Abstract
The secreted frizzled-related protein 2 (SFRP2) plays a pivotal role in the Wnt pathway, however, it functions as an agonist or an antagonist is still controversial. We profiled SFRP2 expression in several lung cancer cell lines, and found that A549 and 95-D exhibited the lowest and the highest level of SFRP2, respectively. Then we employed the SFRP2-overexpressing plasmid and siRNA to transfect A549 and 95-D cells, respectively. Through MTT assays, we found that SFRP2 knockdown inhibited cell proliferation, and halted the 95-D cells in G1 phase of the cell cycle by downregulation of CCND1 and CDK4, indicating that SFRP2 has the ability of promoting lung cancer cell proliferation. We further checked the cell properties of migration and invasion, using wound scratch assay and Transwell assays. The data showed decreased migrated and invasive 95-D cells after SFRP2 knockdown, and the observations were the opposite in the overexpressing model, implying that SFRP2 promoted lung cancer cell invasion. Moreover, the canonical Wnt pathway was also studied through detection of β-catenin by western blotting. In the SFRP2 overexpressing model, A549 cells presented stronger expression of β-catenin compared with controls, while it was the opposite in 95-D cells. These results suggested that SFRP2 serves as a Wnt agonist in lung cancer cells. Together, the findings of this study implied that SFRP2 is not only an agonist of Wnt pathway, but also a cancer promoting protein for lung cancer, indicating SFRP2 as a promising therapeutic target for lung cancer treatment.
Keywords: secreted frizzled-related protein 2, cell proliferation, cell invasion, lung cancer, Wnt pathway
Introduction
The secreted frizzled-related protein 2 (SFRP2) belongs to a large family of SFRPs consisting of SFRP1-5, and is homologous to the extracellular portion of the Wingless-type (Wnt) receptor Frizzled (1). The Wnt signaling pathway plays important roles in carcinogenesis, embryonic development and neurodegenerative disease (2), and has been extensively studied in human cancer. It is reported to be aberrantly activated in various human cancers, such as non-small cell lung cancer (NSCLC) (3), colorectal cancer (4), melanoma (5) and leukemia (6).
The secreted ligands of Wnt pathway possesses 19 highly conserved and hydrophobic glycoproteins (7), and the receptor of this signaling is a protein complex consisting of Frizzled and lipoprotein receptor-related protein (LRP) (1). There are canonical and non-canonical Wnt signaling (1). The former is dependent on β-catenin, while the latter is not and may proceed through calcium flux, G proteins, and JNK (8). In the activated state of canonical pathway, the ligand binds to Frizzled/LRP receptor complex and released cytosolic β-catenin from a degradation complex consisting of adenomatous polyposis coli (APC), Axin and glycogen synthase kinase-3β (GSK-3β) (1,9). Consequently, β-catenin accumulates in the cytoplasm and subsequently translocates to the nucleus, where it interacts with T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors and turns on the downstream genes, including CCND1, Axin2, c-myc and MMP-7 (10).
Considering the homology of SFRP2 and extracellular portion of Frizzled, it is reported that SFRP2 is an antagonist to Wnt pathway by competing for Wnt binding to Frizzled (11). However, there are reports claiming that SFRP2 is an agonist of the Wnt pathway (12–17). Recently, investigators have shown that SFRP2 inhibition reduced tumor volume of breast cancer to 46% compared with control mice (18), suggesting that SFRP2 is a tumor-promoting protein. Consistent with this, another recent report showed that SFRP2 intensity increased with tumor size in murine angiosarcoma (19).
For the purpose of clarifying whether SFRP2 is an oncoprotein or a tumor suppressor in lung cancer, we assessed the effects of SFRP2 on Wnt signaling pathway as well as on lung cancer cell proliferation, apoptosis, migration and invasion. The molecular mechanisms underlying the functions of SFRP2 in lung cancer were also investigated.
Materials and methods
Cell lines and culture conditions
Human lung cancer cell lines 95-D, SPCA-1, and A549 were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All these cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin (Life Technologies, Carlsbad, CA, USA).
Cell transfection
Three pairs of siRNAs were synthesized by Life Technologies (Shanghai, China). The expression plasmid containing SFRP2 coding region was purchased from ViGene BioSciences (Shandong, China) and confirmed by DNA sequencing. The day before transfection, cells were plated into 6-well plates and cultured overnight. Cell transfection experiments were performed according to the transfection reagent manufacturer's instructions. In brief, 2 µg of SFRP2 siRNA or expression plasmid and their respective control DNAs were re-suspended in Opti-MEM (Life Technologies, Carlsbad, CA, USA) to 100 µl in a tube; meanwhile, 10 µl X-tremeGENE siRNA transfection reagent (Roche, Indianapolis, IN, USA) was added to 90 µl Opti-MEM in another tube. Transfection reagent in the second tube was then added into the first tube containing DNA, and mixed and incubated at room temperature for 20 min. After that, the complex was added to the cells in 6-well plates and the cells were cultured for an additional 24–48 h.
