Abstract
Defects in the DNA mismatch repair (MMR) proteins, result in a phenotype called microsatellite instability (MSI), occurring in up to 15% of sporadic colorectal cancers. Approximately one quarter of colon cancers with deficient MMR (dMMR) develop as a result of an inherited predisposition syndrome, Lynch syndrome (formerly known as HNPCC). It is essential to identify patients who potentially have Lynch syndrome, as not only they, but also family members, may require screening and monitoring. Diagnostic criteria have been developed, based primarily on Western populations, and several methodologies are available to identify dMMR tumours, including immunohistochemistry and microsatellite testing. These criteria have provided evidence supporting the introduction of reflex testing. Yet, it is becoming increasingly clear that tests have a limited sensitivity and specificity and may yet be superseded by next generation sequencing. In this review, the limitations of diagnostic criteria are discussed, and current and emerging screening technologies explained. There is now useful evidence supporting the prognostic and predictive value of dMMR status in colorectal tumours, but much less is known about their value in extracolonic tumours, that may also feature in Lynch syndrome. This review assesses current literature relating to dMMR in endometrial, ovarian, gastric and melanoma cancers, which it would seem, may benefit from large-scale clinical trials in order to further close the gap in knowledge between colorectal and extracolonic tumours.
Keywords: Lynch syndrome, deficient mismatch repair, microsatellite instability, prognostic, predictive
1. Introduction
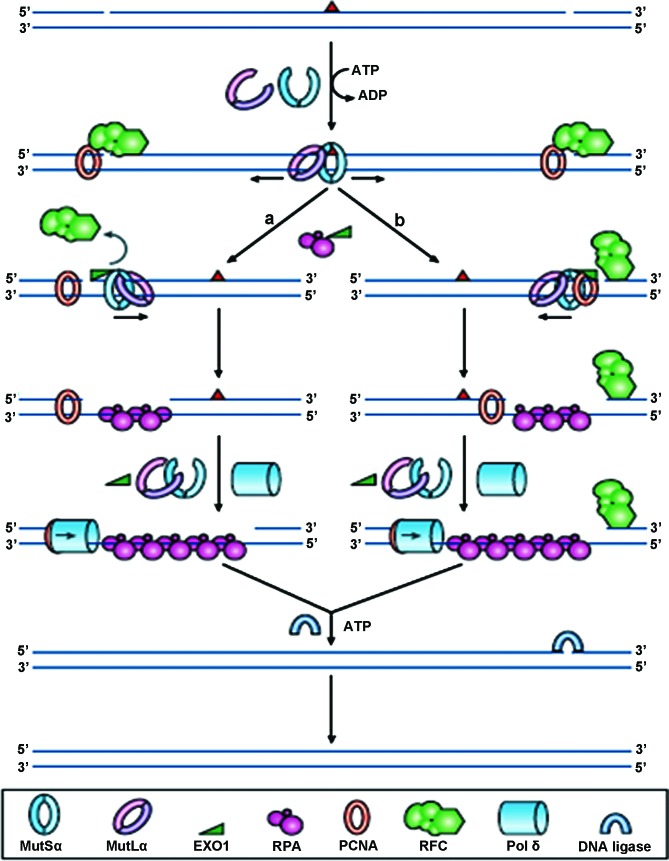
DNA mismatch repair (MMR) is a very highly conserved cellular process, involving many proteins, resulting in the identification, and subsequent repair of mismatched bases, likely to have arisen during DNA replication, genetic recombination or chemical or physical damage (Fig. 1). The MMR genes play additional roles in double-strand break repair, apoptosis and recombination. The four key genes identified to date are mutL homologue 1 (MLH1), mutS homologue 2 (MSH2), mutS homologue 6 (MSH6) and postmeiotic segregation increased 2 (PMS2), so named because of their homology to the E. coli MMR genes. The MSH2 and MSH6 proteins form a heterodimeric complex (mutSα) which is involved in the initial identification of mismatched bases, and initiates DNA repair. Binding to the mismatch results in an ATP-dependent conformational change, which subsequently recruits mutLα, a heterodimer comprising of MLH1 and PMS2. Other proteins are recruited to complete the DNA repair, but are not discussed further in this review. The repair complexes ensure that it is the newly synthesised strand of DNA which is targeted for repair, not the parental strand.
Figure 1.
Reprinted by permission from Macmillan Publishers Ltd., Nature Reviews Molecular Cell Biology, 7 (5): 335–346, copyright 2006. DNA mismatch repair. In normal cells, any mismatched base pairs (or incorrect insertion or deletion loops) are repaired by the complex machinery which forms the DNA mismatch repair process. MSH2 and MSH6 form a heterodimeric complex, called mutSα, which identifies and binds to the error, resulting in an ATP-dependent conformational change, which recruits mutLα, a heterodimer consisting of MLH1 and PMS2. The resultant complex undergoes an ATP-driven conformational alteration, releasing it from the error site. If it diffuses upstream, it displaces replication factor C (RFC) and loads exonuclease-1 (EXO1). This degrades the strand in the 5′→3′ direction. Replication factor A (RPA) then stabilises the single-stranded DNA, while a complex of DNA polymerase Pol δ (Pol δ) and proliferating cell nuclear antigen (PCNA) fills the gap and finally DNA ligase seals the remaining nick to finalise the repair. If the mutSα/mutLα complex diffuses downstream, EXO1 is recruited and degrades the region of the DNA strand, up to the RFC complex. As stated before, the single-strand is stabilised by bound RPA, which also inhibits EXO1 activity. Pol δ fills the gap and finally DNA ligase I seals the remaining nick to finalise the repair.
When the MMR system develops a functional error or defect, this results in a particular phenotype called microsatellite instability (MSI). This is characterised by the insertion or deletion of short, repetitive sequences of DNA, resulting in mutations in cancer-related genes. The increase in the rate of mutations in cells exhibiting deficient mismatch repair (dMMR), may confer a Darwinian survival advantage. The cause of the dMMR system is different depending upon whether the tumour is sporadic in origin, or as a result of the autosomal dominant inherited predisposition condition, Lynch syndrome.
2. dMMR in sporadic colorectal cancer (CRC)
Many people wrongly group colon and rectal tumours together as ‘colorectal’, when referring to rates of dMMR. It is in fact very rarely seen in rectal cancers (1) but accounts for between 10 and 15% of sporadic colon cancers. This MSI phenotype is associated with several clinicopathological features such as a proximal primary tumour location, high grade, mucinous pathology, early stage, diploid and the presence of the BRAF p. (V600E) mutation (2). In addition, they tend to also be associated with being female, smoking and older age at onset. Furthermore, most of these sporadic MSI tumours are thought to arise from sessile serrated adenomas or polyps (3). This pathway of colorectal cancer development is different to the Fearon and Vogelstein adenoma-carcinoma pathway (4). In the majority of tumours, the defect in the MMR system is the inactivation of MLH1, through methylation of CpG islands in the promoter, causing transcriptional silencing of the gene. Limited data also suggest that inactivation in a small subset of tumours is caused my mutation of the MLH1 gene itself (5–9).
3. dMMR in Lynch syndrome
Lynch syndrome (10) (formerly known as hereditary non-polyposis colorectal cancer; HNPCC) is the most common hereditary cancer predisposition syndrome, and is associated with a high risk of colorectal cancer and also extra-colonic tumours, particularly endometrial. In fact, the risk of endometrial cancer in women within some affected families may actually be greater than the risk of CRC (11). The average age at onset, of <45, is significantly lower than that for sporadic tumours and the cause of the defect in the MMR system in Lynch syndrome is constitutional mutations of the MLH1 or MSH2 genes, rather than methylation-induced inactivation of MLH1. The InSiGHT database (12) has been developed to record all mutations observed in patients with Lynch syndrome, and data from this suggest that mutations in MLH1 account for 42% of Lynch syndrome, mutations in MSH2 account for 33% and the remainder are found in MSH6 (18%) and PMS2 (7%).
A very small subset of Lynch syndrome patients is characterised by the presence of ‘constitutional epimutations’ of MLH1. These are characterised by promoter methylation and transcriptional silencing of a single allele of a gene in normal tissues, in an otherwise intact gene. Since they appear to confer a similar phenotype to that caused by sequence mutations, they are considered to be an alternative aetiological mechanism for Lynch syndrome (13). This phenomenon was first recognised in 2002 by Gazzoli et al (14). Several more recent studies (15–17) screened constitutive DNA samples for MLH1 methylation, in CRC patients who had lost MLH1 expression in their tumours, without deleterious germline mutations in MLH1. Each study found low levels of constitutional MLH1 epimutations, but Ward et al suggest expanding screening programmes to include such patients, since testing of relatives identified paternal transmission (16).
In 2009, Ligtenberg et al proposed an alternative mechanism causing a defect in the MMR system in a subset of Lynch syndrome families (18). The study of patients from Dutch and Chinese families identified tumours which were deficient in MSH2 as a result of the presence of heterozygous germ-line deletions of the 3′ exons of the epithelial cell adhesion molecule (EPCAM; also known as TACSTD1) gene. Such deletions in EPCAM cause transcriptional read-through, which silences MSH2, and has been termed MSH2 ‘epimutation’. In 2011, Kloor et al suggested that loss of EPCAM protein expression, as assessed by immunohistochemistry (IHC) may be a suitable method of identifying Lynch syndrome patients with EPCAM germline deletions, as the majority of tumours with EPCAM germline deletions also showed loss of protein expression (19). Further to this study, Huth et al hypothesised that, as loss of expression did not always correlate with the presence of EPCAM germline deletions, that it was potentially the actual type of second somatic hit that determined EPCAM protein expression. Using multiplex ligation-dependent probe amplification (MLPA) to assess allelic deletion status, tumours with loss of EPCAM expression showed biallelic deletions, whereas tumours retaining EPCAM expression demonstrated monoallelic retention of the EPCAM gene. The group therefore concluded that EPCAM protein expression is dependent upon the actual localisation of the second somatic hit that inactivates MSH2 (20). More recently, a study by Musulen et al, showed a high specificity between the presence of EPCAM germline mutations and loss of EPCAM expression, and recommended the addition of EPCAM IHC into diagnostic Lynch syndrome testing, in patients with MSH2-negative tumours (21).
