Abstract
The aim of our study was to analyze the consequences of non-synonymous SNPs in Slc11a2 gene using bioinformatic tools. There is a current need of efficient bioinformatic tools for in-depth analysis of data generated by the next generation sequencing technologies. SNPs are known to play an imperative role in understanding the genetic basis of many genetic diseases. Slc11a2 is one of the major metal transporter families in mammals and plays a critical role in host defenses. In this study, we performed a comprehensive analysis of the impact of all non-synonymous SNPs in this gene using multiple tools like SIFT, PROVEAN, I-Mutant and PANTHER. Among the total 124 SNPs obtained from amplicon sequencing of Slc11a2 gene by Ion Torrent PGM involving 10 individuals of Gir cattle and Murrah buffalo each, we found 22 non-synonymous. Comparing the prediction of these 4 methods, 5 nsSNPs (G369R, Y374C, A377V, Q385H and N492S) were identified as deleterious. In addition, while tested out for polar interactions with other amino acids in the protein, from above 5, Y374C, Q385H and N492S showed a change in interaction pattern and further confirmed by an increase in total energy after energy minimizations in case of mutant protein compared to the native.
Abbreviations: ATM, ataxia telangiectasia mutated; BRAF, B-Raf; CFTR, cystic fibrosis transmembrane conductance regulator; GalNAc-T1, N-acetylgalactosaminyltransferase 1; GATK, Genome Analysis Tool Kit; HBB, hemoglobin beta; HMM, Hidden Markov Model; IGF1R, insulin-like growth factor 1 receptor; NCBI, National Center for Biotechnology Information; PANTHER, Protein Analysis Through Evolutionary Relationships; PolyPhen, Polymorphism Phenotyping; PROVEAN, Protein Variation Effect Analyzer; RMSD, root-mean-square deviation; SIFT, sorting intolerant from tolerant; Slc11a2, solute carrier family 11 member 2; SNP, single nucleotide polymorphism; TMDs, transmembrane domains; TYRP1, tyrosinase-related protein 1
Keywords: Non-synonymous, PANTHER, Ion torrent PGM, SIFT, Protein
Highlights
-
•
22 nsSNPs were predicted to decrease the stability of protein based on I-Mutant.
-
•
From these SNPs, 5 was identified as deleterious by SIFT, PROVEAN, and PANTHER.
-
•
Y374C, Q385H and N492S were found to be damaging.
1. Introduction
Single-nucleotide polymorphisms (SNPs) play a major role in understanding the genetic basis of many complex diseases and it is still a major challenge to identify the functional SNPs in a disease-related gene. Non-synonymous SNPs (nsSNPs) cause changes in the amino acid residues and are important factors contributing to the functional diversity of the encoded proteins [1]. Non-synonymous SNPs affect gene regulation by altering DNA and transcriptional binding factors and maintaining the structural integrity of cells and tissues. Also, nsSNPs affect the functional roles of proteins involved in signal transduction of visual, hormonal, and other stimulants [2], [3].
The advents in computational algorithms are useful for predicting the impact of amino-acid substitutions on protein structure and function. The computational tools like SIFT, PolyPhen, I-Mutant, PANTHER are used nowadays for detecting impact of amino acid substitution especially in coding exonic region [2], [4], [5]. Earlier reports have shown that the computational tools precisely predicted the consequences of nsSNPs associated with genes such as IGF1R[6], ATM[7], HBB[8], CFTR[9], BRAF[10], TYRP1[11], and GalNAc-T1.[12].
The mammalian Slc11a1 and Slc11a2 proteins are a large family of secondary metal transporters. Slc11a1 and Slc11a2 function as pH-dependent divalent cation transporters that play a critical role in host defenses against infections and in Fe2 + homeostasis respectively [13]. Slc11a1 is expressed primarily in macrophages and Slc11a2 has a much broader range of tissue expression. The mechanism by which these proteins exert their antimicrobial activity is uncertain. However observation that these proteins transport Fe2 + down a proton gradient suggests that their antimicrobial activity is due to the removal of Fe2 + (or other divalent metals) from the acidic phagosome and bacterial death due to essential micronutrients starvation [14]. Slc11a2is a 90–100 kD transmembrane protein with intracellular N- and C-termini with an even number of 12 transmembrane domains. First ten TMDs constitute the main functional unit of this family of transporters and TMD1 is a highly conserved sequence motif (residues 384–403), in which alterations abrogate transport [15]. The Slc11a2 gene is comprised of 17 exons and spans more than 36 kb. It contains an additional 5′ exon and intron (exon and intron 1) and an additional 3′ exon (exon 17) and intron (intron 16).Slc11a2 proteins play a central role in iron homeostasis and transport is electrogenic caused by proton movement through the transporter(substrate-dependent and substrate-independent H + leak) [16], [17]. A loss-of-function mutation (G185R) is reported to cause very severe microcytic anemia in the mk mouse and in the Belgrade rat.In addition, a number of loss-of-function missense (R416C, G212V, delV114) and splicing mutations have been detected in the human Slc11a2 gene in patients suffering from hypochromic microcytic anemia with serum and liver iron overload [18], [19]. The aim of our study was to identify functional and structural impact of nsSNPs of Slc11a2. From amino acid sequence retrieved from NCBI, 3D model of this protein was constructed using RaptorX protein modeling tool and visualized in PyMOL. SNPs were inserted in the native sequence of protein and its consequences were checked using several computational tools.