RNA isolation and real-time RT-qPCR
Total RNA was isolated by TRIzol (Life Technologies, Carlsbad, CA, USA), and then quantified using the NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). For RT-qPCR, 2 µg of total RNA and primers were mixed with regents supplied by Quant One Step RT-qPCR (SYBR Green I) kit (Tiangen, Beijing, China) according to the instructions. Specific primers for mRNA detection are indicated in Table I. The qPCR was carried out using the Bio-Rad CFX96 Real-Time PCR system (Bio-Rad, Hercules, CA, USA), using the following reaction conditions: the first step at 95°C for 5 min, followed by 35 amplification cycles composed of 95°C for 10 sec, 59°C for 20 sec and 72°C for 20 sec. All reactions were performed in triplicate and included negative controls (sterilized water as template). Results were processed and exported using the Bio-Rad CFX Manager software, version 1.1 (Bio-Rad). The expression of target mRNA was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, and determined using the 2−ΔΔCT method. All the experiments were repeated at least three times.
Table I.
Primers for real-time RT-qPCR.
| Gene | Forward primers (5′→3′) | Reverse primers (5′→3′) |
|---|---|---|
| CDK4 | CACAGTTCGTGAGGTGGCTTTA | GATGTGGCACAGACGTCCATC |
| CDK6 | TCATTCAAAATCTGCCCAACC | TCCTGGAAGTATGGGTGAGACA |
| CCNE1 | GACTGCCTTGAATTTCCTTATGGT | TCGCACCACTGATACCCTGAA |
| CCND1 | GCATCTACACCGACAACTCCA | CCAGGTTCCACTTGAGCTTG |
| p27 | AGAAGCCTGGCCTCAGAAGAC | CCATTCCATGAAGTCAGCGATA |
| GAPDH | GAAGGTCGGAGTCAACGGAT | CTGGAAGATGGTGATGGGATT |
| SFRP2 | ATGATGATGACAACGACATAATG | GAGCCACAGCACCGATTT |
| WIF-1 | AATGCCAATGTCAAGAAGG | GATGTCGGAGTTCACCAGA |
| APC | CCAACAAGGCTACGCTAT | CTGCTCGCCAAGACAAAT |
| Wnt5a | GAGTGCTCGCATCCTCAT | GCATGTCTTCAGGCTACA |
Protein extraction and western blotting
Cell lysates were obtained in lysis buffer (20 mM Tris-HCl, pH 7.5, 1% NP-40, 5 mM sodium pyrophosphate, 5 mM EDTA, pH 8.0, 1 mM sodium vanadate, 150 mM NaCl, 10 µg/ml aprotinin, 10 µg/ml leupeptin and 1 µM PMSF). After quantification and denaturation, equal proteins of each sample were subjected to 10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad). The membrane was first blocked with 5% non-fat dry milk for 1 h at room temperature, and then incubated with specific primary antibodies overnight at 4°C, such as antibodies against SFRP2, Cyclin D1, Cyclin E1, CDK4, CDK6, p27, Vimentin, E-cadherin, β-catenin, P-β-catenin and β-actin (Abcam, Cambridge, MA, USA, 1:1,000). Next, the membrane was washed and incubated with the secondary antibodies against rabbit or mouse for 1 h at room temperature. Lastly, the membrane was exposed to enhanced electrochemiluminescence (ECL) following the detection kit (KeyGen Biotechnology, Nanjing, Jiangsu, China) and photographed by the ChemiDoc XRS gel documentation system (Bio-Rad). All the experiments were repeated at least three times.
Cell proliferation assay
The cells were seeded at 8×103 cells/well into 96-well plates and incubated overnight prior to transfection. At 0, 24, 48 or 72 h after transfection, MTT (5 mg/ml in phosphate-buffered saline) was added to each well and incubated for 4 h, and then the supernatants were removed from the wells by aspiration. The cells were solubilized in 200 µl of dimethyl sulfoxide (DMSO) and the absorbance was recorded on a microplate reader at the wavelength of 490 nm. This assay was repeated at least three times.
Wound scratch assay
After transfection, cells were allowed to recover for 24 h, and then cells were cultured in FBS-free RPMI-1640 medium for starvation overnight. The next day, the cell monolayers were gently scratched with a pipette tip across the diameter of the well, rinsed with media to remove cellular debris, and photographed by microscopy as 0 h timepoint. After 24 h of culture, images were taken at the same position as 0 h time-point. The healing distances between the wound was established by calculating the ratio of the wound surface distance change from 0 to 24 h over the wound surface distance at 0 h. This assay was repeated at least three times.
Transwell assays
Cell suspension at 1×105 cells in 100 µl RPMI-1640 medium was added to the top chambers with 8-µm pore sized filter inserts of 24-well Transwell plates (Costar, Cambridge, MA, USA). For invasion assay, the top surface of filter membranes were coated with 30% of BD Matrigel matrix (BD Biosciences, San Jose, CA, USA) before cell seeding, and placed for 4 h in an incubator. For migration assay, the top surface of filter membranes was not coated with Matrigel. For both cell migration and invasion assays, the following procedure was the same. The bottom chambers were soaked by 600 µl RPMI-1640 medium supplemented with 10% FBS. After overnight incubation, the non-migrated or non-invaded cells on top surface of the filter membrane were removed by cotton swabs, and the migrated or invaded cells under the filter membrane were kept. Then the filter membranes were submerged in methanol for 15 min at room temperature to fix the cells. Subsequently, the cells were stained with crystal violet for another 15 min and then PBS was used to remove the excess stain. After the filter membranes were dried, the numbers of migrated or invaded cells in more than three fields were counted under a microscope, and the mean of the migrated or invaded cells for each membrane was determined. The experiments were repeated for a minimum of three times.