4. Who (and how) to test for mismatch repair deficiencies?
Diagnostic criteria and guidelines
Amsterdam criteria
The identification of a patient with a colorectal or endometrial tumour raises the question of whether to screen for the presence of Lynch syndrome. Various criteria have been in place for the past 35 years, to help guide this decision. In 1991, the Amsterdam criteria arose from a meeting of the International Collaborative Group on Hereditary Non-Polyposis Colon Cancer (ICG-HNPCC) where an attempt was made to standardise international criteria for identifying HNPCC patients for research purposes (22). These criteria were known as the ‘3-2-1 rule’: a) at least three relatives should have histologically confirmed CRC, with one being a first degree relative of the other two; b) there must be two successive generations affected; and c) one or more relatives must be diagnosed by the age of 50.
The Amsterdam criteria was later renamed Amsterdam criteria I, following the subsequent identification of the genes involved, which lead to the expansion of the criteria and its renaming Amsterdam Criteria II.
Amsterdam Criteria II
Based upon further research identifying the fact that Lynch syndrome tumours were not confined to the colon or rectum, the criteria were further expanded and updated in 1998, and renamed the Amsterdam II criteria (23). These new criteria added in the fact that at least three relatives should have a histologically confirmed HNPCC-associated cancer (colorectal, endometrial, small bowel, ureter or renal pelvis), rather than just a colorectal tumour.
Bethesda Guidelines
At around the same time, the National Cancer Institute of the USA published its own set of guidelines (24). These included the following criteria: a) individuals with cancer in families that meet the Amsterdam criteria; b) individuals with two HNPCC-related cancers, including synchronous and metachronous colorectal cancers or associated extra-colonic cancers (endometrial, ovarian, gastric, hepatobiliary, or small bowel cancer or transitional cell carcinoma of the renal pelvis or ureter); c) individuals with colorectal cancer and a first-degree relative with colorectal cancer and/or HNPCC-related extra-colonic cancer and/or a colorectal adenoma: one of the cancers diagnosed by age 45, and the adenoma diagnosed by age 40; d) individuals with colorectal cancer or endometrial cancer diagnosed by age 45; e) individuals with right-sided colorectal cancer with an undifferentiated pattern (solid/cribriform) on histopathology diagnosed by age 45; f) individuals with signet-ring-cell-type colorectal cancer diagnosed by age 45; and g) individuals with adenomas diagnosed by age 40.
Revised Bethesda Guidelines
In 2004, the NCI revised these guidelines, and went on to publish the Revised Bethesda Guidelines (25). These remain the most recent clinical diagnostic criteria upon which a patient is identified as likely having Lynch syndrome; a) individuals with CRC diagnosed by age 50; b) individuals with synchronous or metachronous CRC, or other HNPCC-associated tumours regardless of age; c) individuals with CRC and MSI-H histology diagnosed by age 60; d) individuals with CRC and more than 1 first degree relative with an HNPCC-associated tumour, with one cancer diagnosed by age 50; and e) individuals with CRC and more than 2 first degree relatives or second degree relatives with an HNPCC-associated tumour, regardless of age.
Jerusalem criteria
In 2009, the ‘Jerusalem criteria’ were published, recommending that either dMMR IHC or MSI testing be carried out on every colorectal tumour, where the patient is under the age of 70 at diagnosis (26). The idea behind this broader screening programme was to identify potential Lynch syndrome patients with an MSH6 or PMS2 mutation, who tend to present at a later age, and would not be included for screening, under the revised Bethesda guidelines.
5. Does ‘one size’ really fit all?
All of the above criteria for selecting patients for screenings have been based upon North American and European populations. In order for these criteria to be used worldwide, this makes the assumption that there are no population-specific differences. A study by Yan et al, has questioned this very point in relation to a Chinese population, where there is a strict one child policy (27). The resultant large number of small families makes it almost impossible to meet all the specified criteria regarding the number of affected relatives. As a result this increases the likelihood of overlooking and not screening a high proportion of potential or actual Lynch syndrome patients.
A second factor bringing the relevance of using the Amsterdam or Bethesda criteria in Asian populations into question, is the fact that gastric and hepatocellular cancers are the most common extracolonic tumours seen in Chinese patients with Lynch syndrome, rather than endometrial tumours as seen in the West. Furthermore, it becomes difficult to gauge how specific this is for Lynch syndrome, when the rates of gastric and hepatocellular carcinoma (HCC) are so high in Asia due to Helicobacter pylori (H. pylori) and chronic hepatitis B virus (HBV) infections respectively. An H. pylori infection induces an inflammatory response, in addition to causing genetic changes which result in genetic instability (28). The oncogenic effects of HBV such as genomic instability result from its integration into the host genome (29).
A third factor is that several studies have reported a predominance of left-sided CRC in Asian populations, which is different to what is seen in Western patients, where there is a predominance of right-sided tumours. Wang et al (30) noted that 60.6% of 60 Lynch syndrome patients under study had distal colorectal tumours. Chew et al (31) undertook a study of 6,736 CRC patients, who underwent surgery for their disease at Singapore General Hospital between 1989 and 2005; 52 (0.8%) fulfilled the Amsterdam I or Amsterdam II criteria, so were included for analysis and 69% of these patients had left-sided tumours, the majority of which were located in the sigmoid colon (31). In a very recent study of 116 Chinese families with suspected Lynch syndrome, 32 of whom had confirmed MLH1 or MSH2 germline mutations, 56.5% of the colorectal tumours were left-sided (32). These observations could be as a result of the fact that rectal cancers are more prevalent in Asian populations, or simply the fact that this is a feature of Asian Lynch syndrome.
In Western populations, we know that 10–15% of sporadic CRC tumours are dMMR. This figure may be much higher in Asian populations, based upon a study carried out in Singapore on 240 CRC patients, under the age of 50 at presentation. MMR IHC was performed and 21% of patients showed loss of expression of at least one of the MMR proteins. The authors identified the fact that, had selection for screening been based solely on the Amsterdam criteria, a staggering 86% of patients would have not been identified as high risk of Lynch syndrome, and would thus not have been screened (33). This provides further evidence for the introduction of population-specific diagnostic screening criteria.
6. Reflex testing
In essence, reflex testing is the routine screening of all newly diagnosed colorectal tumours for dMMR, to increase the likelihood of identifying Lynch syndrome patients. Obviously early diagnosis will result in increased surveillance, thus hopefully reducing morbidity and mortality, not only for the affected individual, but also family members.
Several studies have proven the cost-effectiveness of such a screening approach (34–38). A Dutch study by Sie et al (39) recommended increasing the cut-off age for testing all CRC from 50 to 70 years old, and still found this strategy cost-effective. However, in spite of the potential financial savings, reflex testing is proving difficult to implement, with areas requiring attention being highlighted at a multidisciplinary working group meeting of the Centers for Disease Control and Prevention in the US (40). The group identified the lack of primary care provider knowledge of Lynch syndrome and testing issues, as the main barrier to implementation. Furthermore, it was recognised that there is a requirement for a strategy to ensure that at-risk relatives are identified and counselling offered. There is also very limited data available on the feasibility of carrying out such testing, so one recommendation is for additional ‘real-world’ studies to be carried out to generate such data.
Taking a whistle-stop tour of current practice worldwide, it would appear that much still needs to be done in terms of implementation. In the UK, despite reflex testing being mandated by the Royal College of Pathologists and recommended by the British Society of Gastroenterologists, less than 50% of National Health Service Hospital Trusts currently carry out screening on patients presenting with the disease under the age of 50. This is the case in England, Wales and Scotland, however, all social care trusts in Northern Ireland have successfully implemented screening. The National Services Division Scotland and the Molecular Pathology Consortium are currently trying to implement national screening throughout Scotland, with the rest of the UK hopefully following suit, once this model is in place (data from a Bowel Cancer UK freedom of information request sent out across the UK to establish the level of implementation) (41). The main reason for not screening was put down to the additional financial burden. A further reason given is a current lack of National Institute for Health and Care Excellence (NICE) guidance. NICE is an executive non-departmental public body within the Department of Health in the UK, and publishes guidelines in, amongst other areas, clinical practice. Another rather interesting reason is the potential impact on patients and their families. The fact remains, and must not be overlooked, that patients simply may not wish to undergo genetic testing. There are many negative perceptions of this type of screening, and unless patients are educated appropriately as to the potential benefits, this could remain a barrier to implementation.
A study in Canada by Tomiak et al determined that in order to increase the uptake of genetic services by patients with suspected Lynch syndrome, several areas needed addressing, such as improving health literacy for the general population, newly diagnosed patients, and perhaps a little surprising, healthcare professionals (42). The study highlighted a general lack of awareness of hereditary cancers and a lack of understanding of the need for, and potential benefits of, genetic screening and what is done with, and who has access to, the results. The requirement for psychosocial support was also highlighted as an area to be addressed. Tomiak et al concluded that these gaps need to be filled for the successful implementation of universal screening, planned by the US Office of Public Health Genomics, by 2020.
In 2012, Beamer et al carried out a questionnaire-based review of reflex testing practise across the United States of America, similar in design to that undertaken by Bowel Cancer UK, in the United Kingdom (43). They found that the level of reflex testing implementation was dependent primarily upon the level of cancer program [ranging from Community Hospital Cancer Programs (CHCP), to Community Hospital Comprehensive Cancer Programs (COMP), and finally up to the most complex level of National Cancer Institute-designated Comprehensive Cancer programs (NCI-CCC)]. Seventy-one percent of NCI-CCCs, 36% of COMPS yet only 15% CHCPs had already implemented reflex testing. Another point arising from this study is whether written patient consent is required. Currently this is not the case, presumably because screening a tumour provides phenotypic, rather than genotypic information, but it will be interesting to see whether this aspect becomes a barrier to worldwide reflex testing.
Back in 2008, in the state of Western Australia, routine screening for Lynch syndrome was implemented. All patients under the age of 60 at the time of diagnosis are screened and figures published recently estimate that the majority of Lynch syndrome cases are being identified as a result of this programme (44).