2. Materials and methods
2.1. Variant calling
Genetic variation of the Slc11a2gene was analyzed from data obtained by sequencing of exonic regions of this innate immune gene which was studied to screen SNPs. Ten Bos taurus animals of Gir breed and ten Bubalus bubalis animals of Murrah breed were used for genomic DNA extraction (unpublished data). GATK software tools (version 2.8; http://www.broadinstitute.org) were used for genotype calling with recommended parameters. Genotypes were called by the GATK Unifiedgenotyper tool, and variants were filtered by depth 60.
2.2. Deleterious nsSNP found by the SIFT program
SIFT performs multiple alignments of a number of peptide sequences until a median conservation for the sequence is reached at the default of 3.0 and then it predicts whether substitution with any of the other amino acids is tolerated or deleterious for every position in the submitted sequence [20]. The SIFT prediction was given as a tolerance index (TI) score ranging from 0.0 to 1.0, which was the normalized probability that the amino acid change was tolerated. A nsSNP with a TI score of V0.05 was considered to be deleterious i.e. an amino acids with probabilities < 0.05 were predicted to be deleterious. We submitted the amino acid sequence of Slc11a2 along with nsSNPs with corresponding amino acid positions.
2.3. Validation and functional characterization predicted nsSNPs by PANTHER-cSNP
The functional validation of nsSNPs predicted by SIFT was analyzed by PANTHER (Protein Analysis Through Evolutionary Relationships;www.pantherdb.org/tools/csnp). This tool estimates the likelihood of a particular non-synonymous coding SNP to cause a functional impact on the protein using Hidden Markov Models (HMM) based modeling and evolutionary relationship. It calculates the subPSEC (substitution position-specific evolutionary conservation) score based on an alignment of evolutionarily related proteins [5]. The score of subPESC ≥− 3 was predicted as a less deleterious, while ≤− 3 was predicted as the deleterious effect. Amino acid sequence in FASTA format was uploaded.
2.4. Prediction of functional impact of nsSNPs
PROVEAN (Protein Variation Effect Analyzer) is a tool which predicts the impact of an amino acid substitution or indel on the biological function of a protein (http://provean.jcvi.org/index.php).This algorithm allows for the best balanced separation between the deleterious and neutral amino acids, based on a threshold. The score <− 2.5 indicates that the variant is deleterious and >− 2.5 score is considered as a neutral variant [21]. A query peptide sequence of Slc11a2 was provided in FASTA format to the PROVEAN server for predicting the functional impact of the SNPs.
2.5. Investigation of mutant protein stability by I-Mutant 2.0
I-Mutant2.0 (http://folding.biofold.org/cgi-bin/i-mutant2.0) is a Support Vector Machine-based web server for the automatic prediction of protein stability changes upon single-site mutations. The input FASTA sequence of protein along with the residues change was provided for analysis of DDG value (kcal/mol) [22]. Also the RI value (reliability index) was computed.
2.6. Modeling of native and mutant structure of Slc11a2
RaptorX, a protein structure prediction server,predicts 3D structures for protein sequences lacking close homologs in the Protein Data Bank (PDB).For given FASTA sequence RaptorX predicted its secondary and tertiary structures as well as solvent accessibility and disordered regions. RaptorX also calculates p-value for the relative global quality, GDT (global distance test) and uGDT (un-normalized GDT) for the absolute global quality, and RMSD for the absolute local quality of each residue in the model. The 3D structures were visualized by PyMOL (http://www.pymol.org/) which is an open source molecular isualization tool. Mutant model was also constructed using PyMoL tool.
