecules in the regulation of gene and protein expression was discovered and documented in early 90s. During the last decade, remarkable amount of research efforts have been dedicated towards understanding the role of miRNA regulatory networks in mammalian genetics and human diseases (12,13).
miRNAs are a class of post-transcriptional regulators, which mainly repress expression of genes and proteins. miRNAs form a part of the cellular regulatory network that governs various important biological functions, such as cell growth, proliferation, differentiation, development and apoptosis (14). In the recent past, miRNAs have become a central focus in cancer research because many miRNAs function as either tumor suppressors or oncogenes (Fig. 1). In addition, miRNAs are very promising diagnostic and prognostic biomarkers for human diseases, including pancreatic cancer. They are found in most biological fluids [e.g., blood, amniotic fluid, breast milk, bronchial lavage, cerebrospinal fluid (CSF), colostrum, peritoneal fluid, pleural fluid, saliva, seminal fluid, tears and urine] (15) and are more stable than protein and mRNA (16). Identification of biomarkers in biological fluids is particularly attractive because this represents a quick, non-invasive, and relatively inexpensive method for detecting and diagnosing disease (17). Identification of a specific miRNA profile in body fluids would be invaluable for early diagnosis, therapy and prognosis of pancreatic cancer. Several miRNAs have been shown to play a vital part in regulating pancreatic cancer by influencing pancreatic cancer growth, progression, invasion, metastasis and resistance to therapy (Table I) (18–28). The miRNAs that are specifically involved in pancreatic cancer and their target genes are shown in Table II (29–39).
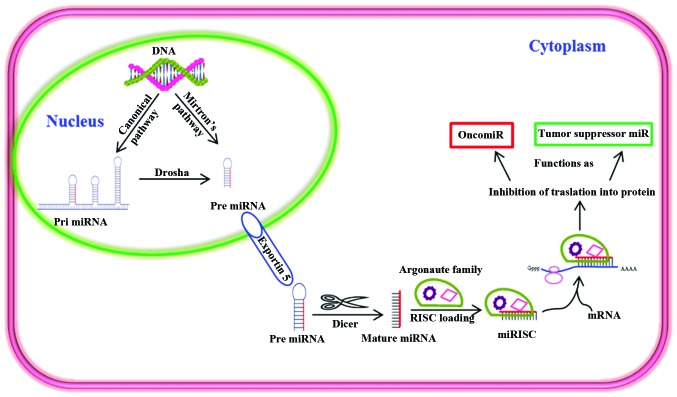
Figure 1.
Biogenesis and function of miRNA in cancer cells. Pre miRNAs are produced in the nucleus through canonical and mirton's pathway. Dicer processes pre miRNAs as they enter the cytoplasm via active exportin 5. miRNA-induced silencing complex (miRISC) is formed by integration of argonaute family proteins that act as guides and allow recognition of the complementary sequences in target mRNA. miRISC subsequently inhibits the translation of target mRNA or degrades the target mRNA. Various miRNAs can function as tumor suppressors or oncomiRs.
Table I.
The reported oncogenic miRNA (OncomiR) and tumor suppressor miRNAs in human pancreatic cancer.
| miRNA | Function | Reported activity for pancreatic cancer | Refs. |
|---|---|---|---|
| miR 21 | OncomiR | Expressed in early PDA | (18) |
| miR181b | OncomiR | miRNA-181b sensitizes PDAC cells to gemcitabine by targeting BCL-2 | (19) |
| miR-221 | OncomiR | Plasma miR-221 concentrations were significantly higher in PCa patients. Useful biomarker for cancer detection, monitoring tumor dynamics and predicting malignant outcomes | (20) |
| miR-27a-3p | OncomiR | Used as a biomarkers for PC diagnosis | (21) |
| miR-100 | OncomiR | Regulates IGF1-receptor expression in metastatic pancreatic cancer cells | (22) |
| miR-34 family | Tumor suppressor miR | Targeting the anti-apoptotic mRNAs such as BCL2, MYCN | (23) |
| miR-124 | Tumor suppressor miR | miR-124 is downregulated and epigenetically silenced in pancreatic cancer | (24) |
| miR-217 | Tumor suppressor miR | Frequently downregulated miR-217 can regulate KRAS and function as a tumor suppressor in PDAC. Useful therapeutic agent for miRNA-based PDAC therapy |
(25) |
| miR-148a | Tumor suppressor miR | An inappropriate therapeutic Tool for PC | (26) |
| miR-138 | Tumor suppressor miR | Regulates IGF1-receptor expression in metastatic pancreatic cancer cells | (22) |
| miR-15a-3p | Tumor suppressor miR | Induces apoptosis | (27) |
| miR-216a | Tumor suppressor miR | Downregulated expression of miR-216a suggests the involvement of miR-216a in the tumorigenesis and development of pancreatic cancer Potentially serve as a novel tumor marker and also a prognostic factor for pancreatic cancer | (28) |
Table II.