7. MMR Immunohistochemistry (IHC)
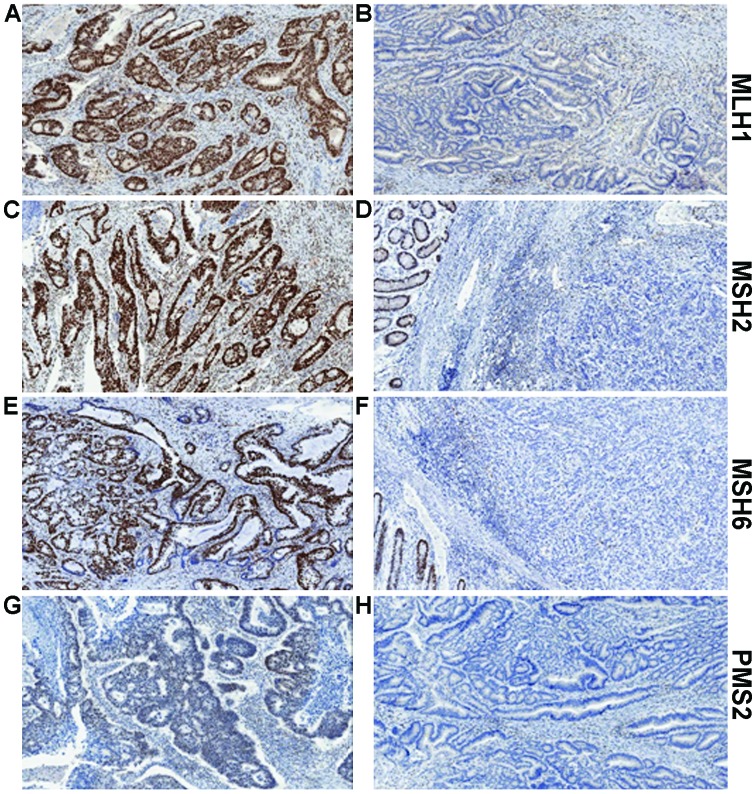
MMR IHC is a quick and relatively simple assay to determine protein expression of MLH1, MSH2, MSH6 and PMS2 (Fig. 2). Tumours with dMMR will usually show complete loss of expression of one or more protein. Assessing all four proteins provides further information to determine the actual defective protein. We know that MLH1 forms a heterodimer complex with PMS2. Loss of expression of PMS2 alone is indicative of a defect in the PMS2 gene. However, combined loss of PMS2 and MLH1 suggests the defect lies in MLH1, as MLH1 is responsible for the stability of PMS2. A similar situation is seen with MSH6 and MSH2, with loss of MSH6 only indicating defective MSH6, whereas loss of expression of both proteins would indicate the defect is within MSH2 (Table I). Based on a recent publication by Mensenkamp et al (45) this may in fact be a real oversimplification of the actual situation. The group sequenced dMMR CRC tumours and endometrial tumours which appeared to have neither a germline mutation in any MMR gene, or hypermethylation of the MLH1 promoter. In more than half of these tumours, somatic mutations were identified as the underlying cause of deficient mismatch repair.
Figure 2.
Examples of MLH1, MSH2, MSH6 and PMS2 immunohistochemistry. (A) Positive MLH1 staining and (B) absence of MLH1 staining in tumour epithelium yet showing the positive internal control staining of lymphocytes in the stroma. (C) Positive MLH2 staining and (B) absence of MLH2 staining in tumour epithelium, yet showing positive staining in the adjacent normal colonic epithelium. (E) Positive MSH6 staining and (F) absence of MSH6 staining in tumour epithelium yet with positive staining in the adjacent normal colonic epithelium. (G) Positive PMS2 staining and (H) absence of PMS2 staining in tumour epithelium yet with positive internal control staining of lymphocytes in the stroma.
Table I.
Loss of MMR protein expression.
| Protein expression lost (determined by IHC) | Interpretation (defective protein) |
|---|---|
| PMS2 | PMS2 |
| MLH1 and PMS2 | MLH1 |
| MSH6 | MSH6 |
| MSH2 and MSH6 | MSH2 |
Due to the heterodimeric nature of the MMR proteins, loss of expression of a particular protein may in fact be due to the loss of expression of its paired protein. For example, loss of PMS2 alone indicates a defect in PMS2, whereas, when expression of both MLH1 and PMS2 are lost, this is likely due to loss of MLH1, as this results in unstable PMS2. The same is true for MSH6 and MSH2, respectively.
Unfortunately, as is often the case with a seemingly straightforward assay, there are exceptions to the rules. Sometimes expression is reduced in intensity, or patchy, rather than completely lost. This may be a result of the expression of a truncated protein with limited stability, and is likely to be accompanied by the predicted normal strong nuclear staining within adjacent stromal cells or lymphocytes. It is often the case that the abnormal staining is seen in both binding partners, i.e., MLH1 and PMS2, or MSH2 and MSH6. Another unusual situation is where staining is seen localised to the cytoplasm, rather than within tumour cell nuclei. This may be caused by a defect in the nuclear localisation signal, and would most likely be reported as dMMR, although PCR-based MSI testing may be requested for confirmation. The single biggest problem in the assessment of MMR IHC is the variability in fixation of the tumour tissue. The actual fixative used, the time in formalin prior to embedding and the uniformity of fixation are all factors which can affect the quality of staining seen. Fadhil and Ilyas compared staining of the four MMR proteins in 30 matched pre-surgical diagnostic biopsy samples and the matched resection tissue, and concluded that not only was the staining in the biopsies identical to that in the resection, but the interpretation was made easier by the staining being more intense and thus easier to interpret (46). This difference was deemed to be a result of more uniform and complete fixation in the biopsy samples, compared to the resection specimens.
A further complication in terms of the interpretation of the MMR IHC was reported by Bao et al in a study of 51 colorectal cancer patients undergoing neoadjuvant chemoradiation (47). Nine of these tumours showed reduced, but not complete loss of MSH6 staining, yet upon MSI analysis, all were microsatellite stable, suggesting that the reduced expression was a result of the chemoradiation treatment.
A slightly contentious issue, worthy of a mention, is whether missense mutations in the MMR genes are associated with reduced or patchy immunohistochemical staining. Missense mutations result in a protein with a single amino acid change, which could lead to no defect at all, or a dysfunctional or ‘pathogenic’ mutation. Difficulty arises in the assessment of the pathogenicity of a missense mutation. Criteria which would have to be met would include: a) the mutation not being present in control subjects; b) the mutation co-segregating with a phenotype in a family; c) the mutation resulting in a nonconservative amino acid alteration; and d) the codon in which the mutation arose being evolutionarily conserved (48,49). PubMed searches for this review failed to identify any studies reporting reduced levels of MMR protein expression, which were attributed to missense mutations. At the present time, this phenomenon may have to remain an ‘urban myth’.
Once an abnormal expression pattern of the MMR proteins has been established, it is vitally important it determine whether the tumour is from a patient with Lynch syndrome. The MMR protein expression profile most commonly associated with Lynch syndrome is loss of both MLH1 and PMS2; however, this would also be seen in a sporadic tumour, if caused by MLH1 methylation. The BRAF p. (V600E) mutation is observed in up to 70% of tumours which have loss of expression of MLH1 and PMS2 or exhibit MLH1 methylation (50,51), but the mutation is almost never seen in Lynch syndrome-associated tumours (52,53). Thus the presence of the BRAF mutation strongly indicates a dMMR tumour of sporadic origin. BRAF mutation testing is currently carried out routinely by traditional sequencing methodologies, such as Sanger sequencing, but in 2011, the first report was published by Capper et al, that used an antibody specific for the V600E mutant protein (VE1), allowing direct immunohistochemical testing of a tumour section (54). Several groups have published data showing very favourable results with the antibody (including refs. 55,56) however, concerns have been voiced regarding the usefulness, and sensitivity of this antibody, particularly when assessing colorectal tumours. Adackapara et al noted a high level of weak staining in wild-type and KRAS mutant tumours, in addition to non-specific nuclear staining. They determined the sensitivity and specificity to be 71 and 74%, respectively, and deemed the antibody not to be a surrogate for standard genotyping (57). A study by Loes et al in 2015 assessed three methods of BRAF mutation detection [IHC, Sanger sequencing and a single probe-based high-resolution melting assay (LightMix) which has clamped wild-type allele amplification] in both melanoma and colorectal tumour samples. Data were available for all three assays in 99 colorectal tumours, of which 63 were wild-type by all methods, 12 were BRAF mutant by all methods, and yet 22 gave discordant results. Using the IHC data alone would have misinterpreted 10 patients as being BRAF mutant, and also failed to detect mutations in a further two patients. The authors conclude that the high level of unexplained, non-specific staining seen in colorectal tumours, much more so than for melanoma tumours, would support that the antibody be used solely as a screening tool, rather than a diagnostic test (58). It is worth noting that the antibody will only identify the specific V600E mutation, so there is always the risk of missing other BRAF mutations, but these are extremely rare, particularly in colorectal tumours.
8. Microsatellite (MSI) testing
As an alternative, or indeed in combination with MMR IHC testing, PCR-based MSI screening may be undertaken. The recommended NCI-reference panel comprises two mononucleotide repeats (BAT-25 and BAT-26) and three dinucleotide repeats (D5S346, D2S123 and D17S250). There is also a commercially available kit, consisting of five mononucleotide markers (BAT-25, BAT-26, MONO-27, NR-21 and NR-24), as data are emerging to suggest that there is a higher level of both sensitivity and specificity in the detection of the MSI-H phenotype when only mononucleotides are used (59). Where available, DNA from normal mucosa is compared to that extracted from the tumour. However, the nature of the mononucleotide markers means that it is not essential to have normal DNA for testing. The tumour is classed into one of three phenotypes; if none of the markers show instability, the tumour is classed as microsatellite stable (MSS). If one of the markers show instability, the tumour is classed as microsatellite-low (MSI-L), and if two or more of the markers show instability, the tumour is classed as microsatellite-high (MSI-H). Often MSS and MSI-L tumours are classified as a single subset, as very few tumours of either phenotype will exhibit loss of expression of any of the MMR proteins. Data surrounding clinical differences between the two tumour phenotypes is still inconclusive (60–63).
IHC or MSI?
There have been several studies carried out to assess the correlation between IHC and MSI-testing, and the overall results seem to suggest that firstly neither test is 100% accurate in the detection of MSI-H tumours and secondly, there is actually a high level of concordance between both technologies. The largest study to date was performed by Cicek et al in 2011, when almost 6,000 tumours from patients in the Colorectal Cancer Family Registry were analysed. The group showed a 90–95% concordance between those cases identified as dMMR by MSI and those detected by IHC. Furthermore, only 2.7% of the 3964 tumours with IHC data available, would have been miscalled, had only these data been used in the initial assessment (64).