2.7. Model quality & structure assessment and RMSD difference
Model quality was checked both of native and altered protein by Ramachandran plot using software RAMPAGE (mordred.bioc.cam.ac.uk/~rapper/rampage.php) which analyzed residue-by-residue geometry and overall structure geometry. PyMOL was used to locate nsSNPs on protein structure and for analyzing RMS deviation by superimposing both native and mutant structures. Amino acids at the position of SNPs were checked for polar interactions with other amino acids in the protein using PyMOL. In addition, total energy after energy minimization was calculated for each altered model using Swiss PDB viewer.
2.8. Binding site and ligand prediction
To find whether these identified nsSNPs are present on any epitope region or any protein binding region, we performed binding site prediction using RaptorX Binding and FT site server which predicted binding site regions in Slc11a2 protein.
3. Results
3.1. Variant calling
Upon variant calling, total 124 SNPs were observed in Slc11a2 gene (Supplementary Table 1). Among these SNPs, 22 (17.74%) and 74 (59.67%) were found to be non-synonymous and synonymous respectively. The remaining 28 (22.58%) were found to be in the non-coding region, 3 in UTR 5′region and 25 in UTR 3′ region.
3.2. Deleterious nsSNP found by the SIFT program
The SIFT identified 8 nsSNPs viz. I114T, G369R, Y374C, A377V, Q385H, M389V, N492S and V497M to be deleterious. Non-synonymous SNPs with SIFT prediction and SIFT score are shown in Table 1.
Table 1.
Functional validations of nsSNPs in Slc11a2 using SIFT, PANTHER-cSNP and PROVEAN.
| Amino acid change | SIFT prediction | SIFTscore | Pantherprediction | Pantherscore(subPSEC) | Pdeleterious | PROVEAN prediction |
PROVEAN score (cutoff = − 2.5) |
|---|---|---|---|---|---|---|---|
| V108I | Tolerated | 0.07 | – | Does not align to HMM | Does not align to HMM | Neutral | − 0.503 |
| I114T | Deleterious | 0.01 | – | Does not align to HMM | Does not align to HMM | Deleterious | − 3.318 |
| V334A | Tolerated | 0.86 | Tolerated | − 1.569 | 0.193 | Neutral | − 1.325 |
| T336K | Tolerated | 0.87 | Tolerated | − 1.268 | 0.150 | Neutral | − 0.385 |
| T343A | Tolerated | 1.00 | Tolerated | − 2.19479 | 0.30891 | Neutral | 0.404 |
| G369R | Deleterious | 0 | Deleterious | − 5.398 | 0.917 | Deleterious | − 7.858 |
| A371S | Tolerated | 0.12 | Tolerated | − 2.564 | 0.393 | Neutral | − 2.409 |
| Y374C | Deleterious | 0 | Deleterious | − 5.197 | 0.9 | Deleterious | − 8.419 |
| A377V | Deleterious | 0.01 | Deleterious | − 3.86405 | 0.70351 | Deleterious | − 3.835 |
| Q385H | Deleterious | 0 | Deleterious | − 4.237 | 0.776 | Deleterious | − 4.899 |
| M389V | Deleterious | 0.02 | Tolerated | − 2.744 | 0.437 | Deleterious | − 3.668 |
| R465Q | Tolerated | 0.39 | Tolerated | − 2.132 | 0.296 | Neutral | − 0.311 |
| W477L | Tolerated | 0.65 | Tolerated | − 1.024 | 0.122 | Neutral | − 2.225 |
| L484V | Tolerated | 0.55 | Tolerated | − 1.540 | 0.187 | Neutral | − 1.12 |
| S490F | Tolerated | 0.19 | Tolerated | − 1.973 | 0.264 | Deleterious | − 3.106 |
| N492S | Deleterious | 0 | Deleterious | − 4.673 | 0.842 | Deleterious | − 4.744 |
| V497M | Deleterious | 0.03 | Tolerated | − 2.734 | 0.434 | Neutral | − 1.549 |
| D502G | Tolerated | 0.34 | Tolerated | − 1.795 | 0.230 | Neutral | − 1.343 |
| V507A | Tolerated | 0.84 | Tolerated | − 1.980 | 0.265 | Neutral | 1.787 |
| V510M | Tolerated | 0.16 | Tolerated | − 2.502 | 0.378 | Neutral | − 1.494 |
| A512V | Tolerated | 0.96 | Tolerated | − 0.927 | 0.118 | Neutral | − 0.17 |
| V517I | Tolerated | 0.90 | Tolerated | − 1.028 | 0.122 | Neutral | − 0.231 |
3.3. Validation and functional characterization predicted nsSNPs by PANTHER-cSNP
The results of SIFT were further confirmed by investigating the effect of nsSNPs on protein function using HMM based PANTHER tool. The analysis of 22 non-synonymous mutations revealed that 5 SNPs (G369R, Y374C, A377V, Q385H and N492S) reflected a subPSEC score >−3, thus PANTHER classified them as deleterious. Remaining SNPs of Slc11a2 had a score <− 3 and were classified as tolerated. Non-synonymous SNPs along with PANTHER score are given in Table 1.
