Abstract
Laryngeal squamous cell carcinoma (LSCC) is one of the most common malignancies of the head and neck tumors Zhang et al., 2013 [1]). Previous studies have associated its occurrence with social activities, such as tobacco and alcohol consumption (Hashibe et al., 2007a [2]; Hashibe et al., 2007b [3]; Shangina et al., 2006 [4]). Here, we performed a genome-wide gene expression profiling in thirty-one patients positively diagnosed for LSCC, in order to investigate new targets involved in tumorigenesis.
Keywords: Laryngeal squamous cell carcinoma tumor, Microarray, Gene expression profile
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Sequencer or array type | Whole Human Genome Oligo Microarray chips (Agilent, G4112F, USA) |
| Data format | Raw: CEL files, normalized data: SOFT, MINIML and TXT |
| Experimental factors | Twenty nine samples of laryngeal squamous cell carcinoma tumor vs. thirteen adjacent non-neoplastic tissue |
| Experimental features | We performed a transcriptome analysis in 31 LSCC patients in order to identify new targets involved in tumorigenesis. |
| Consent | Informed consent was obtained from all patients included in the study. |
| Sample source location | Ribeirao Preto, Sao Paulo, Brazil |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Study population and clinical data
All the samples were collected from patients undergoing surgical ablation of larynx squamous cell carcinoma at the Head and Neck Surgery Division of the Department of Ophthalmology, Otorhinolaryngology and Head & Neck of Ribeirao Preto Medical School, USP (Brazil). Inclusion criteria were: histopathological diagnosis of LSCC, elective surgeries for LSCC in patients without previous treatments and patients' allowance to donate part of their tumor for genetic studies. Exclusion criteria were: doubtful diagnosis of LSCC, unavailable post-surgical follow-up, patients without complete clinical data or signed agreement for collection of samples.
A total of twenty nine LSCC tumor samples and thirteen adjacent non-neoplastic tissues proceed for the transcriptome analysis. Clinical information and TNM staging classification of the LSCC patients can be found in Table 1 and Supplemental Table 1. In our study most of the patients were male (96.8%), all of them were smokers and alcoholic, including both current and former. Thirty five percent of the tumors were originated from glottis, followed by larynx (29.0%) and supraglottis (22.6%). Patients were further classified according to the TNM system: 48.4% of the patients were in early staging and 51.6% in late staging. Both staging groups showed similar proportions of tumor relapse, cured and dead patients. This study was approved by the Ethics Committee of Ribeirao Preto Medical School, University of Sao Paulo (USP) (Proc. No. 9371/2003) and signed informed consent was obtained from all patients.
Table 1.
Clinical information of LSCC patients.
| Characteristics | Staging |
Total (n = 31) | |
|---|---|---|---|
| Early (n = 15) | Late (n = 16) | ||
| Mean ages | |||
| Year (min–max) | 60 (40–78) | 62.2 (49–83) | 61.1 |
| Gender | |||
| Male | 14 (93.3%) | 16 (100%) | 30 (96.8%) |
| Female | 1 (7.7%) | 0 (0%) | 1 (3.2%) |
| Smoking (%) | |||
| Smoker | 11 (73.3%) | 6 (37.5%) | 17 (54.8%) |
| Former smoker | 4 (26.7%) | 10 (62.5%) | 14 (45.2%) |
| Non-smoker | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Alcoholism (%) | |||
| Alcoholic | 10 (66.7%) | 7 (43.75%) | 17 (54.8%) |