9. Next generation sequencing
There is no doubt that sequencing methodologies have been transformed over the past few years, with the advent of next generation sequencing platforms. Several companies are now producing panels and kits, allowing the massive parallel sequencing of MMR genes. This additional depth of sequencing may cause the problem with the identification of variants of unknown significance (VUS). Furthermore, there will undoubtedly be mutations detected at lower levels than previous technologies have allowed. The issue with these is that the clinical significance has not yet been determined, thus with the technology being still in its infancy, there remains the need to validate such panels. Pritchard et al carried out one such validation study of the ColoSeq panel, which correctly identified all 28 previously characterised mutations in MLH1, MSH2 MSH6, PMS2, EPCAM, APC and MUTYH. Two VUS were also detected in 19 samples from patients without cancer (65). The significance of such variants should become apparent once more data are available and they can be related to pathogenicity.
10. Deficient MMR and clinical outcomes
Prognostic value in sporadic colorectal cancer
The majority of the data published recently on the prognostic and predictive value of MMR has been gathered on CRC patients. There is definitely a distinction between the prognostic benefit of dMMR in early (stage II/III) and late (stage IV) disease. Several studies and meta-analyses have shown that dMMR in stage II +/or III tumours is a positive prognostic factor. Back in 2003, a study of 570 stage II or II CRC patients showed that those patients whose tumours were MSI-H had an improved 5-year OS, compared to MSI-L or MSS tumours (HR for death was 0.31 (95% CI, 0.14–0.72, p=0.004) (66). In 2010, a large meta-analysis pooled data from 12,782 CRC patients, including 1,972 MSI-H patients. The odds ratio (OR) for disease-free survival (DFS) was 0.58, 95% CI 0.47–0.72, p<0.0001 and a similar value obtained for OS (OR=0.6, 95% CI 0.53–0.69, p<0.0001) (67). This was confirmed by Sargent et al, in a further meta-analysis of 457 patients, where it was reported that dMMR status was associated with improved DFS (HR, 0.46; 95% CI, 0.22–0.95; p=0.03) and a trend was seen towards improved OS (HR, 0.51; 95% CI, 0.24–1.10; p=0.06) (68). The QUASAR (QUick And Simple And Reliable trial provided a more recent dataset on which to confirm the positive prognostic significance of dMMR. The recurrence rate in the dMMR cohort was 11% (25/218), compared to 26% (438/1695) in the pMMR cohort [risk ratio (RR), 0.53; 95% CI, 0.40–0.70] (69).
Because of the fact that dMMR appears to be a good prognostic marker in early CRC, it stands to reason that prevalence of dMMR would be lower in advanced CRC (aCRC), since these patients should be less likely to develop metastatic disease (70). This has been reported in several studies (71–73). The question remains as to why these tumours appear to metastasise less frequently. This may be as a result of the increased immune response seen in dMMR tumours. Tikidzhieva et al, have suggested a possible mechanism, involving β2-microglobulin (B2M) (74). Mutations in B2M, within microsatellite coding regions, are reported frequently in MSI-H tumours, and result in the inability to present antigens at the cell surface, through HLA-class I molecules. This in turn, may stimulate natural killer (NK) cell-mediated tumour cell death.
In terms of the prognostic value of dMMR in aCRC, a recent large meta-analysis by Venderbosch et al (75) of patients in four randomised clinical trials (CAIRO, CAIRO2, FOCUS and COIN) provides convincing evidence of the negative prognostic effect of dMMR in the metastatic CRC (mCRC) setting. Data on dMMR was gathered on 3,063 patients, recruited into the four clinical trials. PFS and OS were significantly reduced in the dMMR cohort, in comparison to the pMMR cohort (PFS, 6.2 versus 7.6 months respectively; HR, 1.33; 95% CI, 1.12–1.57; p=0.001; and OS, 13.6 versus 16.8 months respectively; HR, 1.35; 95% CI, 1.13–1.61; p=0.001). The analysis also demonstrated the negative prognostic effect of the presence of the BRAF p. (V600E) mutation, but ruled out any interaction between the two poor prognosis markers. The group suggest that the negative value of dMMR is as a result of the mutant BRAF status, since significantly more dMMR tumours also contained the mutation.
Predictive value in colorectal cancer
Since its introduction into clinical practice almost 40 years ago, 5-fluorouracil (5-FU) has, until recently, been the ‘gold standard’ chemotherapy agent in the treatment of CRC. As a result of this, there is much, and it has to be said, conflicting data regarding the predictive value of MMR status and response to 5-FU-based therapy, with some studies reporting benefit from 5-FU (76,77) whilst most reporting no benefit or indeed a dis-benefit (66,68,78,79).
The final results from the MOSAIC trial where 2,246 stage II or II CRC patients were randomised between 5-FU plus leucovorin (LV5FU2) and FOLFOX (LV5FU2 + oxaliplatin), provided convincing evidence that the addition of oxaliplatin resulted in improved 5-year DFS and 6-year OS, and in particular, ought to be given to stage III patients after surgery (80). Following this, studies were performed to assess whether microsatellite status was predictive of response to oxaliplatin; Zaanan et al (81) analysed 233 MSI-H stage III patients, receiving either 5-FU/LV or FOLFOX, and finding that those on FOLFOX had an improved 3-year DFS compared to those on 5-FU/LV. However, in the same year, a study of 135 patients receiving FOLFOX following surgery, found no difference in DFS or OS when patients were stratified for MMR status (82). In the metastatic setting, Muller et al in a 108-patient study, comparing two oxaliplatin and 5-FU-containing regimens, demonstrated a lower rate of disease control in MSI-H patients compared to non-MSI-H patients (p=0.02) (73). Kim et al however, showed that MMR status did not predict response to oxaliplatin-based treatment, when 171 recurrent or mCRC patients were analysed (83).
There is also conflicting data as to the predictive value of MMR status and response to irinotecan. Bertagnolli et al showed that patients with dMMR/MSI-H had improved DFS, compared to MSS patients, when irinotecan was added to standard 5-FU/LV treatment, with this benefit not being seen in patients treated with 5-FU/LV alone (84). However, this was not confirmed by the PETACC-3 study (85) or by a Korean study of almost 300 patients (86), or by the UK MRC FOCUS study (71).
It would be difficult to summarise the prognostic and predictive value of MMR status in both the adjuvant and metastatic CRC settings, based on the data presented above. It is apparent that dMMR/MSI-H in the adjuvant setting is a good prognostic marker, but in the metastatic setting, the evidence suggests the complete opposite effect. As for the predictive value, there are conflicting data regarding each treatment regimen. One can speculate as to why this is the case; perhaps we are seeing population differences, perhaps the method of determining MMR status had differing sensitivities. The small numbers of patients in some of the studies should also be taken into account. It is without doubt safe to say, that one cannot use only MMR status for the prediction of response to therapy.
Prognostic and predictive value in extra-colonic tumours
The majority of published data regarding the role of the mismatch repair system in carcinogenesis, and the resultant prognostic and predictive value, is within colorectal cancer. There are, however, several extra-colonic cancers where there are high percentages of dMMR have been reported, yet little is known of the prognostic or predictive value.
Endometrial cancer
dMMR has been reported in 20–30% of endometrial cancers (87), yet there are scarce data available regarding the prognostic and predictive impact of mismatch repair deficiencies. In a study reported earlier this year, Kato et al analysed 191 endometrial tumours, and found that 40% of them were deficient in at least one of the MMR proteins, as assessed by IHC (88). This cohort displayed differences in tumour grade histology and International Federation of Gynecology and Obstetrics (FIGO) stage, when compared to the proficient MMR tumours. Furthermore, dMMR cases had improved PFS and OS, with MMR status being an independent prognostic factor for OS in endometrial cancers. A further study, admittedly smaller, of 66 patients with endometrial cancer and lymphatic invasion, also reported improved disease specific survival (DSS) (p=0.04) and OS (p=0.03) in dMMR patients, compared to those with pMMR. The authors also reported increased OS particularly in FIGO stage 3C and stage 4 dMMR patients, which may suggest that despite the lymphatic invasion and lymph node metastases, this subgroup has a better prognosis than patients with an intact MMR system. The other factor that cannot be ignored is the effect that adjuvant chemotherapy has contributed to this improved survival (89). A third study, of 477 patients, investigated whether MMR status impacted upon response to chemotherapy or pelvic teletherapy [also known as external beam radiotherapy (EBRT)]. There was no significant difference in PFS or OS between dMMR and pMMR subgroups, when stratified by treatment. However, when patients were stratified between endometrioid and non-endometrioid tumours, significantly improved OS (p=0.003) and PFS (p=0.004) was seen for dMMR/non-endometrioid tumours, receiving teletherapy. The opposite was seen for patients receiving adjuvant chemotherapy, where those with intact MMR showed improved PFS and OS (90). Taking these data together, it would possibly appear that dMMR in endometrial cancers, or at least within subgroups, is a positive biomarker. However, Ruiz et al reported no association between MMR status and survival, in a study of 212 endometrioid tumours (91), and a further study actually reported an increased risk of disease-specific death in dMMR high-grade endometrioid carcinomas (HGEC). Interestingly in this study, dMMR was only seen in these HGEC tumours, and not serous or clear cell tumours, suggesting the use of MMR testing to aid in tumour-type diagnosis (92). Cohn et al reported improved DFS in a cohort of endometrial cancer patients who had retained expression of both MLH1 and MSH2, in comparison to patients who displayed abnormal expression (p=0.035) (93). A large meta-analysis carried out in 2013 summarised very eloquently the lack of concrete evidence of an association between MMR status and clinical outcome, where in a pooled analysis of 23 studies (published between 1980 and 2011), the group failed to show a significant association between MSI and a worse OS (p=0.11) or DFS (p=0.66) (94). The heterogeneous nature of the method of determining MSI status, combined with variability in the study populations, still make it very difficult to determine the usefulness of MMR status in relation to outcome in this disease.