3.4. Prediction of functional impact of nsSNPs
Further confirmation of effect of nsSNPs on protein was done using PROVEAN tool which revealed 8 from 22 nsSNPs (I114T, G369R, Y374C, A377V, Q385H, M389V, S490F and N492S) to be deleterious. The higher the tolerance index is, the less functional impact a particular amino acid substitution is likely to have, and vice versa. Among the 22 nsSNPs, 8 (36.36%) were found to be deleterious, having a tolerance index score of ≤− 2.5 using PROVEAN tool (Table 1).
3.5. Investigation of mutant protein stability by I-Mutant 2.0
To add another layer of confirmation, we also analyzed effect of these nsSNPs using I-Mutant 2.0. It gave result in the form of effect of mutants on stability of protein with reliability index at pH 7.0 and temperature 25 °C. Here in our case, in Slc11a2, all 22 non-synonymous SNPs showed resulting decrease in stability of the protein. All 22 SNPs with reliability index and DDG value are given in Table2.
Table 2.
Investigation of mutant protein stability by I-Mutant 2.0.
| Protein symbol | Amino acid change | Amino acid position | Reliability index (RI) | DDG value (kcal/mol) | Stability prediction |
|---|---|---|---|---|---|
| Slc11a2 | V/I | 108 | 7 | 0.86 | Decrease |
| I/T | 114 | 3 | − 2.03 | Decrease | |
| V/A | 334 | 9 | − 2.13 | Decrease | |
| T/K | 336 | 4 | − 0.48 | Decrease | |
| T/A | 343 | 8 | − 2.05 | Decrease | |
| G/R | 369 | 2 | − 1.01 | Decrease | |
| A/S | 371 | 7 | − 0.81 | Decrease | |
| Y/C | 374 | 1 | 0.71 | Decrease | |
| A/V | 377 | 2 | 0.06 | Decrease | |
| Q/H | 385 | 3 | 0.27 | Decrease | |
| M/V | 389 | 6 | − 0.17 | Decrease | |
| R/Q | 465 | 8 | − 1.88 | Decrease | |
| W/L | 477 | 6 | 1.00 | Decrease | |
| L/V | 484 | 9 | − 1.07 | Decrease | |
| S/F | 490 | 0 | − 0.88 | Decrease | |
| N/S | 492 | 6 | − 0.88 | Decrease | |
| V/M | 497 | 8 | − 1.28 | Decrease | |
| D/G | 502 | 7 | − 2.14 | Decrease | |
| V/A | 507 | 9 | − 2.93 | Decrease | |
| V/M | 510 | 8 | − 2.69 | Decrease | |
| A/V | 512 | 1 | 0.76 | Decrease | |
| V/I | 517 | 9 | − 1.61 | Decrease |
3.5. Analysis of structural model of Slc11a2 protein
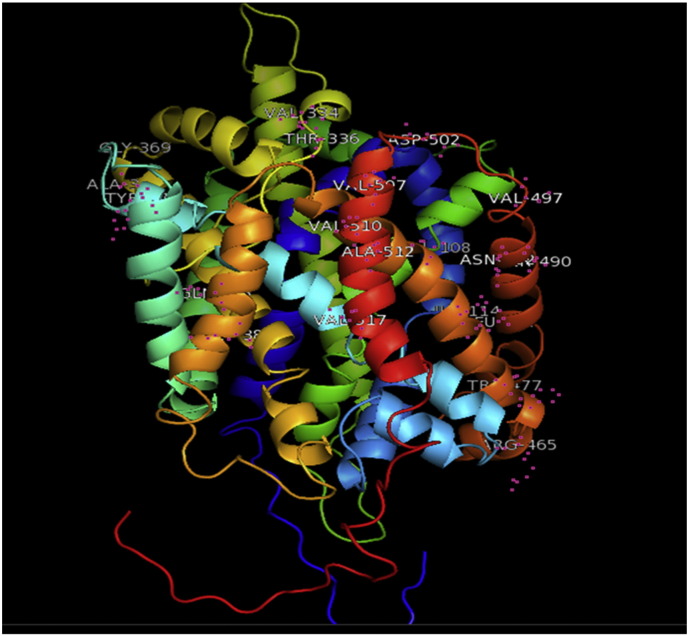
The 3D structure of native model generated through RaptorX was visualized using PyMoL. Slc11a2 is having 568 amino acid residues (Supplementary Fig. 1). From these 480(85%) residues were modeled and 65(11%) positions predicted as disordered. Secondary structures revealed 69% helix, 0% beta sheet and 30% loop structures. The solvent accessibility was divided into three states by 2 cut-off values: 10% and 42%. Value less than 10% was identified as buried, larger than 42% value was identified as exposed and if value was between 10% and 42% was identified as