Abstract
WD repeat domain 5 (WDR5) plays an important role in various biological functions through the epigenetic regulation of gene transcription (Wysocka et al., 2005 [1]; Sandstrom et al., 2014[2]; Ang et al., 2011[3]). Recently, our study found that WDR5 was upregulated in bladder cancer tissues, promoted bladder cancer cell proliferation, self-renewal and chemoresistance to cisplatin in bladder cancer cells in vitro, and tumor growth in vivo (Chen et al., 2015). To gain a molecular understanding of the role of WDR5 in promoting bladder cancer, we performed a genome-wide analysis on WDR5 knockdown by microarray gene expression profiling. Here we provide detailed experimental methods and analysis for the microarray data, which have been deposited into Gene Expression Omnibus (GEO): GSE59132.
Keywords: WDR5, Bladder cancer, Microarrays, Transcriptional profiling
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/human bladder cancer cell line/UM-UC-3(CRL-1749, ATCC) |
| Sex | Male |
| Sequencer or array type | Affymetrix PrimeView Human Gene Expression Array |
| Data format | Raw data: CEL files |
| Experimental factors | Two different siRNA knocked down WDR5 vs. control siRNA in human bladder cancer |
| Experimental features | Microarray gene expression profiling to identify transcripts that are regulated by WDR5 |
| Consent | None necessary, data are publicly available |
| Sample source location | Guangzhou, China |
1. Direct link to deposited data
The deposited data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59132.
2. Experimental design, materials and methods
2.1. Cell culture
The human bladder cancer cell line UM-UC-3 was obtained from ATCC (CRL-1749, ATCC), and cultured in DMEM (Gibco, Shanghai, China). All media were supplemented with 10% FBS (Shanghai ExCell Biology, China) and 1% penicillin/streptomycin. Cells were grown in a humidified atmosphere of 5% CO2 at 37 °C.
2.2. RNA interference
WDR5 is a core component of the MLL/SET1 complexes and mediates H3K4 methylation, which is correlated with gene activation [1]. The present study reveals that WDR5 plays a critical role in undifferentiated progenitor cells of the baboon subventricularzone [2] and embryonic stem cell self-renewal [3]. We identified that WDR5 was upregulated in bladder cancer tissues, and elevated WDR5 protein levels positively correlated with advanced tumor stage and poor survival [4]. To study the role of WDR5 in bladder cancer, we knocked down WDR5 by siRNA. SiRNA oligos that targeted WDR5 (1-GCUGGGAAUAUCCGAUGUATT, 2-GCUCAGAGGAUAACCUUGUTT, 3-CCCAGUCCAACCUUAUUGUTT) were purchased from GenePharma (Shanghai, China). SiRNA transfections were performed with 75 nM siRNA and Lipofectamine RNAimax (Life Technologies) following the manufacturer's instructions. WDR5 was remarkably downregulated in UM-UC-3 cells by the siRNA si-WDR5-2 or si-WDR5-3, but not si-WDR5-1 [4]. To gain a molecular understanding of the mechanisms of WDR5 in bladder cancer cells, UM-UC-3 cells were transfected with a control siRNA, si-WDR5-2 or si-WDR5-3 siRNA respectively for 48 h in three independent experiments.
2.3. RNA isolation and qRT-PCR
We extracted total RNA from nine samples (three biological replicates) of UM-UC-3 cells that had been transfected with the control siRNA or with two siRNA targeting WDR5 expression using TRIzol reagent (Invitrogen) and further purified the RNA using Qiagen RNeasy Mini Kit according to the manufacturer's protocol. The RNA quality was assessed by formaldehyde agarose gel electrophoresis and was quantitated by NanoDrop spectrophotometer. One microgram of total RNA of each sample was used for reverse transcription (PrimeScript RT-PCR kit, TaKaRa Biotechnology, Dalian, China) and was further performed qRT-PCR to detect the expression of WDR5. We confirmed that the expression of WDR5 was obvious downregulated in transfection with si-WDR5-2 or si-WDR5-3 siRNA compared with control siRNA in three biological replicates (Fig. 1). Therefore, we equally mixed the RNA of three biological replicates samples as a sample to further perform the microarray analysis.