Flow cytometry
Cells at 1×106 were prepared, washed, and resuspended in PBS. Then cells were fixed in 70% ice-cold ethanol overnight at 4°C. After washing, cells were incubated for 15 min at 37°C in the dark in 200 µl of PI staining solution containing 20 µg/ml PI, 100 U/ml DNase-free RNase, and 0.1% v/v Triton X-100. All the samples were analyzed by the BD Accuri C6 (BD Biosciences).
Statistical analysis
All experimental data are presented as mean ± standard deviation (SD). Statistical significance was evaluated using the Student's t-test, and the significance was set at P-value <0.05, and highly significant at ≤0.01.
Results
SFRP2 expression profiles in lung cancer cells
To reveal SFRP2 expression profiles in lung cancer cells, we examined the lung cancer cell lines 95-D, A549 and SPCA1. By real-time RT-PCR, we found that 95-D cells expressed the highest level of SFRP2 mRNA, while A549 and SPCA1 cells expressed SFRP2 mRNA at significantly lower level as compared with 95-D cells (5.01±0.00 or 8.14±0.00 vs. 100.00±6.80%, respectively; both pairs P<0.01) (Fig. 1A). These results were further confirmed by the data of western blotting for SFRP2 protein (Fig. 1B), which showed extremely strong band in 95-D cell lysate, and the weakest band in A549 cell lysate. Thus, A549 and 95-D cells were subsequently chosen as cell models based on their remarkable difference in SFRP expression at both transcriptional and translational levels in order to investigate the cellular functions of SFRP2.
Figure 1.
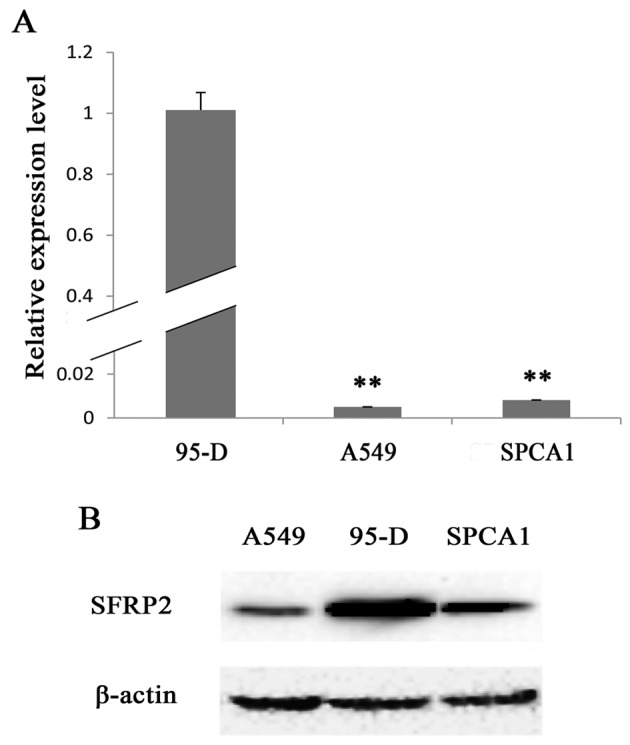
The SFRP2 expression profiles in lung cancer cell lines. (A) Real-time RT-qPCR detection of SFRP2 mRNA levels in the lung cancer cell lines 95-D, A549 and SPCA1. (B) Representative images for western blotting of SFRP2 protein in 95-D, A549 and SPCA1 cells. Data are presented as mean ± SD. Statistical significance was evaluated using the Student's t-test. **P-value ≤0.01.
Cell models for SFRP2 cellular function
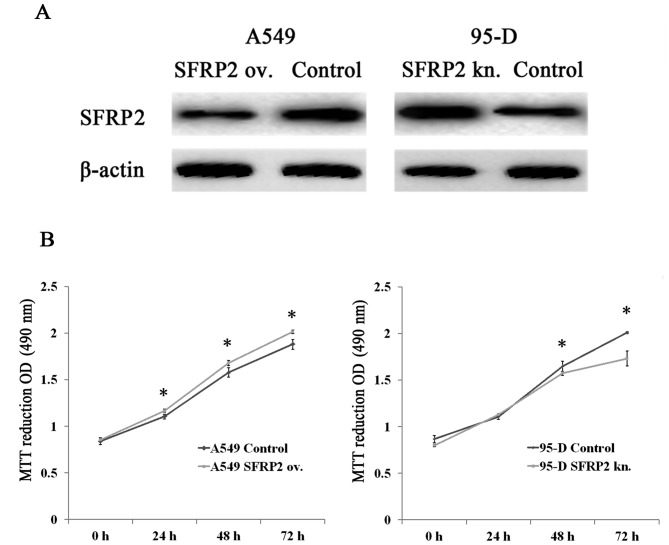
We employed A549 cells transfected with SFRP2 expressing plasmid and 95-D cells transfected with SFRP2 siRNA as two cell models for the following study on SFRP2 cellular functions. After transfection for 48 h, cells were lysed and subjected to western blotting for detecting SFRP2 protein. As indicated in Fig. 2A, SFRP2 protein was upregulated in A549 cells after transfection with the SFRP2 expressing plasmid in comparison with the vector control cells (left panel), whereas the level of SFRP2 protein was remarkably reduced in 95-D cells with siRNA transfection as compared with controls (right panel). These data suggest that two cell models for the overexpression and knockdown of SFRP2 were established and ready to use for subsequent experiments.