The deregulated miRNAs and target genes in PDAC.
| miRNA | Expression profile in tumors | Target genes of miRNA in PDAC | Refs. |
|---|---|---|---|
| miR-126, 217, 96, let-7, 143/145 | Downregulated | K-RAS, CRK, RREB1, Notch-1, AKT and β-catenin | (29,30) |
| miR-21 | Upregulated | PTEN, HNRPK, PDCD4, K-RAS, BTG2, TGFBR2, MARCKS, pellino-1, TM1, SPRY2, TPM1, Maspin, PPAR α, RECK, NFIB, CDC25A, LRRFIP1, BCL2, RTN4 and TAp63 | (31–35) |
| miR-210 | Upregulated | NPTX1, EFNA3, AcvR1b, ISCU, COX10, E2F3, RAD52, ACVR1B, MNT, FGFRL1, HOXA1 and HOXA9 | (31,33,34,36,37) |
| miR-155 | Upregulated | LDOC1, TP53INP1, HIF-1a, VEZF1, FOXP3, FOXO3a, CYR61, RHOA, SMAD1, SMAD5, HIVEP2, CEBPB, RUNX2, MYO10, MyD88, SHIP1, C/EBP β, IFN-gammaR α, JARID2, BACH1, ZIC3, ZNF652, ARID2, GTR1, AT1R, RIPK1, FADD, AID, JARID2 and RHOA | (31–34,36,37) |
| miR-196a | Upregulated | HOXA1, HOXA5, HOXB6, S100A9, KRT5, SPRR2C, HOXA7, HOXB8, HOXC8, HOXD8, HMGA2 and ANXA1 | (31,34,36,37) |
| miR-200b | Upregulated | EP300 and RND3 | (34,35) |
| miR-221 | Upregulated | p27 Kip1, p57 Kip 2, KIT, ER-α, PUMA, PTEN, MDM2, PI3K, ICAM-1, Bmf and Bim | (32–36) |
| miR-222 | Upregulated | P57, TP53INP1, PUMA, PTEN, p27, Bim, MMP1, SOD2 and STAT5A | (33,34,36) |
| miR-196a | Upregulated | HOXA1, HOXA5, HOXB6, S100A9, KRT5, SPRR2C, HOXA7, HOXB8, HOXC8, HOXD8, HMGA2 and ANXA1 | (31,34,36,37) |
| miR-15b | Upregulated | HOXB8, ZEB1, SIP1, Arl2 and BCL2 | (32,34,38) |
| miR-186 | Upregulated | P2X7, HOXB8, ZEB1 and SIP1 | (34) |
| miR-190 | Upregulated | NeuroD and TP53INP1 | (31,34,39) |
All the miRNAs and their targeting genes are involved in the following major 11 signaling pathways of PDAC such as K-Ras signaling, Notch signaling, angiogenesis signaling, cell cycle signaling, apoptosis signaling, invasion and metastasis signaling, Her2/neu signaling, PTEN signaling, insulin receptor signaling, PI3K/AKT signaling, Hedgehog signaling, glioblastoma multiforme signaling and prostate cancer signaling (29,30,39).