medium. Proportions of buried, medium and exposed regions in our protein were 62%, 21% and 15% respectively. Overall uGDT (GDT) value was 143(25%). The uGDT is the unnormalized GDT (global distance test) score. For a protein with > 100 residues, uGDT > 50 is a good indicator. For a protein with < 100 residues, GDT > 50 is a good indicator. If a model has acceptable uGDT (> 50) but lower GDT (< 50), it indicates that only a small portion of the model may be good. P-value is the likelihood of a predicted model being worse than the best of a set of randomly-generated models for this protein (or domain), so P-value evaluates the relative quality of a model. The smaller the p-value, the higher is the quality of the model. For alpha proteins, p-value less than 10− 3 is a good indicator. For manly beta proteins, p-value less than 10− 4 is a good indicator. For this model of Slc11a2, RaptorX predicted p-value of 4.55e − 07. Twenty two mutant models were generated in PyMOL (Supplementary Fig. 2, Supplementary Fig. 3, Supplementary Fig. 4, Supplementary Fig. 5, Supplementary Fig. 6, Supplementary Fig. 7, Supplementary Fig. 8, Supplementary Fig. 9, Supplementary Fig. 10, Supplementary Fig. 11, Supplementary Fig. 12, Supplementary Fig. 13, Supplementary Fig. 14, Supplementary Fig. 15, Supplementary Fig. 16, Supplementary Fig. 17, Supplementary Fig. 18, Supplementary Fig. 19, Supplementary Fig. 20, Supplementary Fig. 21, Supplementary Fig. 22, Supplementary Fig. 23).
Supplementary Fig. 1.
Native protein Slc11a2.
Supplementary Fig. 2.
Altered protein Slc11a2 (V108I).
Supplementary Fig. 3.
Altered protein Slc11a2 (I114T).
Supplementary Fig. 4.
Altered protein Slc11a2 (V334A).
Supplementary Fig. 5.
Altered protein Slc11a2 (T336K).
Supplementary Fig. 6.
Altered protein Slc11a2 (T343A).
Supplementary Fig. 7.
Altered protein Slc11a2 (G369R).
Supplementary Fig. 8.
Altered protein Slc11a2 (A371S).
Supplementary Fig. 9.
Altered protein Slc11a2 (Y374C).
Supplementary Fig. 10.
Altered protein Slc11a2 (A377V).
Supplementary Fig. 11.
Altered protein Slc11a2 (Q385H).
Supplementary Fig. 12.
Altered protein Slc11a2 (M389V).
Supplementary Fig. 13.
Altered protein Slc11a2 (R465Q).
Supplementary Fig. 14.
Altered protein Slc11a2 (W477L).
Supplementary Fig. 15.
Altered protein Slc11a2 (L484V).
Supplementary Fig. 16.
Altered protein Slc11a2 (S490F).
Supplementary Fig. 17.
Altered protein Slc11a2 (N492S).
Supplementary Fig. 18.
Altered protein Slc11a2 (V497M).
Supplementary Fig. 19.
Altered protein Slc11a2 (D502G).
Supplementary Fig. 20.
Altered protein Slc11a2 (V507A).
Supplementary Fig. 21.
Altered protein Slc11a2 (V510M).
Supplementary Fig. 22.
Altered protein Slc11a2 (A512V).
Supplementary Fig. 23.
Altered protein Slc11a2 (V517I).