| Former alcoholic | 5 (33.3%) | 9 (56.25%) | 14 (45.2%) |
| Non-alcoholic | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Tumor site | |||
| Glottis | 6 (40%0) | 5 (31.3%) | 11 (35.5%) |
| Larynx | 3 (20.0%) | 6 (37.5%) | 9 (29.0%) |
| Supraglottis | 3 (20.0%) | 4 (25.0%) | 7 (22.6%) |
| Subglottis & oropharynx | 1 (6.7%) | 0 (0%) | 1 (3.2%) |
| Glottis & supraglottis | 0 (0%) | 1 (6.3%) | 1 (3.2%) |
| Laryngopharynx | 1 (6.7%) | 0 (0%) | 1 (3.2%) |
| Epiglottis | 1 (6.7%) | 0 (0%) | 1 (3.2%) |
| Tumor relapse | 4 (26.7%) | 4 (25.0%) | 8 (25.8%) |
| Metastasis | 1 (6.7%) | 2 (12.5%) | 3 (9.7%) |
| Cured? | |||
| Yes | 7 (46.7%) | 9 (56.3%) | 16 (51.6%) |
| No | 8 (53.3%) | 7 (43.8%) | 15 (48.4%) |
| Patient status | |||
| Alive | 9 (60.0%) | 10 (62.5%) | 19 (61.3%) |
| Dead | 6 (40.0%) | 6 (37.5%) | 12 (38.7%) |
| Follow-up | |||
| Month average | 41 | 46 | 44 |
2.2. Microarray experiments
After LSCC histopathological confirmation and microdissection of the tumors from their non-neoplastic adjacent tissue, the samples were store in liquid nitrogen. Total RNA was extracted with TRIzol (Life Technologies, USA). After extraction, the RNA was purified with RNeasy Kit (Qiagen, USA) and quantified with Nanodrop spectrophotometer (Thermo Scientific, USA). Its quality was evaluated by 1.5% agarose gel electrophoresis (28S and 18S ribosomal RNA detection).
For the microarray analysis it was used the Whole Human Genome Oligo Microarray kit 4 × 44K (Agilent, G4112F, USA). cDNA was hybridized to the microarray chip in the Fluidics Station 450 system (Affymetrix, USA), using the Quick Amp Labeling one-color kit (Agilent, USA). As an internal mRNA control, the One Color RNA Spike-In Kit was used (Agilent, USA). The arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 9.5.3.1) was used to analyze acquired array images.
2.3. Quality control and signal pre-processing
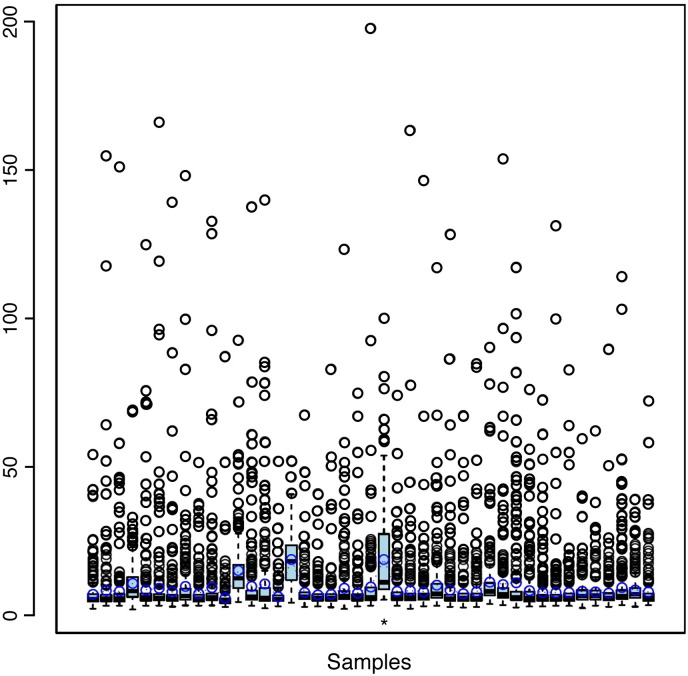
Plain text files were loaded and processed into R with the bioconductor package Agi4 × 44Pre-Process [5]. The annotation package hgug4112a.db [6] was used to assign gene information to each probe. The CV.rep.probes function was used to estimate the percent of coefficient of variation (% CV) for replicated non-control probes, inferring the reproducibility of the arrays (Fig. 1). The gProcessedSignal and the gBGUsed values of each array were loaded as foreground and background signals, respectively into an RGList object. Next, those arrays were renamed according to each sample clinical information. The following flags were used to filter probes: Control, Well Above BG, Is Found, Well Above NEG CTRLS, Is Saturated, Population Outlier, and Non Uniform Outlier. Additionally probe signals with at least 90% of good features in an experimental condition were selected. Logarithmic transformation of base 2 was applied for high quality probes. Quality control was accessed with package arrayQualityMetrics [7]. Outlier detection did not identify experimental problems. It was performed by looking for arrays (a and b), which the sum (S) of the distances (d) to all other arrays, Sa = Σb dab was exceptionally large; calculating the Kolmogorov–Smirnov statistic (Ka) between each array and the pooled data distribution; and computing Hoeffding's statistic (Da) on the joint distribution of A and M for each array (M = log2(I1) − log2(I2); A = 1/2 (log2(I1) + log2(I2)), where I1 is the intensity of the array studied, and I2 is the intensity of a “pseudo”-array that consists of the median across arrays).