Ovarian cancer
Ovarian cancer is the 7th most common cancer worldwide for females, with over 239,000 new cases diagnosed in 2012, and has the highest mortality rate of all the gynaecological cancers (95). Early detection is difficult, and as a result, only 15% of women present with localised disease (96). Women with Lynch syndrome, have a lifetime risk of ovarian cancer of approximately 8% (97–99). As we find in common with other extracolonic cancers, data on MMR is sparse. Several authors have attempted to clarify dMMR or MSI rates through meta-analyses; Xiao et al (100) found disparities between reported rates of MSI frequency, ranging from 5 to 13% (101–103). Murphy and Wentzensen combined results from 22 studies, arriving at a figure for MSI of 10% for unselected ovarian cancer patients (104). This figure was further refined to 9%, when only patients who had been tested for MSI using the five Bethesda markers were analysed. Pal et al also suggest that 10% of ovarian cancers show MSI, analysing data from 18 studies (105). In terms of dMMR as assessed by IHC, larger differences were observed; ranging from 2 to 29% across the 12 studies analysed by Xiao et al (100). One feature common to most studies was the fact that there was an overrepresentation of the non-serous tumours within the MSI cohorts, which parallels the overrepresentation of mucinous and endometrioid histologies in CRC and endometrial cancers respectively. In terms of data relating to the effect of dMMR or MSI on prognosis or response to chemotherapy, very little has been published, and the results are varied. Scartozzi et al found that loss of expression of MLH1 correlated with increased survival in patients with stage III/IV disease, although the study size was only 34 patients (106). Zhia et al assessed 322 tumours for MSH6 expression, and found no correlation with survival. The group did find a correlation between loss of expression and clear cell, mucinous and endometrioid histologies (p<0.007) (107). Another study finding no association between MSI and survival was carried out on a series of Danish patients by Begum et al, who used a panel of 16 dinucleotide markers to assess status (108). In terms of response to therapy, there have been two reports of a correlation between a lack of MSH2 and response to platinum-based chemotherapy; Ercoli et al showed that patients who did not respond to treatment had lower levels of MSH2 than patients who had at least a partial response (109). A report by Marcelis et al described two Lynch syndrome patients, both carrying a deletion in exon 6 of MSH2, who developed a rapid resistance to cisplatin-based therapy (110). Based upon current literature, very little can be reasonably or reliably concluded regarding the role of the MMR proteins in ovarian cancer survival or response. There is clearly a need for large, randomised studies in this disease field, where one can control for factors such as MMR assessment criteria, tumour histology, treatment regimen and sample size.
Melanoma
Malignant melanoma is the 19th most common cancer worldwide, with around 232,000 new cases diagnosed in 2012 (111). MSI has been reported to be present in anywhere between 2 and 30% of primary tumours (112–116) and 20–77% of metastatic lesions (117–123). Castiglia et al suggest that the inactivation of the MMR system, in combination with the deregulation of the Wnt/beta-catenin pathway may act cooperatively to promote the development of melanoma (124). It may be that in melanoma, it is a downregulation of the MMR proteins, rather than a complete loss of expression, or gene inactivation that is important, as seen in a study by Korabiowska et al, who confirmed the downregulation by both IHC and in situ hybridisation in 59 malignant melanomas (125). Alvino et al also reported a reduction in expression of MLH1, MSH2 and PMS2 in primary melanomas compared to benign nevi. Interestingly they also noted the opposite for MSH6, and this increased expression was also associated with increased risk of melanoma mortality (R, 3.76; 95% CI, 1.12–12.70) (126). With such little data available on the MMR proteins in melanoma, the only conclusion that can be reliably drawn is that as the cancer progresses from benign nevus, through primary melanoma to metastatic melanoma, the level of MSI increases. This may, however, only be at an MSI-L level, rather than MSI-H. The significance of this is yet to be determined.
Gastric cancer
Gastric cancer is the 5th most common cancer worldwide, with more than 951,000 new cases diagnosed in 2012 (127). In gastric cancer, MSI exists in approximately 10–20% tumours (128–130). Such tumours are associated with older patients, distal location, lower pTNM stage and intestinal subtype and reduced lymph node involvement. Several large studies have assessed the prognostic effect of the MSI phenotype, all showing that MSI correlates with improved survival; back in 2000, Schneider et al showed that in MSI-H and MSI-L patients, there was an increased median survival time, compared to MSS patients (p=0.027) (131). In 2002, Lee et al analysed 327 consecutive gastric cancers, assessing MSI status with the BAT-26 marker. Patients with MSI had improved overall survival compared to those with MSS tumours (p=0.046) (130). Beghelli et al, determined the MSI status of 510 sporadic gastric cancers, also concluding that MSI correlated with improved survival, but only in stage II disease (p<0.011) (128). In a study of 159 patients, Falchetti et al demonstrated an association between MSI-H phenotype and improved survival at 15 years (p=0.01) (132). Finally Fang et al showed that there was an improved 5-year OS benefit in the MSI-H cohort (p=0.03) and also a trend towards an improved 3-year disease-free survival (p=0.076), when analysing 214 gastric cancer patients (129). However, as one has come to expect in this field, there is conflicting data to suggest that MSI status has no influence on survival; Perez et al found no survival benefit in the MSI patients, compared to the MSS patients, however, it must be noted that there were only 24 patients in this study (133). In a slightly larger study of 83 patients, An et al also did not find an association between MSI status and survival (134). Given the disparity between sample sizes, the evidence is pointing to the direction that gastric cancer patients with an MSI-H tumour are likely to have improved survival compared to patients whose tumours are MSS. Looking at MSI status and its predictive value in terms of response to 5-FU-based chemotherapy, there is yet again conflicting data; a large study by An et al, of 1990 patients, identified an MSI-H rate of 8.5%. The group determined that MSI status was not prognostic, as DFS between MSI-H and the MSI-L/MSS groups was not significantly different, even taking each disease stage separately. However, DFS was improved in the MSI-L/MSS group treated with 5-FU-based chemotherapy (p=0.008) (135). Oki et al, determined that there was no correlation between MSI status and survival following 5-FU-therapy, in their study of 240 patients, collected over a 9-year period (136). Clearly the gastric cancers with MSI form a distinct subset, and as such, are likely to be driven by slightly different signalling pathways. It still remains to be determined, how to identify and best and treat these patients.
11. Conclusion
Deficiencies in the DNA mismatch repair system have been identified in many unrelated cancer types. These deficiencies may be the result of either the inactivation of MLH1, through methylation, as seen in sporadic cancers, or through germline mutations of MLH1 or MSH2, as seen in inherited cancers. Despite it being almost 50 years since the initial observations by Henry Lynch, which subsequently lead to the term ‘Lynch syndrome’, there are still gaps in our knowledge of the role of dMMR in cancer. Progress is being made, however, particularly in the field of colorectal cancer. We now have evidence that the prognostic role of dMMR is stage-dependent, and steps are beginning to be implemented, to ensure that every patient who may require screening actually has access to this service. In terms of identifying dMMR or MSI patients, there is now some standardisation of IHC and adoption of the use of the Bethesda marker panel, but with the recent introduction of next generation screening, the additional depth of sequence data, may complicate the situation as more VUS are identified. Furthermore, the clinical significance of low-level variants is yet to be elucidated, adding a further layer to complexity to the use of this emerging technology. Extracolonic cancers trail far behind in terms of what is known of the prognostic and predictive value of MMR, and, our understanding will remain limited unless large controlled trials are performed.