Figure 2.
The influence of SFRP2 on the proliferation in lung cancer cells. (A) Representative images for SFRP2 protein detection in SFRP2-overexpressing A549 cells (left panel) and SFRP2-knockdown 95-D cells (right panel). (B) The OD490 values in A549 cells with SFRP2 overexpression (left panel) and 95-D cells with SFRP2-knockdown (right panel), along with their respective controls at 24h, 48h and 72h after transfection by MTT assays. ov. and kn. represent overexpression and knockdown, respectively. Data are presented as mean ± SD. Statistical significance was evaluated using the Student's t-test. *P-value <0.05.
Effects of SFRP2 up- or down-regulation on lung cancer cell proliferation
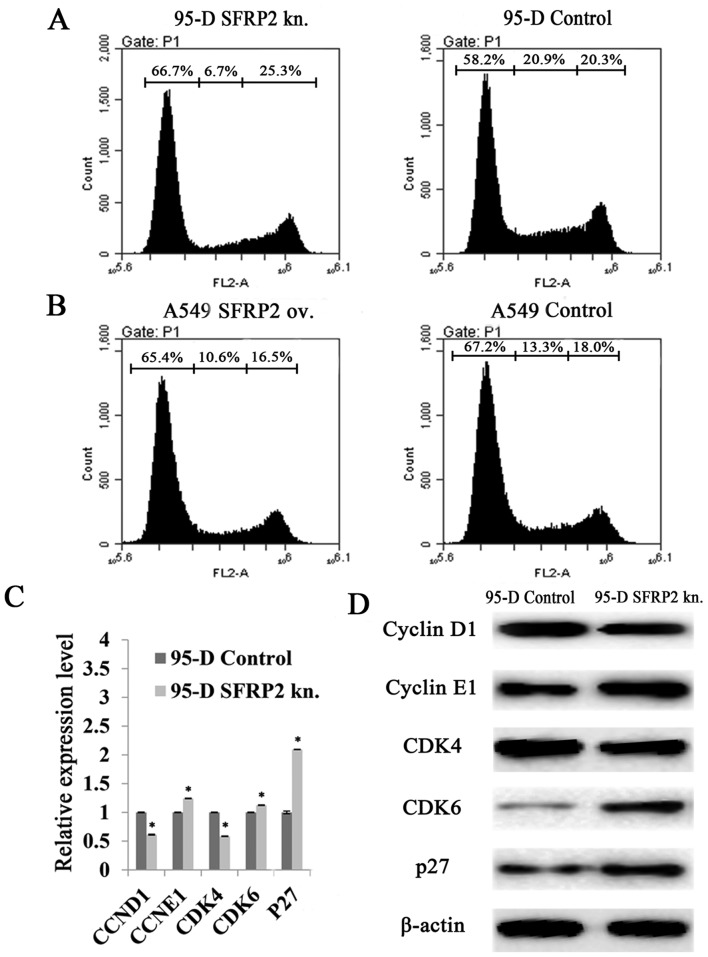
The two cell models established were applied to MTT assay, which showed significantly higher OD490 values at 24h, 48h and 72h after SFRP2 induction in A549 cells, compared with controls (Fig. 2B, left panel). In contrast, the readouts of OD490 declined significantly in 95-D cells with SFRP2 downregulation at 48h and 72h compared with 95-D control cells (Fig. 2B, right panel). These results indicated that SFRP2 had the potential of promoting cell proliferation in lung cancer cells. Sequentially, flow cytometry analysis demonstrated that the cell cycle was retarded in SFRP2 downregulated 95-D cells. The percentage of cells in G1 phase rose to 66.7% after SFRP2 knockdown, with an increase of 14.60% compared to 95-D controls (Fig. 3A), while no significant change was found in SFRP2 overexpressing A549 cells (Fig. 3B). In addition, the molecules related to G1 phase processing were further investigated by real-time RT-PCR. In SFRP2 downregulated 95-D cells, the mRNA levels of cyclin E1 (CCNE1), CDK6 and p27 were significantly increased compared with 95-D control cells (124.26±0.49 vs. 100.00±0.45%, 112.51±0.53 vs. 100.00±0.49%, and 209.46±0.37 vs. 100.00±2.32%, respectively; all pairs P<0.05) (Fig. 3C). The mRNA levels of cyclin D1 (CCND1) and CDK4 were reduced when compared with 95-D control cells (61.27±0.80 vs. 100.00±0.63% and 58.51±0.45 vs. 100.00±0.51%, respectively; all pairs P<0.05) (Fig. 3C). The protein levels of these G1 phase-related molecules were also evaluated by western blotting. As shown in Fig. 3D, cyclin D1 and CDK4 exhibited much weaker bands after SFRP2 knocked down in 95-D cells, while cyclin E1, CDK6 and p27 with stronger bands (Fig. 3D).