Fig. 1.
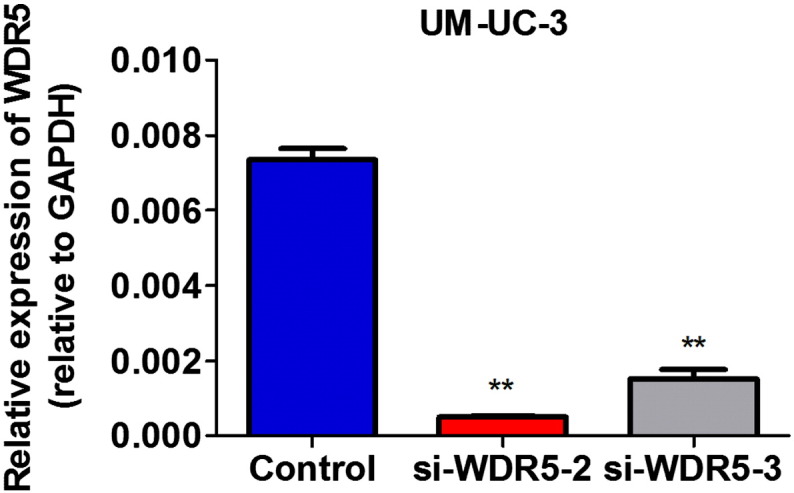
Efficiency of WDR5 knockdown in UM-UC-3 cells by siRNA was verified by qRT-PCR. The results are presented as the means ± SD of values obtained in three independent experiments. Statistical significance was calculated using the ANOVA analysis. **p < 0.01.
2.4. Microarray and quality control
The Affymetrix PrimeView Human Gene Expression Array was used in this study and performed by CapitalBio Corporation (Beijing, China). 100 ng of total RNA was used to synthesize double-stranded cDNA. The bio-tagged cRNA was produced by using the MessageAmp™ Premier RNA Amplification Kit and were fragmented to strands of 35–200 bases in length according to the protocols from Affymetrix. The fragmented cRNA was hybridized to PrimeView Human Gene Expression Array containing about 36,000 transcripts. Hybridization was performed at 45 °C with rotation for 16 h (Affymetrix GeneChip Hybridization Oven 640). The GeneChip arrays were washed and then stained (streptavidin-phycoerythrin) on an Affymetrix Fluidics Station 450 followed by scanning on a GeneChip Scanner 3000.
The scanned images were assessed and analyzed to generate raw data files saved as CEL files using Affymetrix GeneChip Operating software (GCOS 1.4). The quality of each CEL file was assessed using Affymetrix Expression Console Software according to the Affymetrix standard protocol. In order to monitor labeling and hybridization quality, we used polyA-control RNAs (Lys, Phe, Thr and Dap) and bacterial spike-in controls (BioB, BioC, BioD and Cre), respectively.
2.5. Data normalization and analysis
All samples were normalized through the Robust Multi-array Average (RMA) using DNA-chip analyzer (dChip). Differentially expressed genes were first identified with fold change of ≥ 2 or ≤ 0.5 in the group of control siRNA comprised with si-WDR5-2 or si-WDR5-3 siRNA. To further investigate the target genes of WDR5, differentially expressed genes were identified with both fold change of ≥ 1.5 and at least one group fold change of ≥ 2 in both siRNA compared with control siRNA. There were 136 upregulated genes and 42 downregulated genes in both siRNAs of WDR5 compared with control siRNA [4]. GO analysis and pathway analysis were performed by using Molecule Annotation System (MAS 3.0, CapitalBio Corporation, Beijing, China).The WDR5 regulated genes were mainly associated with regulation of transcription, cell cycle, anti-apoptosis and cell adhesion (Fig. 2).