Abbreviations
- BRAF
v-Raf murine sarcoma viral oncogene homolog B
- CRC
colorectal cancer
- DFS
disease-free survival
- dMMR
deficient mismatch repair
- EPCAM
epithelial cell adhesion molecule
- EXO1
exonuclease-1
- HNPCC
hereditary non-polyposis colorectal cancer
- IHC
immunohistochemistry
- mCRC
metastatic colorectal cancer
- MLH1
mutL homologue 1
- MMR
mismatch repair
- MSH2
mutS homologue 2
- MSH6
mutS homologue 6
- MSI
microsatellite instability
- MSI-H
microsatellite instability-high
- MSI-l
microsatellite instability-low
- MSS
microsatellite stable
- NGS
next generation sequencing
- OS
overall survival
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- PFS
progression-free survival
- pMMR
proficient mismatch repair
- PMS2
post-meiotic segregation increased 2
- Pol δ
DNA polymerase δ
- RFA
replication factor A
- RFC
replication factor C
- VUS
variants of unknown significance
References
- 1.Nilbert M, Planck M, Fernebro E, Borg A, Johnson A. Microsatellite instability is rare in rectal carcinomas and signifies hereditary cancer. Eur J Cancer. 1999;35:942–945. doi: 10.1016/S0959-8049(99)00045-3. [DOI] [PubMed] [Google Scholar]
- 2.Sinicrope FA, Rego RL, Foster N, Sargent DJ, Windschitl HE, Burgart LJ, Witzig TE, Thibodeau SN. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006;101:2818–2825. doi: 10.1111/j.1572-0241.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Thibodeau SN, French AJ, Cunningham JM, Tester D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, et al. Microsatellite instability in colorectal cancer: Different mutator phenotypes and the principal involvement of hMLH1. Cancer Res. 1998;58:1713–1718. [PubMed] [Google Scholar]
- 6.Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 7.Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, Halling KC, McDonnell SK, Schaid DJ, Walsh Vockley C, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 9.Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, Young J, Jenkins MA, Hopper JL, Baron JA, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch HT, Shaw MW, Magnuson CW, Larsen AL, Krush AJ. Hereditary factors in cancer. Study of two large midwestern kindreds. Arch Intern Med. 1966;117:206–212. doi: 10.1001/archinte.1966.03870080050009. [DOI] [PubMed] [Google Scholar]
- 11.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plazzer JP, Sijmons RH, Woods MO, Peltomäki P, Thompson B, Den Dunnen JT, Macrae F. The InSiGHT database: Utilizing 100 years of insights into Lynch syndrome. Fam Cancer. 2013;12:175–180. doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 13.Hitchins MP, Ward RL. Constitutional (germline) MLH1 epimutation as an aetiological mechanism for hereditary non-polyposis colorectal cancer. J Med Genet. 2009;46:793–802. doi: 10.1136/jmg.2009.068122. [DOI] [PubMed] [Google Scholar]
- 14.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 15.Crucianelli F, Tricarico R, Turchetti D, Gorelli G, Gensini F, Sestini R, Giunti L, Pedroni M, Ponz de Leon M, Civitelli S, et al. MLH1 constitutional and somatic methylation in patients with MLH1 negative tumors fulfilling the revised Bethesda criteria. Epigenetics. 2014;9:1431–1438. doi: 10.4161/15592294.2014.970080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward RL, Dobbins T, Lindor NM, Rapkins RW, Hitchins MP. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med. 2013;15:25–35. doi: 10.1038/gim.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pineda M, Mur P, Iniesta MD, Borràs E, Campos O, Vargas G, Iglesias S, Fernández A, Gruber SB, Lázaro C, et al. MLH1 methylation screening is effective in identifying epimutation carriers. Eur J Hum Genet. 2012;20:1256–1264. doi: 10.1038/ejhg.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 19.Kloor M, Voigt AY, Schackert HK, Schirmacher P, von Knebel Doeberitz M, Bläker H. Analysis of EPCAM protein expression in diagnostics of Lynch syndrome. J Clin Oncol. 2011;29:223–227. doi: 10.1200/JCO.2010.32.0820. [DOI] [PubMed] [Google Scholar]
- 20.Huth C, Kloor M, Voigt AY, Bozukova G, Evers C, Gaspar H, Tariverdian M, Schirmacher P, von Knebel Doeberitz M, Bläker H. The molecular basis of EPCAM expression loss in Lynch syndrome-associated tumors. Mod Pathol. 2012;25:911–916. doi: 10.1038/modpathol.2012.30. [DOI] [PubMed] [Google Scholar]
- 21.Musulen E, Blanco I, Carrato C, Fernandez-Figueras MT, Pineda M, Capella G, Ariza A. Usefulness of epithelial cell adhesion molecule expression in the algorithmic approach to Lynch syndrome identification. Hum Pathol. 2013;44:412–416. doi: 10.1016/j.humpath.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 23.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/S0016-5085(99)70510-X. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: Meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 25.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boland CR, Shike M. Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology. 2010;138:2197 e2191–2197. doi: 10.1053/j.gastro.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan HL, Hao LQ, Jin HY, Xing QH, Xue G, Mei Q, He J, He L, Sun SH. Clinical features and mismatch repair genes analyses of Chinese suspected hereditary non-polyposis colorectal cancer: A cost-effective screening strategy proposal. Cancer Sci. 2008;99:770–780. doi: 10.1111/j.1349-7006.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148:719–731 e713. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringelhan M, O'Connor T, Protzer U, Heikenwalder M. The direct and indirect roles of HBV in liver cancer: Prospective markers for HCC screening and potential therapeutic targets. J Pathol. 2015;235:355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]
- 30.Wang XL, Yuan Y, Zhang SZ, Cai SR, Huang YQ, Jiang Q, Zheng S. Clinical and genetic characteristics of Chinese hereditary nonpolyposis colorectal cancer families. World J Gastroenterol. 2006;12:4074–4077. doi: 10.3748/wjg.v12.i25.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chew MH, Koh PK, Ng KH, Lim JF, Ho KS, Ooi BS, Tang CL, Eu KW. Phenotypic characteristics of hereditary non-polyposis colorectal cancer by the Amsterdam criteria: An Asian perspective. ANZ J Surg. 2008;78:556–560. doi: 10.1111/j.1445-2197.2008.04570.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Yang L, Zhou X, Sheng W, Cai S, Liu L, Nan P, Xu Y. Clinicopathological and genetic features of Chinese hereditary nonpolyposis colorectal cancer (HNPCC) Med Oncol. 2014;31:223. doi: 10.1007/s12032-014-0223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew MH, Koh PK, Tan M, Lim KH, Carol L, Tang CL. Mismatch repair deficiency screening via immunohistochemical staining in young Asians with colorectal cancers. World J Surg. 2013;37:2468–2475. doi: 10.1007/s00268-013-2134-2. [DOI] [PubMed] [Google Scholar]
- 34.Ladabaum U, Wang G, Terdiman J, Blanco A, Kuppermann M, Boland CR, Ford J, Elkin E, Phillips KA. Strategies to identify the Lynch syndrome among patients with colorectal cancer: A cost-effectiveness analysis. Ann Intern Med. 2011;155:69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 2010;12:93–104. doi: 10.1097/GIM.0b013e3181cd666c. [DOI] [PubMed] [Google Scholar]
- 36.Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, Frayling I, Hyde C. A systematic review and economic evaluation of diagnostic strategies for Lynch syndrome. Health Technol Assess. 2014;18:1–406. doi: 10.3310/hta18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schofield L, Grieu F, Amanuel B, Carrello A, Spagnolo D, Kiraly C, Pachter N, Goldblatt J, Platell C, Levitt M, et al. Population-based screening for Lynch syndrome in Western Australia. Int J Cancer. 2014;135:1085–1091. doi: 10.1002/ijc.28744. [DOI] [PubMed] [Google Scholar]
- 38.Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, Frayling I, Hyde C. A model-based assessment of the cost-utility of strategies to identify Lynch syndrome in early-onset colorectal cancer patients. BMC Cancer. 2015;15:313. doi: 10.1186/s12885-015-1254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sie AS, Mensenkamp AR, Adang EM, Ligtenberg MJ, Hoogerbrugge N. Fourfold increased detection of Lynch syndrome by raising age limit for tumour genetic testing from 50 to 70 years is cost-effective. Ann Oncol. 2014;25:2001–2007. doi: 10.1093/annonc/mdu361. [DOI] [PubMed] [Google Scholar]
- 40.Bellcross CA, Bedrosian SR, Daniels E, Duquette D, Hampel H, Jasperson K, Joseph DA, Kaye C, Lubin I, Meyer LJ, et al. Implementing screening for Lynch syndrome among patients with newly diagnosed colorectal cancer: summary of a public health/clinical collaborative meeting. Genet Med. 2012;14:152–162. doi: 10.1038/gim.0b013e31823375ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. [Accessed April 15, 2015];Data briefing: Reflex testing for Lynch syndrome in people diagnosed with bowel cancer under the age of 50. http://www.bowelcanceruk.org.uk/media/426888/lynch_syndrome_briefing_final.pdf.
- 42.Tomiak E, Samson A, Spector N, Mackey M, Gilpin C, Smith E, Jonker D, Allanson J, Asmis T. Reflex testing for Lynch syndrome: If we build it, will they come? Lessons learned from the uptake of clinical genetics services by individuals with newly diagnosed colorectal cancer (CRC) Fam Cancer. 2014;13:75–82. doi: 10.1007/s10689-013-9677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beamer LC, Grant ML, Espenschied CR, Blazer KR, Hampel HL, Weitzel JN, MacDonald DJ. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol. 2012;30:1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schofield L, Grieu F, Goldblatt J, Amanuel B, Iacopetta B. A state-wide population-based program for detection of Lynch syndrome based upon immunohistochemical and molecular testing of colorectal tumours. Fam Cancer. 2012;11:1–6. doi: 10.1007/s10689-011-9494-2. [DOI] [PubMed] [Google Scholar]
- 45.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, Goossens M, Ouchene H, Hendriks-Cornelissen SJ, Kwint MP, Hoogerbrugge N, Nagtegaal ID, Ligtenberg MJ. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–646 e648. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Fadhil W, Ilyas M. Immunostaining for mismatch repair (MMR) protein expression in colorectal cancer is better and easier to interpret when performed on diagnostic biopsies. Histopathology. 2012;60:653–655. doi: 10.1111/j.1365-2559.2011.04021.x. [DOI] [PubMed] [Google Scholar]