Figure 3.
Effects of SFRP2 on cell cycle in lung cancer cells. (A) Representative images of the cell cycle assays using flow cytometry in 95-D cells with or without SFRP2 knockdown at 48 h after transfection. (B) Representative images of cell cycle assays using flow cytometry in A549 cells with or without SFRP2 knock-in at 48 h after transfection. (C) Real-time RT-PCR detection of the cell cycle-related molecules in 95-D cells with or without SFRP2 knockdown. (D) Representative results of western blotting on Cyclin D1, Cyclin E1, CDK4, CDK6, p27 and β-actin in 95-D cells transfected by SFRP2 siRNA for 48 h. ov. and kn. represent overexpression and knockdown, respectively. Data are presented as mean ± SD. Statistical significance was evaluated using the Student's t-test. *P-value <0.05.
Influences of SFRP2 on lung cancer cell migration and invasion
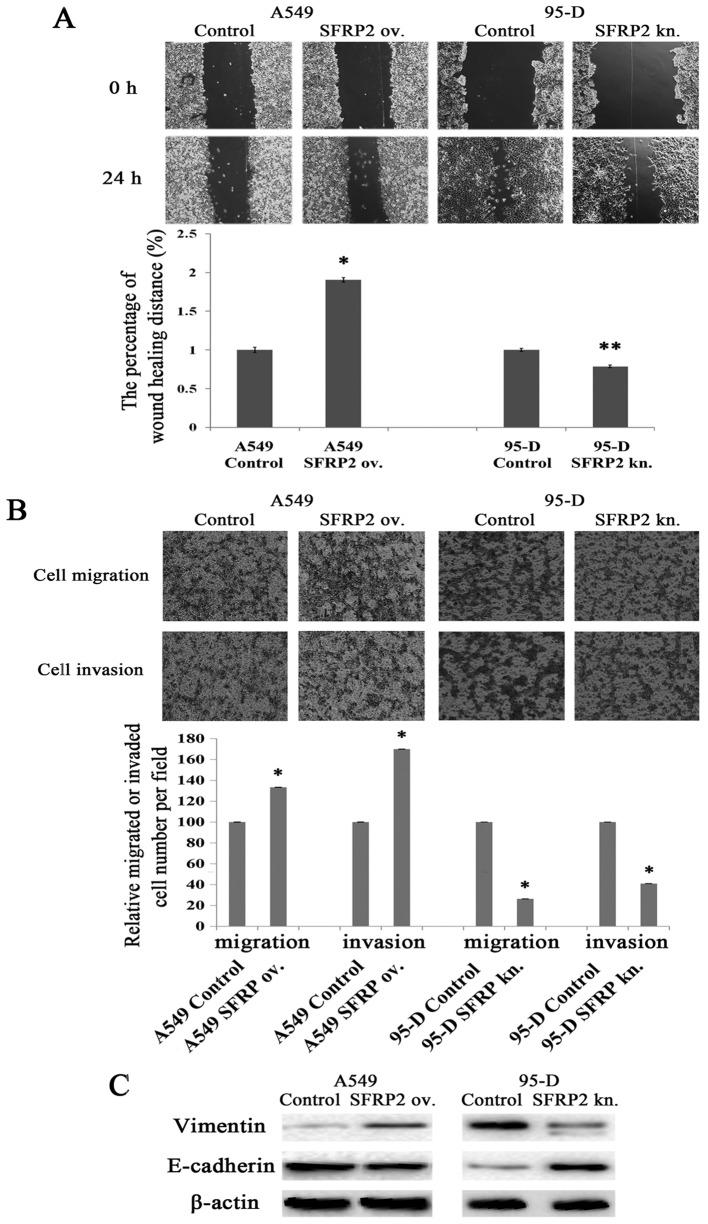
Wound scratch assays and Transwell assays were performed to evaluate the influences of SFRP2 on lung cancer cell migration and invasion. As shown in Fig. 4A, the distance changes between 0 and 24 h was calculated by defining the change of control cells as 100%, as presented Fig. 4A. A549 cells migrated faster after SFRP2 overexpression (190.34±3.04 vs. 100.00±3.53%; P<0.05) (Fig. 4A), which indicates that SFRP2 promotes lung cancer cell migration. There was also significant difference on migration distance between 95-D cells with and without SFRP2 knockdown (78.62±1.66 vs. 100.00±1.97%; P<0.001) (Fig. 4A). Furthermore, Transwell assays with or without Matrigel were used to evaluate lung cancer cell properties of invasion and migration. As shown in Fig. 4B, the numbers of migrated and invaded A549 cells significantly increased after SFRP2 overexpression compared with controls (133.33±0.12 vs. 100.00±0.19%, 170.00±0.08 vs. 100.00±0.19%, respectively; both pairs P<0.05). While the numbers of migrated and invaded 95-D cells were reduced after the knockdown of SFRP2 by siRNA transfection as compared to controls (26.32±0.12 vs. 100.00±0.01% and 40.94±0.10 vs. 100.00±0.08%, respectively; both pairs P<0.05). Taken together, these findings demonstrated the involvement of SFRP2 in promoting lung cancer cell migration and invasion. We next probed the proteins related to the epithelial to mesenchymal transition (EMT). As shown in Fig. 4C, cytoskeletal protein vimentin remarkably increased, but E-cadherin significantly decreased in SFRP2 overexpressing A549 cells (left panel). In the model of 95-D cells with SFRP2 knockdown, vimentin was weaker, while E-cadherin increased (Fig. 4C, right panel).