Fig. 1.
Boxplot shows the percent of the coefficient of variation for non-control probes to each array. The asterisk represents the technical replicate array.
2.4. Normalization
Stagewise normalization is recommended when data include technical and biological replicates producing a better median chip [8]. Normalization steps were done with the R bioconductor package limma [9]. Technical replicates were normalized between arrays using the smooth function “cyclicloess”. Then, quantile normalization methods were applied, within groups and subsequently between all arrays. Each set of replicated non-control probes has been collapsed into a single value computed as the median of the probes intensities belonging to the same set.
2.5. Differentially expressed genes and pathways analysis
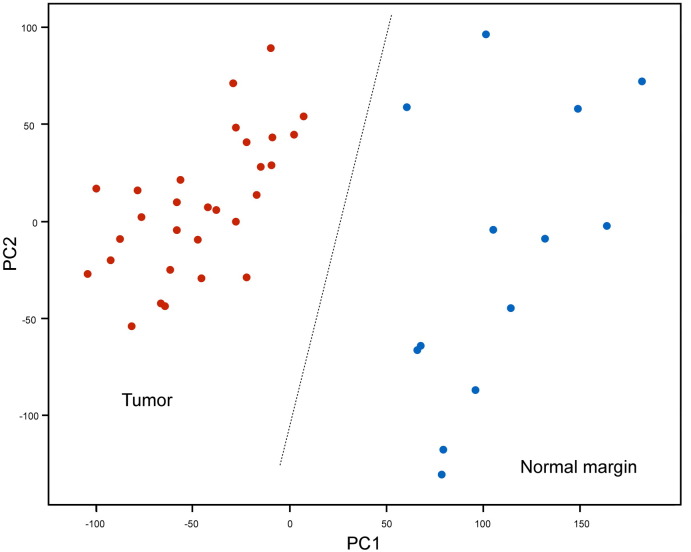
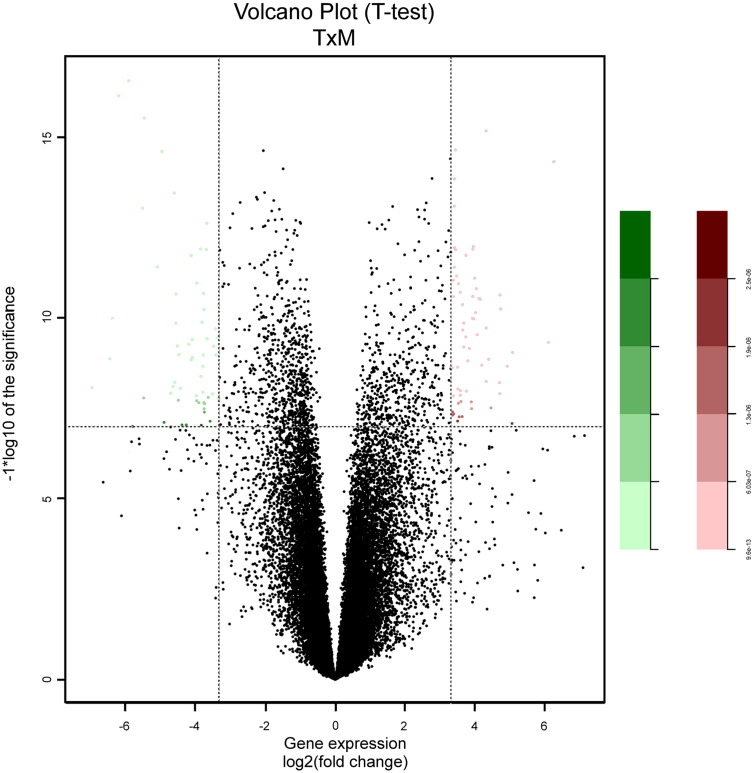
Principal component analysis (PCA) was applied to classify samples by tumor and adjacent non-neoplastic tissue as demonstrated on Fig. 2. We compared the gene expression between tumor and adjacent non-neoplastic tissue using a non-paired-T test analysis. False discovery rate (FDR) adjusted p-value < 1e − 07 and fold change (FC) > 10 were accepted to consider genes to be differentially expressed, identifying a total of 81 probes above this cut-off (Fig. 3). Then we investigated the biological process of upregulated and downregulated genes in tumor compared to normal samples with Web-based Gene Set Analysis Toolkit (WebGestalt) [10]. P-value was calculated using hypergeometric distribution and FDR as multiple hypothesis test correction method. The cut-off was set at < 0.05. Tissue morphogenesis (GO: 0009888) and differentiation (GO: 0032332) are the main biological process in upregulated genes while cytokines and growth factor genes are highly expressed in adjacent non-neoplastic patients, suggesting a role on LSCC tumorigenesis.