3. miRNAs as biomarkers for pancreatic cancer diagnosis
The overall sensitivity for the detection of pancreatic cancer by established gastrointestinal tumor markers, such as carcinoembryonic antigen (CEA) and CA 19-9 varies depending on tumor grade. However, the sensitivity and specificity of CA 19-9 for accurate diagnosis of pancreatic cancer using serum is debatable. Therefore, conventional serum antigen markers are inappropriate tools for diagnosing pancreatic cancer (40,41). There is a great need to develop better diagnostic markers for pancreatic cancer. Some progress has been made towards delineating the roles of miRNAs in the pathogenesis of pancreatic cancer. Blood is one of the easiest and most preferable types of biofluid for diagnosis and detection of most diseases. Plasma levels of miRNA-16 and miRNA196a in combination with CA 19-9 has been shown to work very efficiently for screening early pancreatic cancer (85.2%) (42). In addition, Wang et al (43) studied the expression levels of miR-21, miR-210, miR-155 and miR-196a in plasma from PDAC patients in comparison to healthy individuals, which showed that miR-155 overexpression was an early biomarker for pancreatic neoplasia, while miR-196a expression correlated with the progression of PDAC (43). In another study, overexpression of miR-155 was found in 80% of early pancreatic lesions (stage II) in microdissected panIN tissues (44). In addition, blood samples collected from pancreatic cancer patients had higher expression levels of miR-200a, 200b and 210 (15). Furthermore, the combination of miR-196a and miR-217 expression patterns differentiated PDAC from healthy controls and chronic pancreatitis cases (45). Using a similar approach, another group of researchers also observed much higher levels of circulating miR-18a in the plasma of 36 pancreatic cancer patients with compared to 30 healthy volunteers (46). These reports indicate the importance of miRNAs as potential biomarkers for the diagnosis of pancreatic cancer.
4. miRNAs as therapeutic targets in pancreatic cancer
The widely used chemotherapeutic treatment for pancreatic cancer is gemcitabine, which shows a moderate tumor suppression response rate of ~12% (12). Therefore, the development of new and improved therapies for the treatment of pancreatic cancer is crucial. Clinical studies have demonstrated the efficacy of miRNA as a therapeutic tool in the management of PDAC (12,47). Tremendous efforts have been made in vitro and in vivo using preclinical models of cancer, to inhibit oncogenic miRNAs with antagomiRs (48,49). AntagomiRs exhibit great potential as miRNA-based therapeutics for cancer treatment. However, optimization and execution of miRNA-based therapeutics lag behind other current therapies (48,49). Additional research is needed in order for miRNA-based therapy to become a standard anticancer therapy. Based on current data, intense research efforts are required to improve outcome for successful pancreatic cancer treatment.
The role of potential target, hsa-miR-155, which is upregulated in PDAC was reported to be a regulator of the putative tumor suppressor SEL1L. According to this report, inhibition of this aberrantly upregulated miRNA in human pancreatic ductal adenocarcinoma would serve as a potential therapeutic strategy for PDAC by increasing expression of SEL1L (50). In another report, the therapeutic efficacy of miR- 34b was demonstrated using pancreatic tissues from 64 pancreatic cancer patients. miR-34b was shown to act as a tumor metastasis suppressor through negative modulation of oncogenic SMAD3 (51). Another miRNA with possible therapeutic potential in pancreatic cancer is miR-142-3p. Triptolide, a diterpene triepoxide isolated from the Chinese herb Tripterygium wilfordii inhibits the proliferation of pancreatic cancer cells by upregulating miR-142-3p which negatively regulates HSP70 expression in PDAC cell lines (52). Similarly, Qazi et al (9) showed that enforced expression of miR-101, enhanced the expression of E-cadherin levels and reduced the pancreatic tumor growth rate in SCID mouse xenograft model. Thus, miR-101 has demonstrated a potential therapeutic target of PDAC (9).
However, despite these promising studies, it is important to consider that not all patients respond the same way to a given anticancer therapy. Therefore, the best treatment approach is the one in which the therapy can be tailored to each individual patient. miRNAs provide the foundation for developing tailored and targeted treatment strategies against pancreatic cancer, because both specific miRNAs and antagomiRs can be identified easily and quickly in blood or other bodily fluids to determine the best treatment strategy. Thus, instead of concentrating on one miRNA or antagomiR, a more effective approach would be a combination of therapies, in which a panel of miRNAs/antagomiRs in conjunction with chemotherapy is tailored to meet the needs of each patient.