- 47.Bao F, Panarelli NC, Rennert H, Sherr DL, Yantiss RK. Neoadjuvant therapy induces loss of MSH6 expression in colorectal carcinoma. Am J Surg Pathol. 2010;34:1798–1804. doi: 10.1097/PAS.0b013e3181f906cc. [DOI] [PubMed] [Google Scholar]
- 48.Genuardi M, Carrara S, Anti M, Ponz de Leòn M, Viel A. Assessment of pathogenicity criteria for constitutional missense mutations of the hereditary nonpolyposis colorectal cancer genes MLH1 and MSH2. Eur J Hum Genet. 1999;7:778–782. doi: 10.1038/sj.ejhg.5200363. [DOI] [PubMed] [Google Scholar]
- 49.Cravo M, Afonso AJ, Lage P, Albuquerque C, Maia L, Lacerda C, Fidalgo P, Chaves P, Cruz C, Nobre-Leitão C. Pathogenicity of missense and splice site mutations in hMSH2 and hMLH1 mismatch repair genes: Implications for genetic testing. Gut. 2002;50:405–412. doi: 10.1136/gut.50.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouzourene H, Hutter P, Losi L, Martin P, Benhattar J. Selection of patients with germline MLH1 mutated Lynch syndrome by determination of MLH1 methylation and BRAF mutation. Fam Cancer. 2010;9:167–172. doi: 10.1007/s10689-009-9302-4. [DOI] [PubMed] [Google Scholar]
- 51.McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 52.Capper D, Voigt A, Bozukova G, Ahadova A, Kickingereder P, von Deimling A, von Knebel Doeberitz M, Kloor M. BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer. 2013;133:1624–1630. doi: 10.1002/ijc.28183. [DOI] [PubMed] [Google Scholar]
- 53.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 54.Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, Pusch S, Mechtersheimer G, Zentgraf H, von Deimling A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 55.Colomba E, Hélias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, Surel S, Côté JF, Peschaud F, Capper D, et al. Detection of BRAF p.V600E mutations in melanomas: Comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagn. 2013;15:94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adackapara CA, Sholl LM, Barletta JA, Hornick JL. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology. 2013;63:187–193. doi: 10.1111/his.12154. [DOI] [PubMed] [Google Scholar]
- 58.Loes IM, Immervoll H, Angelsen JH, Horn A, Geisler J, Busch C, Lønning PE, Knappskog S. Performance comparison of three BRAF V600E detection methods in malignant melanoma and colorectal cancer specimens. Tumour Biol. 2015;36:1003–1013. doi: 10.1007/s13277-014-2711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xicola RM, Llor X, Pons E, Castells A, Alenda C, Piñol V, Andreu M, Castellví-Bel S, Payá A, Jover R, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99:244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 60.Bapat B, Lindor NM, Baron J, Siegmund K, Li L, Zheng Y, Haile R, Gallinger S, Jass JR, Young JP, et al. The association of tumor microsatellite instability phenotype with family history of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:967–975. doi: 10.1158/1055-9965.EPI-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graham T, Halford S, Page KM, Tomlinson IP. Most low-level microsatellite instability in colorectal cancers can be explained without an elevated slippage rate. J Pathol. 2008;215:204–210. doi: 10.1002/path.2351. [DOI] [PubMed] [Google Scholar]
- 62.Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62:53–57. [PubMed] [Google Scholar]
- 63.Tomlinson I, Halford S, Aaltonen L, Hawkins N, Ward R. Does MSI-low exist? J Pathol. 2002;197:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- 64.Cicek MS, Lindor NM, Gallinger S, Bapat B, Hopper JL, Jenkins MA, Young J, Buchanan D, Walsh MD, Le Marchand L, et al. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumors results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13:271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritchard CC, Smith C, Salipante SJ, Lee MK, Thornton AM, Nord AS, Gulden C, Kupfer SS, Swisher EM, Bennett RL, et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 70.Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–3839. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 71.Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, Barrett JH, Selby P, Meade AM, Stephens RJ, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: Results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 72.Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ, van Krieken JH. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Müller CI, Schulmann K, Reinacher-Schick A, Andre N, Arnold D, Tannapfel A, Arkenau H, Hahn SA, Schmoll SH, Porschen R, et al. AIO Colorectal Study Group. Predictive and prognostic value of microsatellite instability in patients with advanced colorectal cancer treated with a fluoropyrimidine and oxaliplatin containing first-line chemotherapy. A report of the AIO Colorectal Study Group. Int J Colorectal Dis. 2008;23:1033–1039. doi: 10.1007/s00384-008-0504-2. [DOI] [PubMed] [Google Scholar]
- 74.Tikidzhieva A, Benner A, Michel S, Formentini A, Link KH, Dippold W, von Knebel Doeberitz M, Kornmann M, Kloor M. Microsatellite instability and Beta2-microglobulin mutations as prognostic markers in colon cancer: Results of the FOGT-4 trial. Br J Cancer. 2012;106:1239–1245. doi: 10.1038/bjc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, Kaplan R, Quirke P, Seymour MT, Richman SD, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: A pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 77.Elsaleh H, Shannon B, Iacopetta B. Microsatellite instability as a molecular marker for very good survival in colorectal cancer patients receiving adjuvant chemotherapy. Gastroenterology. 2001;120:1309–1310. doi: 10.1053/gast.2001.23646. [DOI] [PubMed] [Google Scholar]
- 78.Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 79.Hong SP, Min BS, Kim TI, Cheon JH, Kim NK, Kim H, Kim WH. The differential impact of microsatellite instability as a marker of prognosis and tumour response between colon cancer and rectal cancer. Eur J Cancer. 2012;48:1235–1243. doi: 10.1016/j.ejca.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 80.André T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 81.Zaanan A, Cuilliere-Dartigues P, Guilloux A, Parc Y, Louvet C, de Gramont A, Tiret E, Dumont S, Gayet B, Validire P, et al. Impact of p53 expression and microsatellite instability on stage III colon cancer disease-free survival in patients treated by 5-fluorouracil and leucovorin with or without oxaliplatin. Ann Oncol. 2010;21:772–780. doi: 10.1093/annonc/mdp383. [DOI] [PubMed] [Google Scholar]
- 82.Kim ST, Lee J, Park SH, Park JO, Lim HY, Kang WK, Kim JY, Kim YH, Chang DK, Rhee PL, et al. Clinical impact of microsatellite instability in colon cancer following adjuvant FOLFOX therapy. Cancer Chemother Pharmacol. 2010;66:659–667. doi: 10.1007/s00280-009-1206-3. [DOI] [PubMed] [Google Scholar]
- 83.Kim ST, Lee J, Park SH, Park JO, Lim HY, Kang WK, Kim JY, Kim YH, Chang DK, Rhee PL, et al. The effect of DNA mismatch repair (MMR) status on oxaliplatin-based first-line chemotherapy as in recurrent or metastatic colon cancer. Med Oncol. 2010;27:1277–1285. doi: 10.1007/s12032-009-9374-x. [DOI] [PubMed] [Google Scholar]
- 84.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 86.Kim JE, Hong YS, Ryu MH, Lee JL, Chang HM, Lim SB, Kim JH, Jang SJ, Kim MJ, Yu CS, et al. Association between deficient mismatch repair system and efficacy to irinotecan-containing chemotherapy in metastatic colon cancer. Cancer Sci. 2011;102:1706–1711. doi: 10.1111/j.1349-7006.2011.02009.x. [DOI] [PubMed] [Google Scholar]
- 87.Matthews KS, Estes JM, Conner MG, Manne U, Whitworth JM, Huh WK, Alvarez RD, Straughn JM, Jr, Barnes MN, Rocconi RP. Lynch syndrome in women less than 50 years of age with endometrial cancer. Obstet Gynecol. 2008;111:1161–1166. doi: 10.1097/AOG.0b013e31817051d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kato M, Takano M, Miyamoto M, Sasaki N, Goto T, Tsuda H, Furuya K. DNA mismatch repair-related protein loss as a prognostic factor in endometrial cancers. J Gynecol Oncol. 2015;26:40–45. doi: 10.3802/jgo.2015.26.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Terada KY, Black M, Terada LH, Davis J, Shimizu DM. Survival of endometrial cancer patients with lymphatic invasion and deficient mismatch repair expression. Gynecol Oncol. 2013;129:188–192. doi: 10.1016/j.ygyno.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 90.Resnick KE, Frankel WL, Morrison CD, Fowler JM, Copeland LJ, Stephens J, Kim KH, Cohn DE. Mismatch repair status and outcomes after adjuvant therapy in patients with surgically staged endometrial cancer. Gynecol Oncol. 2010;117:234–238. doi: 10.1016/j.ygyno.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiz I, Martín-Arruti M, Lopez-Lopez E, Garcia-Orad A. Lack of association between deficient mismatch repair expression and outcome in endometrial carcinomas of the endometrioid type. Gynecol Oncol. 2014;134:20–23. doi: 10.1016/j.ygyno.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 92.Nelson GS, Pink A, Lee S, Han G, Morris D, Ogilvie T, Duggan MA, Köbel M. MMR deficiency is common in high-grade endometrioid carcinomas and is associated with an unfavorable outcome. Gynecol Oncol. 2013;131:309–314. doi: 10.1016/j.ygyno.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 93.Cohn DE, Frankel WL, Resnick KE, Zanagnolo VL, Copeland LJ, Hampel H, Kelbick N, Morrison CD, Fowler JM. Improved survival with an intact DNA mismatch repair system in endometrial cancer. Obstet Gynecol. 2006;108:1208–1215. doi: 10.1097/01.AOG.0000239097.42987.0c. [DOI] [PubMed] [Google Scholar]
- 94.Diaz-Padilla I, Romero N, Amir E, Matias-Guiu X, Vilar E, Muggia F, Garcia-Donas J. Mismatch repair status and clinical outcome in endometrial cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2013;88:154–167. doi: 10.1016/j.critrevonc.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 95.CRUK. [Accessed April 15, 2015];Ovarian cancer incidence statistics. http://www.cancer-researchuk.org/cancer-info/cancerstats/types/ovary/incidence/
- 96.Surveillance, Epidemiology, and End Results Program - National Cancer Institute. [Accessed April 15, 2015];SEER Stat Fact Sheets: Ovary Cancer. http://seer.cancer.gov/statfacts/html/ovary.html.
