Abstract
Continued progress in addressing challenges associated with detection and management of tuberculosis requires new diagnostic tools. These tools must be able to provide rapid and accurate information for detecting resistance to guide selection of the treatment regimen for each patient. To achieve this goal, globally representative genotypic, phenotypic, and clinical data are needed in a standardized and curated data platform. A global partnership of academic institutions, public health agencies, and nongovernmental organizations has been established to develop a tuberculosis relational sequencing data platform (ReSeqTB) that seeks to increase understanding of the genetic basis of resistance by correlating molecular data with results from drug susceptibility testing and, optimally, associated patient outcomes. These data will inform development of new diagnostics, facilitate clinical decision making, and improve surveillance for drug resistance. ReSeqTB offers an opportunity for collaboration to achieve improved patient outcomes and to advance efforts to prevent and control this devastating disease.
Keywords: Mycobacterium tuberculosis, drug resistance, database
Tuberculosis remains a global threat to public health, with an estimated 9 million new cases and 1.5 million deaths in 2013 [1]. Of these, 480 000 cases, and 210 000 deaths, were estimated to be due to multidrug resistant (MDR) tuberculosis, defined as tuberculosis caused by Mycobacterium tuberculosis (Mtb) that is resistant to at least rifampin and isoniazid, 2 of the most effective first-line drugs. MDR tuberculosis is complicated to treat, requiring lengthy durations of therapy with second-line drugs that are more expensive, less effective, and more toxic than first-line antituberculosis drugs [2]. Alarming reports of extensively drug-resistant tuberculosis—that is, MDR tuberculosis isolates with additional resistance to at least 1 fluoroquinolone and 1 second-line injectable—have revealed deficiencies in testing capabilities and health systems [3–5]. Efforts to control the spread of tuberculosis are hindered by insufficient diagnostic capacity, gaps in infection control procedures, and the emergence of drug resistance [6, 7]. Although global progress has been made in the rapid bacteriologic detection of MDR tuberculosis as a result of broad implementation of World Health Organization (WHO)–endorsed technologies such as the Xpert MTB/RIF [8] and line-probe assays [9], continued advancement will require new diagnostic tools. These tools must be capable of rapidly providing accurate and actionable information about drug resistance, including new regimens, in the context of the health systems that must have the capacity to provide effective treatment. The data necessary to support development of these tools should be available in a globally representative standardized database; however, to date, this type of data platform has not been developed for Mtb and is a critical need [10].
Currently, phenotypic drug susceptibility testing (DST) is utilized to detect or confirm drug resistance or the emergence of resistance, to guide the choice of therapy that will provide the best chance of cure, and to monitor the prevalence of primary and acquired drug resistance within a community [11]. DST has long been considered the gold standard. However, DST is not feasible when cultures are contaminated, and its turnaround times are long because of the slow growth rate of Mtb and the delays that are added by shipping to a laboratory with the capacity for DST. Additionally, the reproducibility of DST results for some drugs is poor because of inherent difficulties with the methods. For the majority of tuberculosis cases, DST is not available because it requires complex biosafety infrastructure and highly skilled laboratory technicians. Globally in 2013, only 8.5% of new reported tuberculosis cases and 17% of retreatment cases had DST results [1]. For some cases, DST was only performed after repeated treatment failure. In the Global End TB strategy endorsed by WHO, focus is placed on universal DST for tuberculosis patients, because detection and treatment of drug-susceptible and -resistant tuberculosis are essential for preventing ongoing transmission and strengthening programmatic efforts [12, 13]. Optimally, all tuberculosis patients would be served by healthcare systems with primary access to rapid molecular tests for the detection of mutations associated with resistance to both first-and second-line drugs. Such access to rapid results would provide the basis for individualized treatment regimens that would start during the first encounter, thus avoiding delays and loss to follow-up and offering the best chance for cure. The realization of these improvements that are based on technological advancements can only be fully realized if the treatment cascade is also strengthened. Rapid tests are needed not only to improve care for tuberculosis patients, but also to facilitate rational use of new drug regimens and a more accurate understanding of the burden of drug-resistant tuberculosis at the global and national levels.