3.6. Model quality & structure assessment and RMSD difference
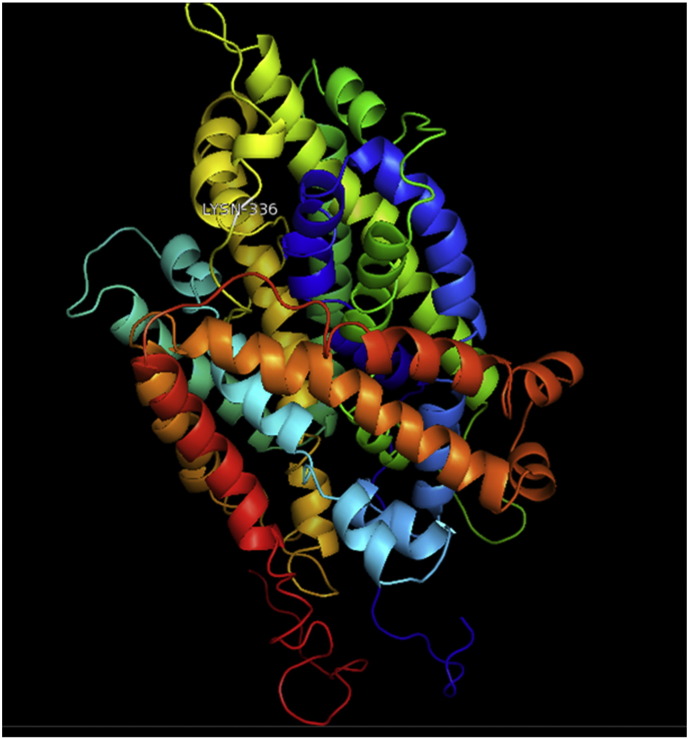
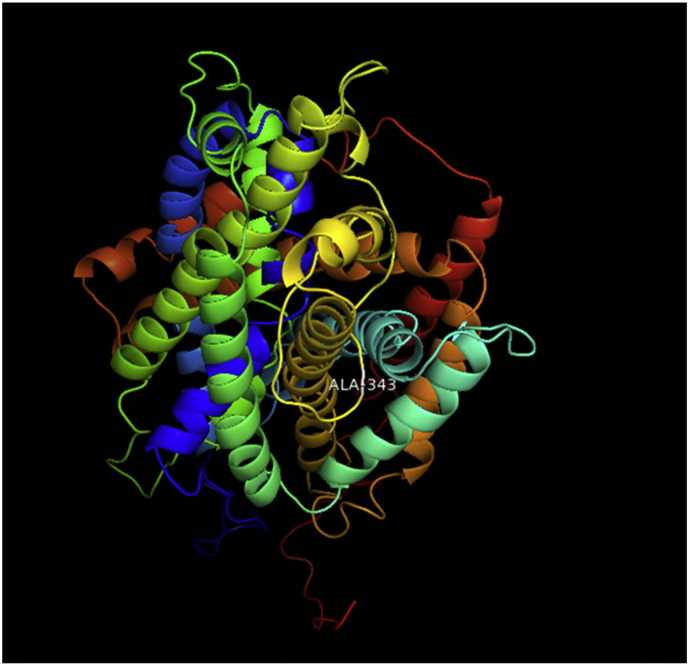