5. Role of miRNAs in the prognosis of pancreatic cancer
Global miRNA microarray profiling may discriminate miRNA expression in normal vs. pancreatic cancer tissues and serve as a potential prognostic predictor of disease. High expression of miR-452, miR-105, miR-127, miR-518a-2, miR-187, and miR-30a-3p correlated with increased survival rates of more than two years (53). Notably, deregulated levels of miRNAs, miR-21, miR-155, and miR-196a in plasma, and miR-141 in the sera were observed in pancreatic cancer patients who had a poor overall survival rate (54). Furthermore, another study also reported that the levels of miR-196a were shown to be elevated in sera of the PDAC patients (55) in correlation with poor survival and advanced disease stage (55,56). In addition, it has been suggested that miR-196a expression is a more specific indicator of PDAC progression (45). Overexpression of miR-196a-2 and miR-219 are also associated with reduced survival. The median survival for patients with miR-196a-2 overexpression was 14.3 months compared with 26.5 months for those with low expression (57). The median survival for those with miR-219 overexpression was 13.6 months compared with 23.8 for those with low expression (58).
Comparative analysis of 98 normal and 88 PDAC patients revealed miR-21 and miR-155 as good biomarkers for predicting tumor stage and prognosis (58). Low expression of miR-21 was associated with increased survival. On the contrary, high expression of miR-34a and miR-30d was correlated with increased survival in patients who did not receive surgical tumor resection. Patients who underwent surgery for PDAC are expected to have reduced overall survival if they have high expression of miR-212 and miR-675 and low expression of miR-148a, miR-187 and let-7g (59). Compared with ductal epithelia from normal pancreatic tissues of organ donors, expression of miR-21 was upregulated by >1000-fold in microdissected pancreatic cancer tumors (60). Overexpression of miR-21 was associated with worse pancreatic cancer progression in gemcitabine-treated PDAC patients (60). miR-10b is another important miRNA marker that could predict successful treatment response and suitability for surgery in PDAC patients (56). In a study, 106 patient samples examined for the levels of expression of miR-10b, which revealed low levels of miR-10b in epithelial cells from benign lesions compared to those in pancreatic cancer lesions. The authors proposed that decreased miR-10b could improve the response to multimodality neoadjuvant therapy (61,62).
Two independent sets of samples obtained from patients with resected PDAC (with subsets of 19 and 60 patients) were subjected to miRNA microarray analysis. The analysis confirmed that miR-211 expression was the most predictive biomarker for treatment outcome in PDAC patients with significant associations with both overall survival and disease-free survival (63). Schultz and others (59) studied the expression levels of miRNAs in tissue samples from 225 patients. The results strongly suggested that miRNAs, miR-212 and miR-675 were upregulated and miR-148a, miR-187 and let-7g were downregulated and these miRNAs could function as independent predictors for reduced overall survival in PDAC patients (59). Thus, altered expression of miRNAs are strongly associated with poor survival, decreased response to treatment, and increased metastatic disease in PDAC (Table III) (16,64–68).
Table III.
MicroRNAs predicted to be involved in pancreatic cancer patient survival.
| miR in pancreatic cancer | Survival status | Expression status | Refs |
|---|---|---|---|
| miR-10b, miR-219, miR-196a and 196a-2p | Poor survival | High expression | (33,55,64) |
| miR-21 | Worse survival | High expression | (65) |
| miR-142-5p, 204 and miR-200c | Improved survival | High expression | (66,67) |
| miR-34a | Better survival | High expression | (67) |
| miR-216 and -217 | Poor survival | Reduced expression | (68) |
6. miRNA induces chemoresistance in pancreatic cancer
Chemotherapy is the most common treatment option for many cancers, including pancreatic cancer. However, it offers little therapeutic value in many cases due to rapid development of chemoresistance. Pancreatic cancer is a notoriously chemoresistant cancer, which renders chemotherapy highly ineffective in a majority of patients. Various miRNAs, notably oncomiR-21, miR-34a, miR-196a, miR-221 and miR-214, have been implicated in the induction of chemoresistance in PDAC (56,69). miR-10b, miR-34a and members of the miR-200 family were reported to have roles in gemcitabine chemoresistance. Upregulation of miR-21 was implicated in the induction of chemoresistance in PANC-1 cells (70). In contrast, downregulation of miR-21 increased the sensitivity of PANC-1 cells to gemcitabine and indole-3-carbinol combination therapy. Similarly, miR-10b has been shown to be responsible for chemoresistance in many tumor types. Cancer stem cells (CSCs) are highly resistance to chemotherapeutic drugs; miRNAs are shown to be involved in developing resistance to chemotherapy by CSCs (71). In this regard, the roles of miRNAs and stem cells in the development of PDAC chemoresistance need to be examined. Pancreatic cancer stem cells sorted from the BxPC3 cell line exhibited much higher chemoresistance upon holoclone formation, which coincided with high levels of miR-214, miR-21, miR-221, miR-222 and miR-155 and lower levels of Let-7a, miR-30c, miR-30b and miR-30a. Holoclone-forming stem cells may contribute to pancreatic cancer chemoresistance. Therefore, the stem cell holoclones represent a useful model for studying pancreatic cancer chemoresistance and for designing treatment strategies to effectively combat the development of chemoresistance (72).