- 97.Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, Guimbaud R, Buecher B, Bignon YJ, Caron O, et al. French Cancer Genetics Network. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304–2310. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- 98.Engel C, Loeffler M, Steinke V, Rahner N, Holinski-Feder E, Dietmaier W, Schackert HK, Goergens H, von Knebel Doeberitz M, Goecke TO, et al. Risks of less common cancers in proven mutation carriers with Lynch syndrome. J Clin Oncol. 2012;30:4409–4415. doi: 10.1200/JCO.2012.43.2278. [DOI] [PubMed] [Google Scholar]
- 99.Watson P, Vasen HF, Mecklin JP, Bernstein I, Aarnio M, Järvinen HJ, Myrhøj T, Sunde L, Wijnen JT, Lynch HT. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer. 2008;123:444–449. doi: 10.1002/ijc.23508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao X, Melton DW, Gourley C. Mismatch repair deficiency in ovarian cancer - molecular characteristics and clinical implications. Gynecol Oncol. 2014;132:506–512. doi: 10.1016/j.ygyno.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 101.Catasús L, Bussaglia E, Rodrguez I, Gallardo A, Pons C, Irving JA, Prat J. Molecular genetic alterations in endometrioid carcinomas of the ovary: Similar frequency of beta-catenin abnormalities but lower rate of microsatellite instability and PTEN alterations than in uterine endometrioid carcinomas. Hum Pathol. 2004;35:1360–1368. doi: 10.1016/j.humpath.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 102.Gras E, Catasus L, Argüelles R, Moreno-Bueno G, Palacios J, Gamallo C, Matias-Guiu X, Prat J. Microsatellite instability, MLH-1 promoter hypermethylation, and frameshift mutations at coding mononucleotide repeat microsatellites in ovarian tumors. Cancer. 2001;92:2829–2836. doi: 10.1002/1097-0142(20011201)92:11<2829::AID-CNCR10094>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 103.Jensen KC, Mariappan MR, Putcha GV, Husain A, Chun N, Ford JM, Schrijver I, Longacre TA. Microsatellite instability and mismatch repair protein defects in ovarian epithelial neoplasms in patients 50 years of age and younger. Am J Surg Pathol. 2008;32:1029–1037. doi: 10.1097/PAS.0b013e31816380c4. [DOI] [PubMed] [Google Scholar]
- 104.Murphy MA, Wentzensen N. Frequency of mismatch repair deficiency in ovarian cancer: A systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int J Cancer. 2011;129:1914–1922. doi: 10.1002/ijc.25835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pal T, Permuth-Wey J, Kumar A, Sellers TA. Systematic review and meta-analysis of ovarian cancers: Estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin Cancer Res. 2008;14:6847–6854. doi: 10.1158/1078-0432.CCR-08-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scartozzi M, De Nictolis M, Galizia E, Carassai P, Bianchi F, Berardi R, Gesuita R, Piga A, Cellerino R, Porfiri E. Loss of hMLH1 expression correlates with improved survival in stage III–IV ovarian cancer patients. Eur J Cancer. 2003;39:1144–1149. doi: 10.1016/S0959-8049(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 107.Zhai QJ, Rosen DG, Lu K, Liu J. Loss of DNA mismatch repair protein hMSH6 in ovarian cancer is histotype-specific. Int J Clin Exp Pathol. 2008;1:502–509. [PMC free article] [PubMed] [Google Scholar]
- 108.Begum FD, Høgdall CK, Kjaer SK, Blaakaer J, Christensen L, Ryan A, Jacobs IJ, Høgdall EV. Distribution of microsatellite instability in Danish ovarian tumor patients and the prognostic value in ovarian cancer patients. Oncol Res. 2008;17:43–49. doi: 10.3727/096504008784046090. [DOI] [PubMed] [Google Scholar]
- 109.Ercoli A, Ferrandina G, Raspaglio G, Marone M, Maggiano N, Del Mastro P, Benedetti Panici P, Mancuso S, Scambia G. hMSH2 and GTBP expression in advanced stage epithelial ovarian cancer. Br J Cancer. 1999;80:1665–1671. doi: 10.1038/sj.bjc.6690579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marcelis CL, van der Putten HW, Tops C, Lutgens LC, Moog U. Chemotherapy resistant ovarian cancer in carriers of an hMSH2 mutation? Fam Cancer. 2001;1:107–109. doi: 10.1023/A:1013865323890. [DOI] [PubMed] [Google Scholar]
- 111.CRUK. [Accessed April 15, 2015];Skin cancer incidence statistics. http://www.cancer-researchuk.org/cancer-info/cancerstats/types/skin/incidence/
- 112.Birindelli S, Tragni G, Bartoli C, Ranzani GN, Rilke F, Pierotti MA, Pilotti S. Detection of microsatellite alterations in the spectrum of melanocytic nevi in patients with or without individual or family history of melanoma. Int J Cancer. 2000;86:255–261. doi: 10.1002/(SICI)1097-0215(20000415)86:2<255::AID-IJC16>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 113.Peris K, Keller G, Chimenti S, Amantea A, Kerl H, Höfler H. Microsatellite instability and loss of heterozygosity in melanoma. J Invest Dermatol. 1995;105:625–628. doi: 10.1111/1523-1747.ep12323809. [DOI] [PubMed] [Google Scholar]
- 114.Quinn AG, Healy E, Rehman I, Sikkink S, Rees JL. Microsatellite instability in human non-melanoma and melanoma skin cancer. J Invest Dermatol. 1995;104:309–312. doi: 10.1111/1523-1747.ep12664612. [DOI] [PubMed] [Google Scholar]
- 115.Richetta A, Silipo V, Calvieri S, Frati L, Ottini L, Cama A, Mariani-Costantini R. Microsatellite instability in primary and metastatic melanoma. J Invest Dermatol. 1997;109:119–120. doi: 10.1111/1523-1747.ep12276783. [DOI] [PubMed] [Google Scholar]
- 116.Talwalkar VR, Scheiner M, Hedges LK, Butler MG, Schwartz HS. Microsatellite instability in malignant melanoma. Cancer Genet Cytogenet. 1998;104:111–114. doi: 10.1016/S0165-4608(97)00452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alvino E, Marra G, Pagani E, Falcinelli S, Pepponi R, Perrera C, Haider R, Castiglia D, Ferranti G, Bonmassar E, et al. High-frequency microsatellite instability is associated with defective DNA mismatch repair in human melanoma. J Invest Dermatol. 2002;118:79–86. doi: 10.1046/j.0022-202x.2001.01611.x. [DOI] [PubMed] [Google Scholar]
- 118.Hussein MR. Genetic pathways to melanoma tumorigenesis. J Clin Pathol. 2004;57:797–801. doi: 10.1136/jcp.2003.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hussein MR, Sun M, Tuthill RJ, Roggero E, Monti JA, Sudilovsky EC, Wood GS, Sudilovsky O. Comprehensive analysis of 112 melanocytic skin lesions demonstrates microsatellite instability in melanomas and dysplastic nevi, but not in benign nevi. J Cutan Pathol. 2001;28:343–350. doi: 10.1034/j.1600-0560.2001.280702.x. [DOI] [PubMed] [Google Scholar]
- 120.Palmieri G, Ascierto PA, Cossu A, Colombino M, Casula M, Botti G, Lissia A, Tanda F, Castello G. Assessment of genetic instability in melanocytic skin lesions through microsatellite analysis of benign naevi, dysplastic naevi, and primary melanomas and their metastases. Melanoma Res. 2003;13:167–170. doi: 10.1097/00008390-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 121.Palmieri G, Cossu A, Ascierto PA, Botti G, Strazzullo M, Lissia A, Colombino M, Casula M, Floris C, Tanda F, et al. Melanoma Cooperative Group. Definition of the role of chromosome 9p21 in sporadic melanoma through genetic analysis of primary tumours and their metastases. Br J Cancer. 2000;83:1707–1714. doi: 10.1054/bjoc.2000.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Richetta A, Ottini L, Falchetti M, Innocenzi D, Bottoni U, Faiola R, Mariani-Costantini R, Calvieri S. Instability at sequence repeats in melanocytic tumours. Melanoma Res. 2001;11:283–289. doi: 10.1097/00008390-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 123.Tomlinson IP, Beck NE, Bodmer WF. Allele loss on chromosome 11q and microsatellite instability in malignant melanoma. Eur J Cancer. 1996;32A:1797–1802. doi: 10.1016/0959-8049(96)00198-0. [DOI] [PubMed] [Google Scholar]
- 124.Castiglia D, Bernardini S, Alvino E, Pagani E, De Luca N, Falcinelli S, Pacchiarotti A, Bonmassar E, Zambruno G, D'Atri S. Concomitant activation of Wnt pathway and loss of mismatch repair function in human melanoma. Genes Chromosomes Cancer. 2008;47:614–624. doi: 10.1002/gcc.20567. [DOI] [PubMed] [Google Scholar]
- 125.Korabiowska M, Cordon-Cardo C, Jaenckel F, Stachura J, Fischer G, Brinck U. Application of in situ hybridization probes for MLH-1 and MSH-2 in tissue microarrays of paraffin-embedded malignant melanomas: Correlation with immunohistochemistry and tumor stage. Hum Pathol. 2004;35:1543–1548. doi: 10.1016/j.humpath.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 126.Alvino E, Passarelli F, Cannavò E, Fortes C, Mastroeni S, Caporali S, Jiricny J, Cappellini GC, Scoppola A, Marchetti P, et al. High expression of the mismatch repair protein MSH6 is associated with poor patient survival in melanoma. Am J Clin Pathol. 2014;142:121–132. doi: 10.1309/AJCPCX2D9YULBBLG. [DOI] [PubMed] [Google Scholar]
- 127.CRUK. [Accessed April 15, 2015];Stomach cancer statistics. http://www.cancerresearchuk.org/cancer-info/cancerstats/types/stomach/
- 128.Beghelli S, de Manzoni G, Barbi S, Tomezzoli A, Roviello F, Di Gregorio C, Vindigni C, Bortesi L, Parisi A, Saragoni L, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery. 2006;139:347–356. doi: 10.1016/j.surg.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 129.Fang WL, Chang SC, Lan YT, Huang KH, Chen JH, Lo SS, Hsieh MC, Li AF, Wu CW, Chiou SH. Microsatellite instability is associated with a better prognosis for gastric cancer patients after curative surgery. World J Surg. 2012;36:2131–2138. doi: 10.1007/s00268-012-1652-7. [DOI] [PubMed] [Google Scholar]
- 130.Lee HS, Choi SI, Lee HK, Kim HS, Yang HK, Kang GH, Kim YI, Lee BL, Kim WH. Distinct clinical features and outcomes of gastric cancers with microsatellite instability. Mod Pathol. 2002;15:632–640. doi: 10.1038/modpathol.3880578. [DOI] [PubMed] [Google Scholar]
- 131.Schneider BG, Bravo JC, Roa JC, Roa I, Kim MC, Lee KM, Plaisance KT, Jr, McBride CM, Mera R. Microsatellite instability, prognosis and metastasis in gastric cancers from a low-risk population. Int J Cancer. 2000;89:444–452. doi: 10.1002/1097-0215(20000920)89:5<444::AID-IJC8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 132.Falchetti M, Saieva C, Lupi R, Masala G, Rizzolo P, Zanna I, Ceccarelli K, Sera F, Mariani-Costantini R, Nesi G, et al. Gastric cancer with high-level microsatellite instability: Target gene mutations, clinicopathologic features, and long-term survival. Hum Pathol. 2008;39:925–932. doi: 10.1016/j.humpath.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 133.Perez RO, Jacob CE, D'Ottaviano FL, Alvarenga C, Ribeiro AS, Ribeiro U, Jr, Bresciani CJ, Zilberstein B, Krieger JE, Habr-Gama A, et al. Microsatellite instability in solitary and sporadic gastric cancer. Rev Hosp Clin Fac Med Sao Paulo. 2004;59:279–285. doi: 10.1590/S0041-87812004000500010. [DOI] [PubMed] [Google Scholar]
- 134.An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, Ajani JA, Rashid A, Hamilton SR, Wu TT. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656–663. [PubMed] [Google Scholar]
- 135.An JY, Kim H, Cheong JH, Hyung WJ, Kim H, Noh SH. Microsatellite instability in sporadic gastric cancer: Its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int J Cancer. 2012;131:505–511. doi: 10.1002/ijc.26399. [DOI] [PubMed] [Google Scholar]
- 136.Oki E, Kakeji Y, Zhao Y, Yoshida R, Ando K, Masuda T, Ohgaki K, Morita M, Maehara Y. Chemosensitivity and survival in gastric cancer patients with microsatellite instability. Ann Surg Oncol. 2009;16:2510–2515. doi: 10.1245/s10434-009-0580-8. [DOI] [PubMed] [Google Scholar]