NEED FOR A TUBERCULOSIS RELATIONAL SEQUENCING DATA PLATFORM
As molecular technologies improve and become increasingly affordable and accessible to the global community, the understanding of the correlation of genotypic markers with phenotypic drug resistance, and ultimately patient outcomes, must keep pace. The majority of drug resistance in Mtb results from chromosomal mutations in genetic loci associated with the drug target, enzymes modifying the target or drug, or proteins needed for conversion of prodrugs to their active form [14]. Although numerous studies have contributed to the current understanding of the genetic basis of resistance in Mtb, most of these studies have examined a limited number of specific loci, and gaps in our knowledge remain, especially for correlating genotypic with phenotypic data [15–17]. We urgently need to advance our understanding of the genetic basis for antimicrobial resistance in Mtb. This pathway leads to the development of rapid molecular tests that can facilitate clinical decisions and guide the selection of treatment regimens.
One powerful and rapidly evolving molecular technology is next-generation sequencing, which provides a breakthrough in whole-genome sequencing (WGS) of Mtb [18, 19]. Increasingly, researchers are reporting results from WGS analysis of diverse sets of drug-resistant and -susceptible isolates of Mtb in correlation with phenotypic results [20–22]. These studies are valuable for improving our as-yet incomplete understanding of drug resistance, which includes questions surrounding cross-resistance, heteroresistance, additive effects of compensatory mechanisms of resistance, and discrepant analysis. However, the massive quantities of data produced by Mtb researchers using next-generation sequencing currently are stored in disparate data sets, thereby limiting the potential for integrating these data into precise predictors of resistance that could be incorporated into new diagnostic tools. Now is the time to meet this need through the development of a globally representative data-sharing platform for Mtb. This platform would not seek to replace but rather to build on the efforts of other available databases including the Tuberculosis Drug Resistance Mutation Database [17], the Pathosystems Resource Integration System [23], and the MUBII-TB-DB [24] to provide a comprehensive, standardized, and curated resource for clinically relevant genetic mutations and the associated metadata (eg, geographic location, methodology, DST result) with ongoing updates. Similar to the well-established HIV Drug Resistance Database [25, 26], this platform would provide a centralized location for aggregation of data to better understand the correlation between genetic patterns, phenotypic DST results, and clinical outcomes. A data platform of this scope has not been developed for Mtb yet, which is a major barrier to understanding the relationship between Mtb genetic mutations and drug resistance. Overcoming this barrier aids in advancing new diagnostic methods rapidly and efficiently.
Recognizing this need, international partners including the Bill & Melinda Gates Foundation (BMGF), Critical Path Institute, the Foundation for Innovative New Diagnostics, the New Diagnostics Working Group, the US Centers for Disease Control and Prevention, the WHO, and the National Institute of Allergy and Infectious Diseases, under the auspices of the Critical Path to TB Drug Regimens (CPTR) Rapid DST (RDST) Consortium, have established a Platform Leadership Team for the development of a comprehensive new system, the Relational Sequencing Tuberculosis Data Platform (ReSeqTB). Funding for the development of ReSeqTB is provided by BMGF. The CPTR-RDST Consortium has focused on accelerating the development of 1 or more clinically useful in vitro rapid DST assays that can be approved by regulators and endorsed by the WHO. ReSeqTB supports this objective by providing a centralized and curated data platform with clinically relevant information. One of the primary uses of the data platform is to guide the development of new diagnostic tests for the rapid detection of drug-specific resistance, for drugs in current use and those in development, thereby facilitating clinical decisions such that patients receive the most effective therapy as quickly as possible. In addition, these data will aid in making the best use of our current molecular tests by providing clarity regarding interpretive criteria for resistance-associated mutations.
BUILDING THE ReSeqTB DATA PLATFORM
The development of an effective data-sharing platform will require data from multiple global contributors. Unlike the tuberculosis databases that have been compiled using data from peer-reviewed literature, ReSeqTB will serve as a centralized repository for the continuous collation, active management, and evidence-based validation of retrospective and prospective data on Mtb drug resistance correlations in a user-friendly standard format. The intent is to collect a large volume of quality-assured data that are globally representative to reveal potential geographic variations in predominant mutations, naturally occurring polymorphisms that may be lineage specific, and novel mutations potentially associated with specific clinical or programmatic practices such as use of standard treatment regimens.