7. Importance of model selection for miRNA research in pancreatic cancer
During the past several decades, a great deal of effort and resources have been utilized but only a few promising advances have been made towards more effective prevention and treatment strategies against pancreatic cancer. A major limitation has been a lack of animal models that mimic the human disease. Several chemotherapeutic drugs for pancreatic cancer may be highly effective in two-dimensional (2D) in vitro cell culture models, while many of these drugs fail to show the same effect in clinical trials. A recent study by Longati et al (8) reported the use of three-dimensional (3D) culture models, in which significant increases in chemo- and radiation therapy resistance were observed in comparison with 2D cell culture models. These 3D models were associated with increased expression of matrix proteins, stromal markers, and various miRNAs, which are known to be involved in the multidrug-resistant phenotypes of many pancreatic cancers. Consequently, most drugs tested in 3D cultures showed limited effects in contrast to 2D cultures (8). Thus, the 3D culture system is a more suitable model for predicting responsiveness to a potential drug. Traditional cell culture-based assays and xenograft models consume huge amounts of financial resources but do not necessarily improve the clinical outcome of patients with PDAC. Genetically engineered mouse models (GEMM) are suitable for identifying early genetic alterations in pancreatic precursor lesions and for studying early diagnosis and treatment. A major advantage of using GEMM is the conditional strategy to activate or inactivate expression of therapeutic or pathogenic genes, respectively, temporarily in specific tissues. GEMM also reflect intratumoral genetic heterogeneity, which is one of the hallmarks of PDAC. A lack of intratumoral genetic heterogeneity in xenografts and the cell lines may produce results that are not a true representation of human pancreatic cancer (73). Another approach is a patient-derived xenograft (PDX) model, a key model to translate the effective miRNAs into clinics as potential biomarkers or therapeutic targets for PDAC. PDX is increasingly used to characterize and validate several diagnostic and therapeutic strategies. The resemblance of intra and intertumoral genetic heterogeneity along with complexity of the microenvironment of PDX mimics more accurately the human PDAC. Direct representation of parental tumor with realistic genetic integrity is an advantage of PDX over the cell line-derived xenograft model. Another advantage of clinically relevant PDX is the similar stromal compartments of the parental tumor at the early passages. The key features of orthotopic and subcutaneous PDX are accurate predictability of drug response, identification of tumor specific treatment regimens, and high correlation between pre-clinical and clinical outcome in terms of relative sensitivity, resistance and therapeutic efficacy of the drug with molecular insights (74,75). Overall, PDX seems to be an appropriate and sufficient model to recapitulate both genetic and morphological alterations that leads to the development of PDAC. Together, these findings suggest that advanced models are required to test and validate miRNAs that would be effective in early diagnosis and development of suitable treatment for pancreatic cancer.
8. Future directions
Future research on the roles of miRNAs in the pathogenesis of pancreatic cancer will unfold the mechanisms of this deadly disease. Studies on the biogenesis of miRNAs will help in understanding the regulatory machinery of the overall metabolic state of the tumor. The identification of the key metabolic switches that favor expression of a particular set of miRNAs should help improve therapeutic odds of success in pancreatic cancer.