Fig. 2.
Scatter plot representing the principal component analysis from expression arrays. The dots are colored by tumor (red) and adjacent non-neoplasic tissue samples (blue).
Fig. 3.
Volcano plot based on log2 fold-change against − log10 (p-value) showing genes overexpressed in tumor samples (green dots) and adjacent non-neoplasic tissue samples (red dots).
3. Discussion
We describe detailed technical methods to reproduce the analysis of Whole Human Genome Oligo Microarray kit (Agilent, G4112F, USA) from twenty-nine LSCC tumor samples and thirteen adjacent non-neoplastic tissues. LSCC is the second most common malignancies of the head and neck tumors [1], and this high frequency may be associated with the lifestyle of patients [2], [3], [4]. The high quality gene expression data are concordant with important clinical parameters, demonstrating the relevant expression profile signature of differentially expressed genes. This data can contribute to future investigations examining molecular changes that promote LSCC tumorigenesis.
The following is the supplementary data related to this article.
Tumor sample description.
Acknowledgments
We thank Amélia Goes de Araújo and Patrícia Vianna Bonini Palma for the technical support at Flow Cytometry Core at National Institute of Science and Technology in Stem Cell and Cell Therapy for the technical support. This work was supported by grant #559809/2009-3 and #2012/00588, CNPq/GENOPROT and grant #13/08135-2, Sao Paulo Research Foundation (FAPESP).
References
- 1.Zhang S.Y., Lu Z.M., Luo X.N., Chen L.S., Ge P.J., Song X.H. Retrospective analysis of prognostic factors in 205 patients with laryngeal squamous cell carcinoma who underwent surgical treatment. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashibe M., Boffetta P., Zaridze D., Shangina O., Szeszenia-Dabrowska N., Mates D. Contribution of tobacco and alcohol to the high rates of squamous cell carcinoma of the supraglottis and glottis in Central Europe. Am. J. Epidemiol. 2007;165(7):814–820. doi: 10.1093/aje/kwk066. [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M., Brennan P., Benhamou S., Castellsague X., Chen C., Curado M.P. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J. Natl. Cancer Inst. 2007;99(10):777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 4.Shangina O., Brennan P., Szeszenia-Dabrowska N., Mates D., Fabianova E., Fletcher T. Occupational exposure and laryngeal and hypopharyngeal cancer risk in central and eastern Europe. Am. J. Epidemiol. 2006;164(4):367–375. doi: 10.1093/aje/kwj208. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Romero P. Agi4 × 44PreProcess: PreProcessing of Agilent 4 × 44 array data. 1.22.0 ed.
- 6.Carlson M. hgug4112a.db: Agilent “Human Genome, Whole” annotation data (chip hgug4112a). 3.0.0 ed.
- 7.Kauffmann A., Gentleman R., Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25(3):415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaratunga D., Cabrera J. first ed. Hoboken; New Jersey: 2004. Exploration and Analysis of DNA Microarray and Protein Array Data. [Google Scholar]
- 9.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741-8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tumor sample description.