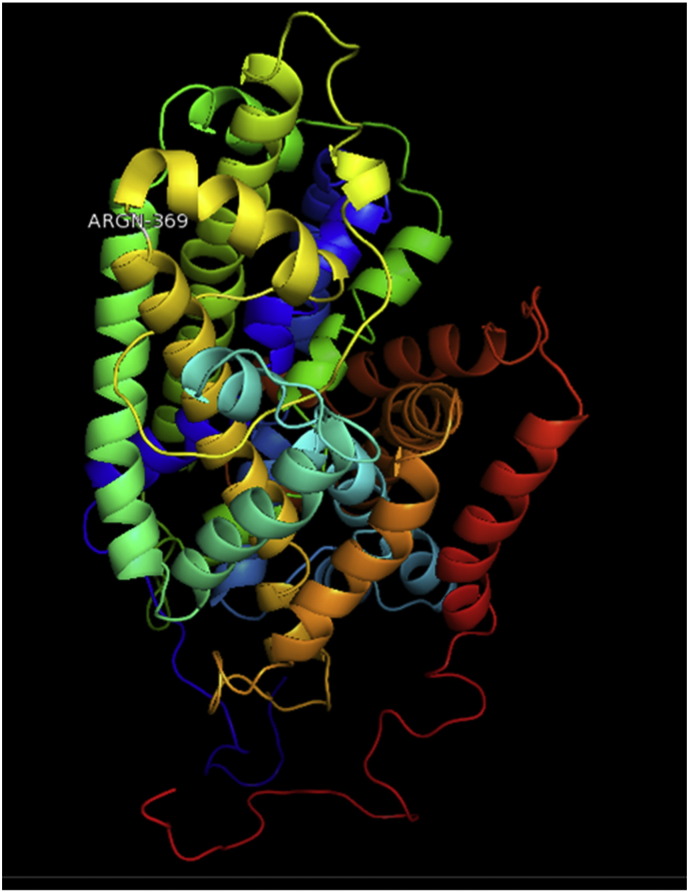
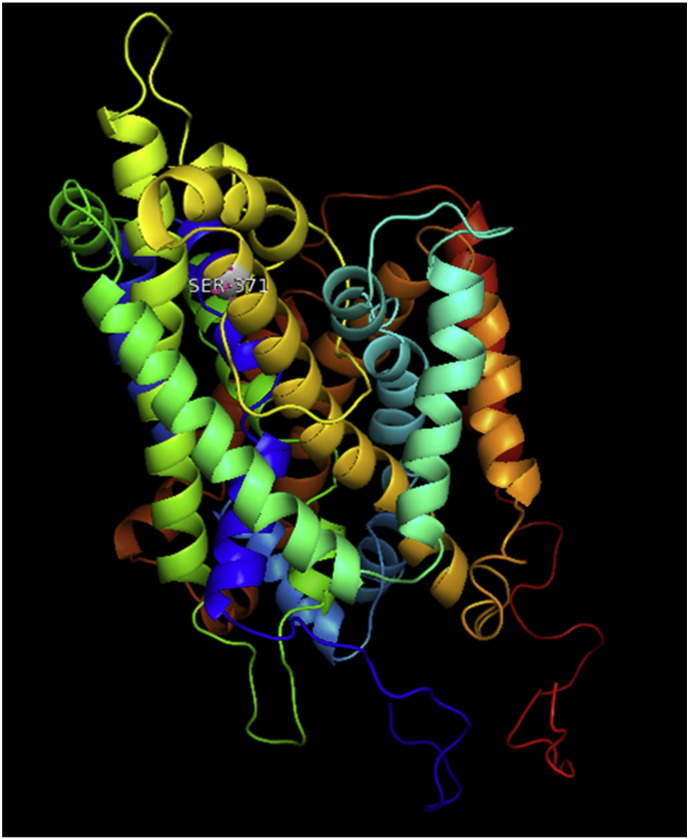
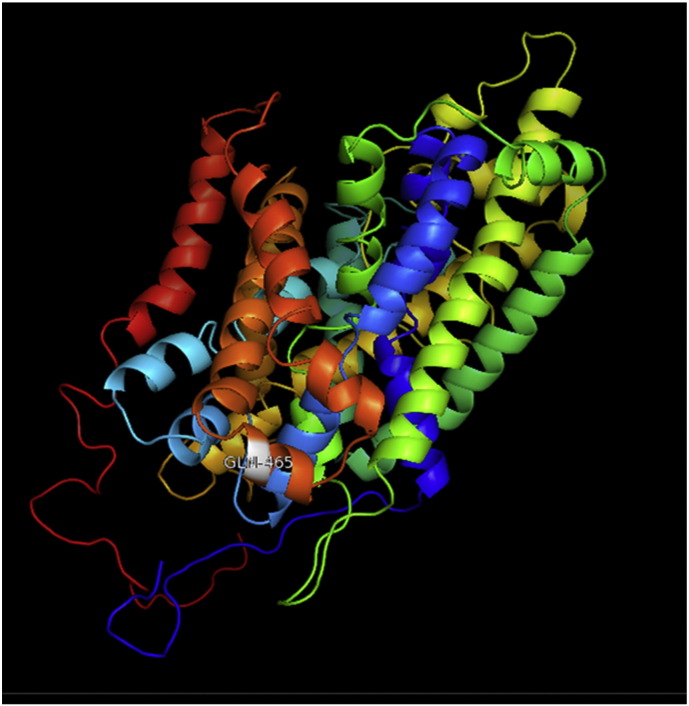