Figure 4.
Effects of SFRP2 on lung cancer cell migration and invasion. (A) Representative images of wound scratch assay at 24 and 48 h, respectively (named as 0 and 24 h correspondingly) in A549 and 95-D cells after transfection of SFRP2-overexpressing plasmid or SFRP2 siRNA (upper panel), and the percentage of wound healing distance is shown as compared with respective control cells (lower panel). (B) Representative images of Transwell assays at 24 h in A549 and 95-D cells after transfection of SFRP2-overexpressing plasmid or SFRP2 siRNA (upper panel), and the number of migrated or invaded cells is shown by comparing with respective control cells (lower panel). (C) Representative results of western blotting on Vimentin, E-cadherin and β-actin. Left panel, lysates from A549 cells transfected with SFRP2 expressing plasmid for 48 h, and right panel, lysates from 95-D cells transfected with SFRP2 siRNA for 48 h. ov. and kn. represent overexpression and knockdown, respectively. Data are presented as mean ± SD. Statistical significance was evaluated using the Student's t-test. *P-value <0.05 and **P-value ≤0.01.
Modulation of Wnt signaling pathway by SFRP2 in lung cancer cells
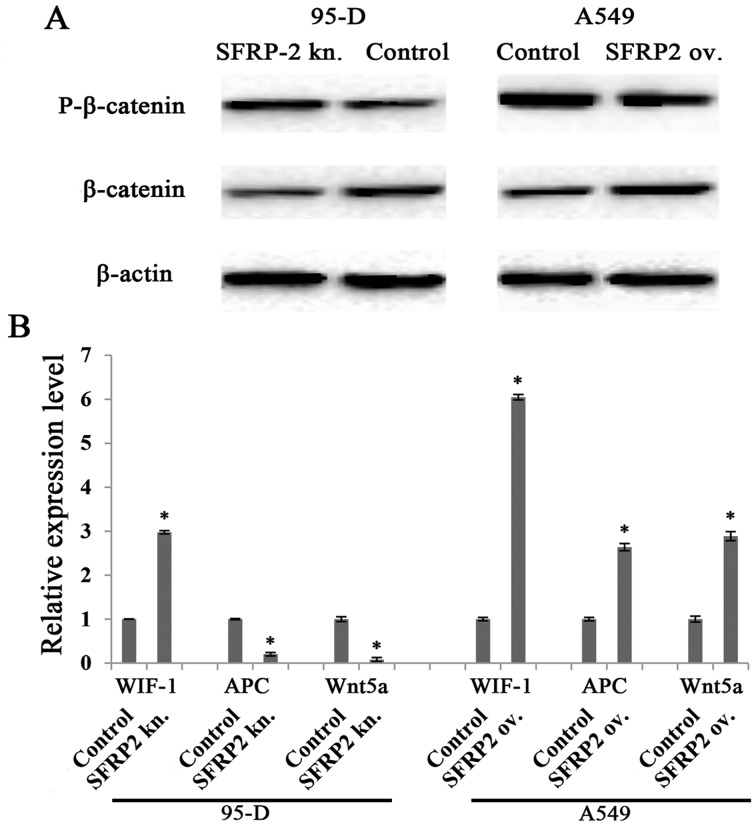
Given that SFRP2 has been reported to be an antagonist of Wnt signaling pathway, we next investigated the pathway variation in our cell line models. We found that phosphorylated β-catenin (degeneration form of β-catenin) was negatively correlated to SFRP2 level in both 95-D and A549 cell models, while β-catenin was positively correlated to SFRP2 level. These observations were supported by the data shown in Fig. 5A, which demonstrated stronger phosphorylated β-catenin band and much fainter β-catenin band in SFRP2 downregulated 95-D cell lysates, whereas the opposite data were revealed in SFRP2 upregulated A549 cells (Fig. 5A, right panel). These findings implied that SFRP2 serves as an agonist for the canonical Wnt signaling pathway. The mRNA levels of several mediators of the Wnt pathway were also checked. In both cell models, WIF-1 increased significantly, while APC and Wnt5a were specifically reduced in 95-D cells with SFRP2 knockdown (Fig. 5B).
Figure 5.