Submissions to the data platform will be sought from academic, governmental, and nonprofit sector researchers as well as clinical laboratories, sponsors of clinical trials, and countries performing drug resistance surveys. Early potential contributors with existing data sources will be identified from a systematic review of scientific literature and publicized work on drug-resistant tuberculosis. Contributions to populate the data platform will include anonymized genotypic (ie, raw sequence files from WGS), correlated phenotypic DST results, and clinical outcomes. Associated minimal and expanded optional metadata will be linked to each submission. Established quality criteria will be monitored as data are received, with iterative feedback to submitters. In addition, data will be curated to provide high confidence in support of the predictive value of the genotypic data. Raw sequence files will be managed and analyzed using a validated pipeline for processing data to produce variant reports categorizing mutations detected after comparison to a reference sequence. Prior to entry of the processed data into an investigational database, variant reports, along with correlated phenotypic and clinical data, will be subjected to robust quality checking and validated using parameters determined by an external panel of subject matter experts. All data will be formatted utilizing Clinical Data Interchange Standards Consortium open data standards for Tuberculosis Therapeutic Area Supplement (version, 1.0, released 29 June 2012) [27]. These data standards are intended to organize the tabulated datasets for the consistent presentation that is necessary for applying for regulatory approval of in vitro diagnostic devices. Ultimately, the criteria for clinical validation of mutations will be established by an internationally recognized external panel, a periodic process for data review will be initiated, and the predictive value of mutations for specific drug resistance of Mtb will be ascertained, regularly updated, and published.
Successful implementation of ReSeqTB requires expertise in database architecture and systems design to ensure quality and security of information and assurance that the standards of law, policy, patient privacy, and intellectual property are considered and addressed. Sufficient capacity for data storage is of utmost concern. A phased approach for the data platform will ensure expandability as the number of whole-genome sequences of Mtb isolates is expected to increase constantly over the next 3–5 years. A security audit of the data sharing platform will be conducted to ensure compliance with regulatory standards and the use of information-security best practices. Additionally, a set of commonly used analytical tools, as recommended by external subject matter experts, will be accumulated over time. User feedback mechanisms will be incorporated into the data platform framework to allow users to provide recommendations about tools and other improvements for future enhancements.
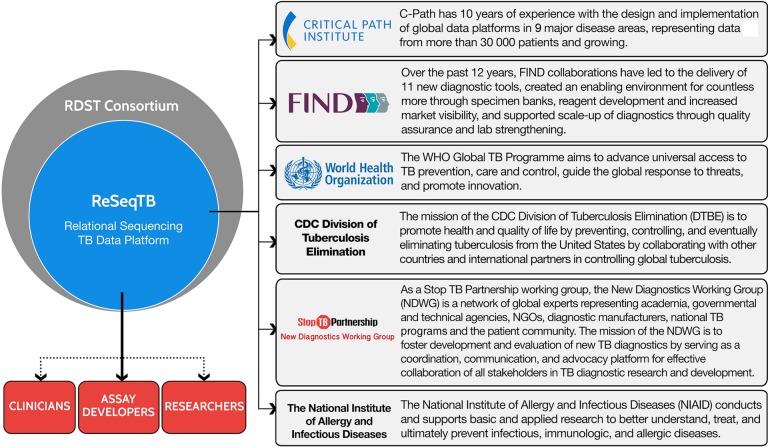
Efficient governance of this complex effort is critical for success and, as described in Figure 1, involves a collaborative partnership consisting of the aforementioned key stakeholders. Although a number of potential logistical obstacles to data sharing exist, including limitations to local data transfer and contributor time and costs in providing data submissions, the global aim of the initiative necessitates involvement of a diverse array of subject-matter experts providing worldwide representation.
Figure 1.
ReSeqTB Collaborative Partnership. The partners comprising the collaborative partnership serve as the group responsible for establishing the purpose and scope of the data platform and the initial platform design. Additionally, representatives from each partner serve as the official decision-making body for the effort. A number of potential end users of ReSeqTB are projected and include researchers, clinicians, assay developers, ministries of health, and national tuberculosis programs. The data housed within the platform could be used for many purposes including writing clinical guidelines, developing diagnostic tests, and investigating drug resistance. Abbreviations: CDC, Centers for Disease Control and Prevention; FIND, Foundation for Innovative New Diagnostics; RDST, Rapid Drug Susceptibility Testing Consortium; TB, tuberculosis; WHO, World Health Organization.
ENCOURAGING DATA CONTRIBUTIONS AND USE
Meeting the goals for ReSeqTB requires a partnership driven by global collaboration. To establish these partnerships, the communication strategies must ensure that potential contributors who could provide data from research studies, clinical trials, clinical laboratory services, or country-level drug resistance surveys are knowledgeable of the policies for data use and access. A key milestone is early access to data from ReSeqTB for use by RDST Consortium members. Data authorized by the contributors to be shared outside of the RDST Consortium will be made available to a broader community of nonmember research participants and other stakeholders.