According to the current scenario, many key oncogenic and tumor suppressor miRNAs and downstream molecular targets have been identified in PDAC. Identification and targeting of pancreatic cancer associated miRNA will have a tremendous impact on non/minimally invasive diagnostics, that may open new doors to improve the rate of curative outcome. The discovery of pancreatic cancer related miRNA as a therapeutic agent is warranted to bring a major breakthrough in many aspects of pancreatic cancer therapy. The development of state-of-the art microRNA delivery methods may increase the potential of miRNAs as therapeutic agents of choice in pancreatic cancer. Future analysis of miRNAs in a larger patient cohort is warranted to verify if this is a useful biomarker for early detection, prognosis, and therapy in a majority of pancreatic cancer patients.
9. Conclusions
Recent studies demonstrate the diverse and significant roles of non-protein-coding genomic regions (miRNAs) in pancreatic cancer progression and metastasis. Therefore, future research efforts that focus on miRNAs must be just as vigorous as the efforts made towards understanding the contributions of the coding genome (individual genes). A great deal of research in this field is required before we can effectively translate this into the clinicals to treat pancreatic cancer. Understanding the roles of miRNAs and tapping into the excellent potential of miRNAs will have a great beneficial impact on the early detection, diagnosis, treatment and prognosis of pancreatic cancer.
Acknowledgements
We thank Dr Rebecca Lopez-Valdez for her professional editing of the manuscript.
Abbreviations
- miRNA
microRNA
- PDAC
pancreatic ductal adeno-carcinoma
References
- 1.Moniri MR, Dai LJ, Warnock GL. The challenge of pancreatic cancer therapy and novel treatment strategy using engineered mesenchymal stem cells. Cancer Gene Ther. 2014;21:12–23. doi: 10.1038/cgt.2013.83. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaud DS. Epidemiology of pancreatic cancer. Minerva Chir. 2004;59:99–111. [PubMed] [Google Scholar]
- 4.Tamburrino A, Piro G, Carbone C, Tortora G, Melisi D. Mechanisms of resistance to chemotherapeutic and anti-angiogenic drugs as novel targets for pancreatic cancer therapy. Front Pharmacol. 2013;4:56. doi: 10.3389/fphar.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 6.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 7.Cheng H1, Shi S, Cai X, Long J, Xu J, Liu C, Yu X. microRNA signature for human pancreatic cancer invasion and metastasis. Exp Ther Med. 2012;4:181–187. doi: 10.3892/etm.2012.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longati P, Jia X, Eimer J, Wagman A, Witt MR, Rehnmark S, Verbeke C, Toftgård R, Löhr M, Heuchel RL. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer. 2013;13:95. doi: 10.1186/1471-2407-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qazi AM1, Gruzdyn O, Semaan A, Seward S, Chamala S, Dhulipala V, Sethi S, Ali-Fehmi R, Philip PA, Bouwman DL, et al. Restoration of E-cadherin expression in pancreatic ductal adenocarcinoma treated with microRNA-101. Surgery. 2012;152:704–711. doi: 10.1016/j.surg.2012.07.020. discussion 711–713. [DOI] [PubMed] [Google Scholar]
- 10.Borja-Cacho D, Jensen EH, Saluja AK, Buchsbaum DJ, Vickers SM. Molecular targeted therapies for pancreatic cancer. Am J Surg. 2008;196:430–441. doi: 10.1016/j.amjsurg.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai ZJ, Gao J, Kang HF, Ma YG, Ma XB, Lu WF, Lin S, Ma HB, Wang XJ, Wu WY. Targeted inhibition of mammalian target of rapamycin (mTOR) enhances radiosensitivity in pancreatic carcinoma cells. Drug Des Devel Ther. 2013;7:149–159. doi: 10.2147/DDDT.S42390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaconstantinou IG, Lykoudis PM, Gazouli M, Manta A, Polymeneas G, Voros D. A review on the role of microRNA in biology, diagnosis, and treatment of pancreatic adenocarcinoma. Pancreas. 2012;41:671–677. doi: 10.1097/MPA.0b013e31823c9d21. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Ping Y, Li X, Zhao H, Wang L, Fan H, Xiao Y, Li X. Prioritizing candidate disease miRNAs by integrating phenotype associations of multiple diseases with matched miRNA and mRNA expression profiles. Mol Biosyst. 2014;10:2800–2809. doi: 10.1039/C4MB00353E. [DOI] [PubMed] [Google Scholar]