Fig. 2.
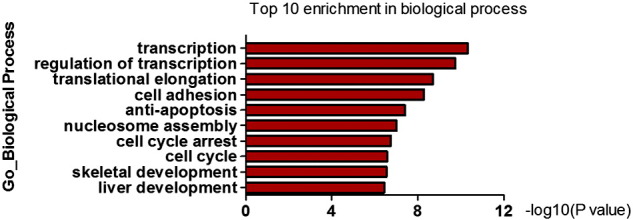
Gene ontology analysis of WDR5 regulated genes in the biological process of bladder cancer.
2.6. Validation of microarray data by qRT-PCR
In order to validate the microarray data, qRT-PCR was performed to detect the differentially expressed genes in UM-UC-3. As shown in Fig. 3, these genes were well validated and the fold changes of them in qRT-PCR were more obvious than microarray.
Fig. 3.
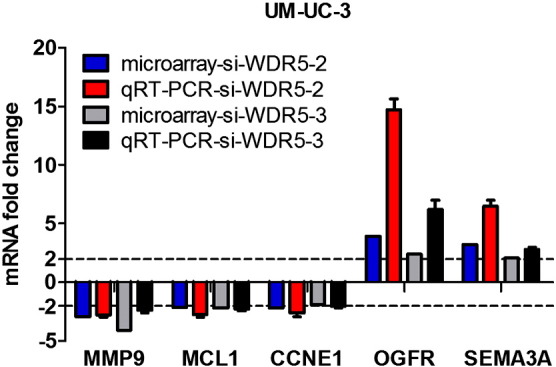
Validation of microarray data by qRT-PCR. Five transcripts were quantified by qRT-PCR both in control and WDR5 siRNA in UM-UC-3 cells. The resulting expression fold change is plotted against the expression fold change obtained from the microarray data.
3. Discussion
We described here a dataset composed of microarray gene expression profiling of WDR5 regulated genes in the human bladder cancer UM-UC-3 cells. With this experiment, we were able to demonstrate that WDR5 directly controls and coordinates the expression of a large number of genes involved in cell cycle and anti-apoptosis [4]. We believe that this dataset would be particularly valuable for investigating and underlying molecular mechanisms of WDR5 in bladder cancer.
Conflict of interest
The authors declare no conflict of interests.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant Nos. U1301221, 81472384, 81372729, 81372883, 81272808, 81172431, 81101935), and Guangdong Province Natural Scientific Foundation (Grant Nos. S2013020012671, 07117336, 10151008901000024), Specialized Research Fund for the Doctoral Program of Higher Education (for Tianxin Lin, 20130171110073), Sun Yat-Sen University Clinical Research 5010 Program (Grant No. 2007018), Elite Young Scholars Program of Sun Yat-sen Memorial Hospital (for Tianxin Lin, J201401), Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, Sun-Yat-Sen University, and Grant [2013]163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology.
Contributor Information
Tianxin Lin, Email: tianxinl@sina.com.
Jian Huang, Email: urolhj@sina.com.
References
- 1.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roederr R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 2.Sandstrom R.S., Foret M.R., Grow D.A., Haugen E., Rhodes C.T., Cardona A.E., Phelix C.F., Wang Y., Berger M.S., Lin C.H. Epigenetic regulation by chromatin activation mark H3K4me3 in primate progenitor cells within adult neurogenic niche. Sci. Rep. 2014;4:5371. doi: 10.1038/srep05371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang Y.S., Tsai S.Y., Lee D.F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., Wang J., Rendl M., Bernstein E., Schaniel C., Lemischka I.R. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Xie W., Gu P., Cai Q., Wang B B., Xie Y., Dong W., He W., Zhong G., Lin L., Huang J. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci. Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]