Influences of SFRP2 on Wnt signaling pathway in lung cancer cells. (A) Representative images of β-catenin and phosphorylated β-catenin by western blotting in SFRP2-overexpressing A549 cells (right panel) and SFRP2-knockdowned 95-D cells (left panel). (B) Real-time RT-PCR detection of molecules related to Wnt signaling pathway in A549 and 95-D cells transfected for 48 h with SFRP2-overexpressing plasmid or SFRP2 siRNA, respectively. ov. and kn. represent overexpression and knockdown, respectively. Data are presented as mean ± SD. Statistical significance was evaluated using the Student's t-test. *P-value <0.05.
Discussion
There is still disagreement on whether SFRP2 possesses cell proliferation promoting or suppressive function. Roth et al observed SFRP2 expressing glioma xenografts were significantly larger than xenografts consisting of control cells (17). In line with this, Sathi et al found that SFRP2 promotes cell proliferation in ameloblastoma (20). In contrast, the overexpression of SFRP2 in oral squamous cell carcinoma cell lines suppressed cell proliferation and arrested the cell cycle in the G1 phase (21). In this study, we investigated SFRP2 functions in two lung cancer cell models. Our data with MTT assay showed a significant inhibition of proliferation at 48h and 72h after SFRP2 knockdown in 95-D cells (Fig. 2B). In contrast, SFRP2 overexpression in A549 cells promoted the lung cancer cell proliferation (Fig. 2B). When SFRP2 was downregulated by siRNA in 95-D cells, the cell proliferation was inhibited at G1 phase of the cell cycle (Fig. 3A) by inducing p27 expression, accompanied by decreasing CDK4 and cyclin D1 expression on both mRNA and protein levels (Fig. 3C and D). It is considered that cyclin D1 binds to its activator CDK4 in order to make cell cycle G1 checkpoint go through. In our SFRP2 knockdown cell model, both CDK4 and cyclin D1 mRNAs and proteins were reduced profoundly (Fig. 3C and D). Therefore, our data suggest that SFRP2 is a protein promoting lung cancer cell proliferation.
The controversy on the tumor functions of SFRP2 still exist. Some researchers considered SFRP2 as a tumorsuppressive protein based on its highly methylated status in gene promoter region in various cancer cells, such as breast cancer (22), oral squamous cell carcinoma (23) and hepatocellular carcinoma (24). Others suggested it is an antagonist to the Wnt pathway as it is homologous to extracellular portion of Frizzled, the Wnt receptor (12–17). However, it has been believed that SFRP2 serves as an oncogene to promote carcinoma development and progression. For example, a recent report declaimed that SFRP2 inhibition reduced tumor volume of breast cancer to 46% compared to control mice (18). As we previously discussed, our findings support the latter hypothesis that SFRP2 is a tumor promoting protein for lung cancer. Metastasis is an ominous feature of malignant solid tumors, and EMT, is a key step of the progression of tumor cell invasion. The speed, site and severity of tumor metastasis vary in different type of cancers. For example, breast tumor undergoes EMT at an early stage (25), and it can be detected with the metastasis to bone, lung or liver for years or decades after the diagnosis or surgery (26). In case of lung adenocarcinoma, however, it has the potential of rapidly spreading to multiple organs, and causes high rate of mortality (27). The high metastatic feature of lung cancer calls for investigators to find the molecules contributing to it. To further investigate the influences of SFRP2 on lung cancer cell invasion, we performed Transwell assays and western blotting, and found that SFRP2 is critical in lung cancer invasion for the observation that both the migration and invasion abilities were positively correlated to the expression level of SFRP2 (Fig. 4A and B). In addition, the expression pattern of EMT related protein, such as E-cadherin and Vimentin, changed according to the variation of cell invasion potential (Fig. 4C). Interestingly, β-catenin, one of the EMT related proteins, also changed in line with alteration of cell invasion ability.
As previously described, in SFRP2 downregulated 95-D cells, the cell proliferation was arrested at the G1 phase (Fig. 3A) along with the decreased cyclin D1 on both mRNA and protein levels (Fig. 3C and D). We also found that the ability of cell invasion was weakened in the same cell model, accompanied by reduced protein level of β-catenin (Fig. 5A). Moreover, β-catenin is also the pivotal molecule in the canonical Wnt signaling pathway, in which, it could be activated and translocated into the nucleus where it binds TCF/LEF-1 to enhance the transcription of specific target genes, such as cyclin D1, c-myc and Axin2 (1). Therefore, these two phenomena were connected as CCND1 is the downstream target of β-catenin. These results implied that SFRP2 knockdown downregulated the canonical Wnt signaling pathway by decreasing β-catenin, and ultimately reduced the transcriptional targets of β-catenin and TCF/LEF-1 complex, such as CCND1. Thus, our data suggest that SFRP2 is an agonist for the Wnt pathway. We also found the changes of several other molecules related to Wnt pathway, such as the agonist Wnt5a, and the antagonist WIF-1 and APC (Fig. 5B). However, whether these molecules function synergistically with SFRP2 is still unknown, and needs to be further investigated.