The ReSeqTB data platform is governed by a robust data policy that meets international ethical standards of research involving human subjects and is informed by guiding principles committed to inclusivity, stewardship, collaboration, respect, and common good aimed at ensuring that access is maximized for researchers and other stakeholders who can directly benefit from the data. This means that access is fair and respectful of data users and contributors through strong security and privacy protections, data quality and integrity, and early access for data contributors.
A tiered access approach will be adopted to incentivize data contributions, and still allow for broader access to the scientific community to support research and innovation. Data will be accessible under specific data use agreements. Data contributors will always be able to access and own their data. Accessibility of the data is key and will be managed via a data sharing process whereby interested users will request access for a documented purpose that is scientifically relevant and consistent with the principles governing the data platform initiative. Upon review and approval of data access, applicants must accept the terms and conditions, including a requirement to reference the ReSeqTB initiative and a key limitation of patent exclusion. A fair and reasonable approach to data access is necessary to accumulate the large number of data contributions key for supporting and expanding this resource to ensure global representation, rapid diagnostic development, and improved understanding of the burden of national and global resistance.
A primary incentive for organizations and individual researchers to contribute data includes early access to the operational, fully populated ReSeqTB data platform for use as a research and development tool for diagnostics. In addition to the benefits of early access to data, researchers could also benefit from collaborative research opportunities and the potential for joint publications should those opportunities arise through the RDST Consortium. At the national level, ministries of health will benefit from having accurate data to make better-informed policy decisions for the diagnosis and treatment of tuberculosis, while eventually clinicians will be aided in the development of targeted and individualized treatment plans for patients. ReSeqTB represents a remarkable opportunity for the members of the global tuberculosis community to collaborate in making this initiative successful.
DISCOVERING FUTURE OPPORTUNITIES AND ALLOWING FOR SUSTAINABILITY
This initiative provides an opportunity to advance scientific knowledge and benefit global health. Future opportunities include the recognition of new research areas that need additional investment, the development of increasingly sophisticated analytic tools as part of the database functionality for end users, and the development of a clinical interface utility. These enhancements could build motivation for data contribution or access and data usage. As described, implementation of the data platform will include capabilities for data analysis within the system consistent with the primary objective to inform diagnostic developers regarding which mutations are predictive of drug resistance. However, feedback will be assessed to project future needs for expanded analysis and bioinformatics support.
Assessment and clinical validation of the mutations derived from ReSeqTB will improve results interpretation for molecular diagnostics detecting drug resistance in Mtb irrespective of the platform. Improving results interpretation enhances a clinical use utility for ReSeqTB that could directly assist healthcare providers in understanding the output of molecular tests to recognize validated resistance mutations and mutations for which less information is known. In addition, output from the data platform could provide simple analyses that include the sensitivity and specificity of user-provided genetic mutations for the prediction of phenotypic drug resistance, including literature citations supporting the analysis. This resource could serve as a tool informing clinical decision making for effective individualized treatment regimens and be used independent of sequencing data by clinicians. To date, a globally representative standardized database of this type does not exist for Mtb and is needed. Without this functionality, laboratories and healthcare providers will have continued reliance on disparate locally derived databases that may have limitations regarding the number and types of drug resistance–associated mutations, thereby hampering data sharing in the scientific community because of the lack of standardization. Adding a clinical-use utility to the data sharing platform could aid the development of standard interpretive criteria and serve to drive global consensus in uptake of molecular diagnostics because universal rules for interpreting output are available. Additionally, this aspect of the data platform could support standards development for the capture of molecular data for global surveillance of drug resistance and reporting language for molecular diagnostics. Clinical utilization of ReSeqTB offers an opportunity to acknowledge these needs and facilitate engagement with partner organizations.
Of primary interest to key stakeholders and, ultimately, the global community is sustainability of a dynamic, well-curated, and actively managed database. Given the multiplicity of benefits, we anticipate that long-term sustainability of ReSeqTB will be secured through continued funding opportunities. The complexity of the database provides an opportunity for a myriad of potential end users and funder collaborations with the ultimate goal of improving patient outcomes and global health.
Notes
Disclaimer. The views expressed in this publication are those of the authors alone and do not necessarily represent the decisions or policies of the World Health Organization. Use of trade names is for identification only and does not constitute endorsement by the US Department of Health and Human Services, the US Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Financial support. This work was supported by the Bill & Melinda Gates Foundation to Foundation for Innovative New Diagnostics (FIND) (grant number OPP1115209) and the Critical Path Institute (grant number OPP1115887).