- 14.Qiu C, Chen G, Cui Q. Towards the understanding of microRNA and environmental factor interactions and their relationships to human diseases. Sci Rep. 2012;2:318. doi: 10.1038/srep00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humeau M, Torrisani J, Cordelier P. miRNA in clinical practice: Pancreatic cancer. Clin Biochem. 2013;46:933–936. doi: 10.1016/j.clinbiochem.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Vlastos G, Verkooijen HM. Minimally invasive approaches for diagnosis and treatment of early-stage breast cancer. Oncologist. 2007;12:1–10. doi: 10.1634/theoncologist.12-1-1. [DOI] [PubMed] [Google Scholar]
- 18.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai B, An Y, Lv N, Chen J, Tu M, Sun J, Wu P, Wei J, Jiang K, Miao Y. miRNA-181b increases the sensitivity of pancreatic ductal adenocarcinoma cells to gemcitabine in vitro and in nude mice by targeting BCL-2. Oncol Rep. 2013;29:1769–1776. doi: 10.3892/or.2013.2297. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY, Wang XL. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila) 2013;6:331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Egger M, Grizzle W, McNally L. MicroRNA-100 regulates IGF1-receptor expression in metastatic pancreatic cancer cells. Biotech Histochem. 2013;88:397–402. doi: 10.3109/10520295.2012.762460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, Chen L, Zhang J, Chen H, Fan J, Wang K, Luo J, Chen Z, Meng Z, Liu L. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2013;33:514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 25.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 26.Delpu Y, Lulka H, Sicard F, Saint-Laurent N, Lopez F, Hanoun N, Buscail L, Cordelier P, Torrisani J. The rescue of miR-148a expression in pancreatic cancer: An inappropriate therapeutic tool. PLoS One. 2013;8:e55513. doi: 10.1371/journal.pone.0055513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Druz A, Chen YC, Guha R, Betenbaugh M, Martin SE, Shiloach J. Large-scale screening identifies a novel microRNA, miR-15a-3p, which induces apoptosis in human cancer cell lines. RNA Biol. 2013;10:287–300. doi: 10.4161/rna.23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou B, Jian Z, Chen S, Ou Y, Li S, Ou J. Expression of miR-216a in pancreatic cancer and its clinical significance. Nan Fang Yi Ke Da Xue Xue Bao. 2012;32:1628–1631. [PubMed] [Google Scholar]
- 29.Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012;7:e32068. doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drakaki A, Iliopoulos D. MicroRNA-gene signaling pathways in pancreatic cancer. Biomed J. 2013;36:200–208. doi: 10.4103/2319-4170.119690. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: From biology to biomarkers of disease. J Biosci. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 32.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mees ST, Mardin WA, Wendel C, Baeumer N, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. EP300 - a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int J Cancer. 2010;126:114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- 36.Szafranska AE1, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 37.Szafranska AE1, Doleshal M, Edmunds HS, Gordon S, Luttges J, Munding JB, Barth RJ, Jr, Gutmann EJ, Suriawinata AA, Marc Pipas J, Tannapfel A, et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54:1716–1724. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Cai X, Huang F, Zhong W, Yu Z. Effect of trichostatin a on viability and microRNA expression in human pancreatic cancer cell line BxPC-3. Exp Oncol. 2008;30:265–268. [PubMed] [Google Scholar]
- 39.Gironella M1, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pawa N, Arulampalam T, Norton JD. Screening for colorectal cancer: Established and emerging modalities. Nat Rev Gastroenterol Hepatol. 2011;8:711–722. doi: 10.1038/nrgastro.2011.205. [DOI] [PubMed] [Google Scholar]
- 41.Lee KJ, Yi SW, Chung MJ, Park SW, Song SY, Chung JB, Park JY. Serum CA 19-9 and CEA levels as a prognostic factor in pancreatic adenocarcinoma. Yonsei Med J. 2013;54:643–649. doi: 10.3349/ymj.2013.54.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131:683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu JK, Hong SM, Karikari CA, Hruban RH, Goggins MG, Maitra A. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10:66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JY, Helm J, Coppola D, Kim D, Malafa M, Kim SJ. MicroRNAs in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2011;17:817–827. doi: 10.3748/wjg.v17.i7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keklikoglou I, Hosaka K, Bender C, Bott A, Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H, et al. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2014 Dec 15; doi: 10.1038/onc.2014.408. http://dx.doi.org/10.1038/onc.2014.408. [DOI] [PMC free article] [PubMed]