In the present study, we examined SFRP2 expression status in several lung cancer cell lines, and studied its roles in two lung cancer cell line models containing SFRP2 overexpression and knockdown, respectively. Our data indicate that SFRP2 is a cancer promoting protein that promotes lung cancer cell proliferation and invasion. Moreover, SFRP2 serves as an agonist for Wnt signaling pathway in lung cancer. These observations are likely to have implications for our understanding of SFRP2 biological function as a critical molecule underlying lung cancer development and progression, and the findings suggest that SFRP2 may serve as a potential target for lung cancer therapy.
Acknowledgments
This study was supported by grants from Guangzhou Medical University (B147048, to J. Tang), the Science and Information Technology Bureau of Guangzhou (2011Y1-00022 and [2012]224-8, to J. Tang), and the Science and Information Technology Bureau of Guangzhou Haizhu District (2012-ZD-02, to J. Tang).
References
- 1.Shi Y, He B, You L, Jablons DM. Roles of secreted frizzled-related proteins in cancer. Acta Pharmacol Sin. 2007;28:1499–1504. doi: 10.1111/j.1745-7254.2007.00692.x. [DOI] [PubMed] [Google Scholar]
- 2.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 3.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: Evidence of dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, Batlle E. EphB/EphrinB receptors and Wnt signaling in colorectal cancer. Cancer Res. 2006;66:2–5. doi: 10.1158/0008-5472.CAN-05-3849. [DOI] [PubMed] [Google Scholar]
- 5.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/S1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 6.Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 8.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/S1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 11.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 12.Courtwright A, Siamakpour-Reihani S, Arbiser JL, Banet N, Hilliard E, Fried L, Livasy C, Ketelsen D, Nepal DB, Perou CM, et al. Secreted frizzle-related protein 2 stimulates angiogenesis via a calcineurin/NFAT signaling pathway. Cancer Res. 2009;69:4621–4628. doi: 10.1158/0008-5472.CAN-08-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirotsou M, Zhang Z, Deb A, Zhang L, Gnecchi M, Noiseux N, Mu H, Pachori A, Dzau V. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc Natl Acad Sci USA. 2007;104:1643–1648. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JL, Chang CJ, Wu SY, Sargan DR, Lin CT. Secreted frizzled-related protein 2 (SFRP2) is highly expressed in canine mammary gland tumors but not in normal mammary glands. Breast Cancer Res Treat. 2004;84:139–149. doi: 10.1023/B:BREA.0000018412.83348.ff. [DOI] [PubMed] [Google Scholar]
- 15.Gehmert S, Sadat S, Song YH, Yan Y, Alt E. The anti-apoptotic effect of IGF-1 on tissue resident stem cells is mediated via PI3-kinase dependent secreted frizzled related protein 2 (Sfrp2) release. Biochem Biophys Res Commun. 2008;371:752–755. doi: 10.1016/j.bbrc.2008.04.151. [DOI] [PubMed] [Google Scholar]
- 16.Lee JL, Chang CJ, Chueh LL, Lin CT. Secreted frizzled related protein 2 (sFRP2) decreases susceptibility to UV-induced apoptosis in primary culture of canine mammary gland tumors by NF-kappaB activation or JNK suppression. Breast Cancer Res Treat. 2006;100:49–58. doi: 10.1007/s10549-006-9233-9. [DOI] [PubMed] [Google Scholar]
- 17.Roth W, Wild-Bode C, Platten M, Grimmel C, Melkonyan HS, Dichgans J, Weller M. Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene. 2000;19:4210–4220. doi: 10.1038/sj.onc.1203783. [DOI] [PubMed] [Google Scholar]
- 18.Fontenot E, Rossi E, Mumper R, Snyder S, Siamakpour-Reihani S, Ma P, Hilliard E, Bone B, Ketelsen D, Santos C, et al. A novel monoclonal antibody to secreted frizzled-related protein 2 inhibits tumor growth. Mol Cancer Ther. 2013;12:685–695. doi: 10.1158/1535-7163.MCT-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuruta JK, Klauber-DeMore N, Streeter J, Samples J, Patterson C, Mumper RJ, Ketelsen D, Dayton P. Ultrasound molecular imaging of secreted frizzled related protein-2 expression in murine angiosarcoma. PLoS One. 2014;9:e86642. doi: 10.1371/journal.pone.0086642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sathi GA, Inoue M, Harada H, Rodriguez AP, Tamamura R, Tsujigiwa H, Borkosky SS, Gunduz M, Nagatsuka H. Secreted frizzled related protein (sFRP)-2 inhibits bone formation and promotes cell proliferation in ameloblastoma. Oral Oncol. 2009;45:856–860. doi: 10.1016/j.oraloncology.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Xiao C, Wang L, Zhu L, Zhang C, Zhou J. Secreted frizzled-related protein 2 is epigenetically silenced and functions as a tumor suppressor in oral squamous cell carcinoma. Mol Med Rep. 2014;10:2293–2298. doi: 10.3892/mmr.2014.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sogabe Y, Suzuki H, Toyota M, Ogi K, Imai T, Nojima M, Sasaki Y, Hiratsuka H, Tokino T. Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma. Int J Oncol. 2008;32:1253–1261. doi: 10.3892/ijo_32_6_1253. [DOI] [PubMed] [Google Scholar]
- 24.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 25.Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmüller G, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]