Supplement sponsorship. This article appears as part of the supplement “Advances in Tuberculosis Research: A Blueprint for Opportunities.” This article was sponsored by the Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and the Critical Path Institute.
Potential conflicts of interest. C. M. D. and D. L. D. are employed by FIND, a nonprofit organization that collaborates with industry partners for the development, evaluation, and demonstration of new diagnostic tests for poverty-related diseases. D. H., E. A., R. L., and M. S. are employed by the Critical Path Institute, a nonprofit organization that supports the Critical Path to TB Drug Regimens, which is a public–private partnership dedicated to delivering a safer, more efficacious, and faster-acting tuberculosis regimen by developing and promoting innovative regulatory science essential for supporting new combination drug development in collaboration with its partners across industry, academia, and government. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Global tuberculosis report 2014. Geneva, Switzerland: WHO, 2014. WHO/HTM/TB/2014.08. [Google Scholar]
- 2.International Union Against Tuberculosis and Lung Disease. Guidelines for clinical and operational management of drug-resistant tuberculosis. Paris, France: International Union Against Tuberculosis and Lung Disease, 2013. [Google Scholar]
- 3.Dheda K, Warren RM, Zumla A, Grobusch MP. Extensively drug-resistant tuberculosis: epidemiology and management challenges. Infect Dis Clin North Am 2010; 24:705–25. [DOI] [PubMed] [Google Scholar]
- 4.Gandhi NR, Weissman D, Moodley P, et al. Nosocomial transmission of extensively drug-resistant tuberculosis in a rural hospital in South Africa. J Infect Dis 2013; 207:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Shao L, Fan X, et al. Evolution and transmission patterns of extensively drug-resistant tuberculosis in China. Antimicrob Agents Chemother 2014; 59:818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah NS, Wright A, Bai GH, et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis 2007; 13:380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma SK, Mohan A. Multidrug-resistant tuberculosis: a menace that threatens to destabilize tuberculosis control. Chest 2006; 130:261–72. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization WHO endorses new rapid tuberculosis test—a major milestone for global TB diagnosis and care. Geneva, Switzerland: WHO, 2010. [Google Scholar]
- 9.World Health Organization Molecular line probe assays for rapid screening of patients at risk of multidrug-resistant tuberculosis (MDR-TB)—policy statement. Geneva, Switzerland: WHO, 2008. [Google Scholar]
- 10.Gessler D, Dye C, Farmer P, et al. Public health. A national tuberculosis archive. Science 2006; 311:1245–6. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva, Switzerland: WHO, 2014. [PubMed] [Google Scholar]
- 12.Cohen T, Jenkins HE, Lu C, McLaughlin M, Floyd K, Zignol M. On the spread and control of MDR-TB epidemics: an examination of trends in anti-tuberculosis drug resistance surveillance data. Drug Resist Updat 2014; 17:105–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. Global strategy and targets for tuberculosis prevention, care and control after 2015. Report by the Secretariat. Geneva, Switzerland: WHO, 2013. WHA65/2012/REC/3. [Google Scholar]
- 14.Smith T, Wolff KA, Nguyen L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol 2013; 374:53–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2011; 55:2032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heysell SK, Houpt ER. The future of molecular diagnostics for drug-resistant tuberculosis. Expert Rev Mol Diagn 2012; 12:395–405. [DOI] [PubMed] [Google Scholar]
- 17.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med 2009; 6:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koser CU, Ellington MJ, Cartwright EJ, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 2012; 8:e1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koser CU, Bryant JM, Becq J, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 2013; 369:290–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guio H, Tarazona D, Galarza M, Borda V, Curitomay R. Genome analysis of 17 extensively drug-resistant strains reveals new potential mutations for resistance. Genome Announc 2014; 2:e00759–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merker M, Kohl TA, Roetzer A, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One 2013; 8:e82551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outhred AC, Jelfs P, Suliman B, et al. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother 2014; 70:1198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wattam AR, Abraham D, Dalay O, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 2014; 42(database issue):D581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flandrois JP, Lina G, Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinformatics 2014; 15:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 2003; 31:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafer RW. Rationale and uses of a public HIV drug-resistance database. J Infect Dis 2006; 194(suppl 1):S51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Critical Path to TB Drug Regimens. Tuberculosis therapeutic area supplement to the study data tabulation model user guide, 2012. Available at: http://www.cdisc.org/therapeutic#tuberculosis. Accessed 15 July 2015.