- 48.Song S, Ajani JA. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2013;10:109–118. doi: 10.1038/nrgastro.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Putative tumor suppressor gene SEL1L was downregulated by aberrantly upregulated hsa-mir-155 in human pancreatic ductal adenocarcinoma. Mol Carcinog. 2013;53:711–712. doi: 10.1002/mc.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, Cheng H, Shi S, Cui X, Yang J, Chen L, Cen P, Cai X, Lu Y, Wu C, et al. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr Mol Med. 2013;13:467–478. doi: 10.2174/1566524011313040001. [DOI] [PubMed] [Google Scholar]
- 52.MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, Subramanian S, Saluja AK. Triptolide induces the expression of miR-142-3p: A negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12:1266–1275. doi: 10.1158/1535-7163.MCT-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun T, Kong X, Du Y, Li Z. Aberrant MicroRNAs in pancreatic cancer: Researches and clinical implications. Gastroenterol Res Pract. 2014;2014:386561. doi: 10.1155/2014/386561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 56.Frampton AE, Krell J, Jacob J, Stebbing J, Jiao LR, Castellano L. microRNAs as markers of survival and chemo-resistance in pancreatic ductal adenocarcinoma. Expert Rev Anticancer Ther. 2011;11:1837–42. doi: 10.1586/era.11.184. [DOI] [PubMed] [Google Scholar]
- 57.Mardin WA, Mees ST. MicroRNAs: Novel diagnostic and therapeutic tools for pancreatic ductal adenocarcinoma? Ann Surg Oncol. 2009;16:3183–9. doi: 10.1245/s10434-009-0623-1. [DOI] [PubMed] [Google Scholar]
- 58.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 59.Schultz NA, Andersen KK, Roslind A, Willenbrock H, Wojdemann M, Johansen JS. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index. World J Surg. 2012;36:2699–2707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 60.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, et al. MicroRNA-21 in pancreatic cancer: Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 61.Bauer AS, Keller A, Costello E, Greenhalf W, Bier M, Borries A, Beier M, Neoptolemos J, Büchler M, Werner J, et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One. 2012;7:e34151. doi: 10.1371/journal.pone.0034151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol. 2012;9:435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- 63.Giovannetti E1, van der Velde A, Funel N, Vasile E, Perrone V, Leon LG, De Lio N, Avan A, Caponi S, Pollina LE, et al. High-throughput microRNA (miRNAs) arrays unravel the prognostic role of MiR-211 in pancreatic cancer. PLoS One. 2012;7:e49145. doi: 10.1371/journal.pone.0049145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, Longnecker DS, Gutmann EJ, Sempere LF, Korc M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17:5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Yu J1, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, Ueda J, Nagai E, Hashizume M, Tanaka M. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18:2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 69.Hasegawa S, Eguchi H, Nagano H, Konno M, Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N, et al. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer. 2014;111:1572–1580. doi: 10.1038/bjc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paik WH, Kim HR, Park JK, Song BJ, Lee SH, Hwang JH. Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. 2013;33:1473–1481. [PubMed] [Google Scholar]
- 71.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM, Fortes P, Garcia-Foncillas J. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 72.Tan L, Sui X, Deng H, Ding M. Holoclone forming cells from pancreatic cancer cells enrich tumor initiating cells and represent a novel model for study of cancer stem cells. PLoS One. 2011;6:e23383. doi: 10.1371/journal.pone.0023383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazur PK, Siveke JT. Genetically engineered mouse models of pancreatic cancer: Unravelling tumour biology and progressing translational oncology. Gut. 2012;61:1488–1500. doi: 10.1136/gutjnl-2011-300756. [DOI] [PubMed] [Google Scholar]
- 74.Choi SY, Lin D, Gout PW, Collins CC, Xu Y, Wang Y. Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv Drug Deliv Rev. 2014:79–80. 222–237. doi: 10.1016/j.addr.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 75.Mazur PK, Herner A, Neff F, Siveke JT. Current methods in mouse models of pancreatic cancer. Methods Mol Biol. 2015;1267:185–215. doi: 10.1007/978-1-4939-2297-0_9. [DOI] [PubMed] [Google Scholar]