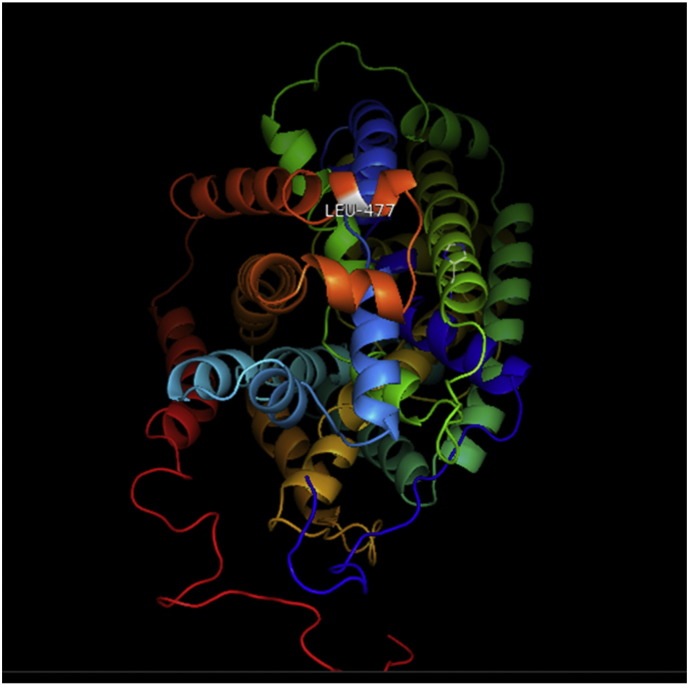
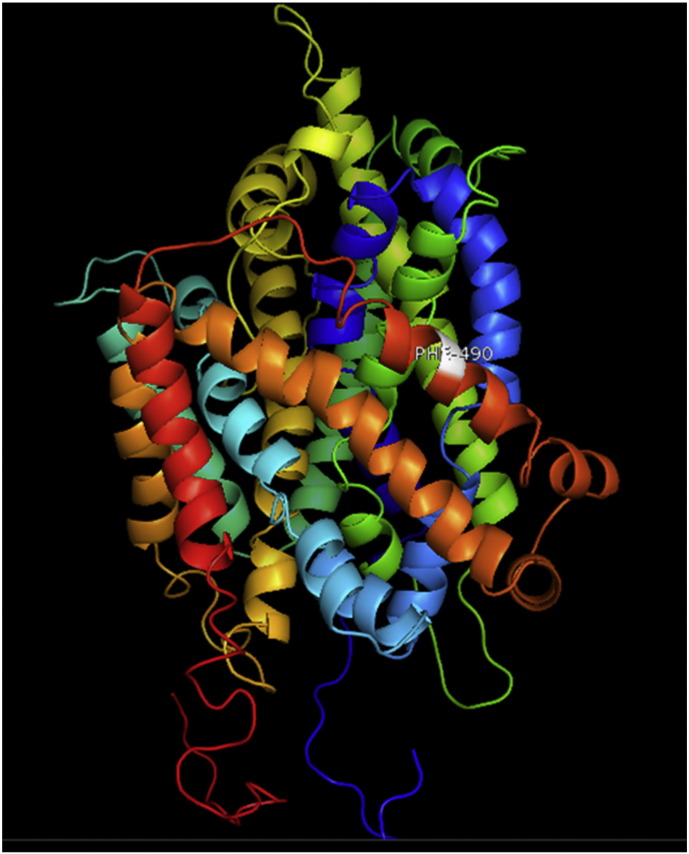
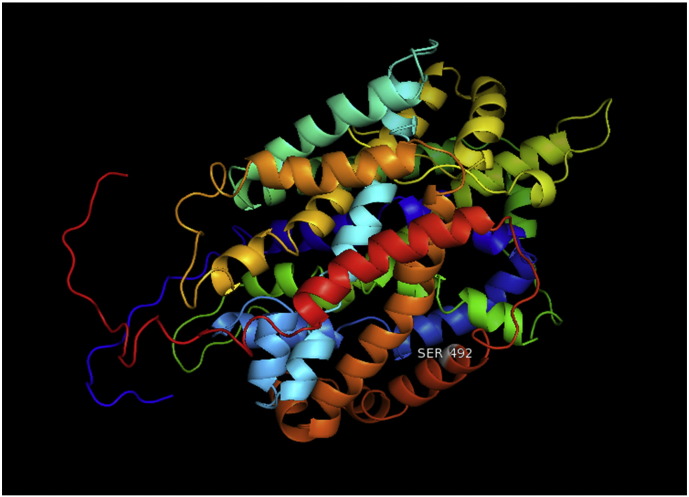
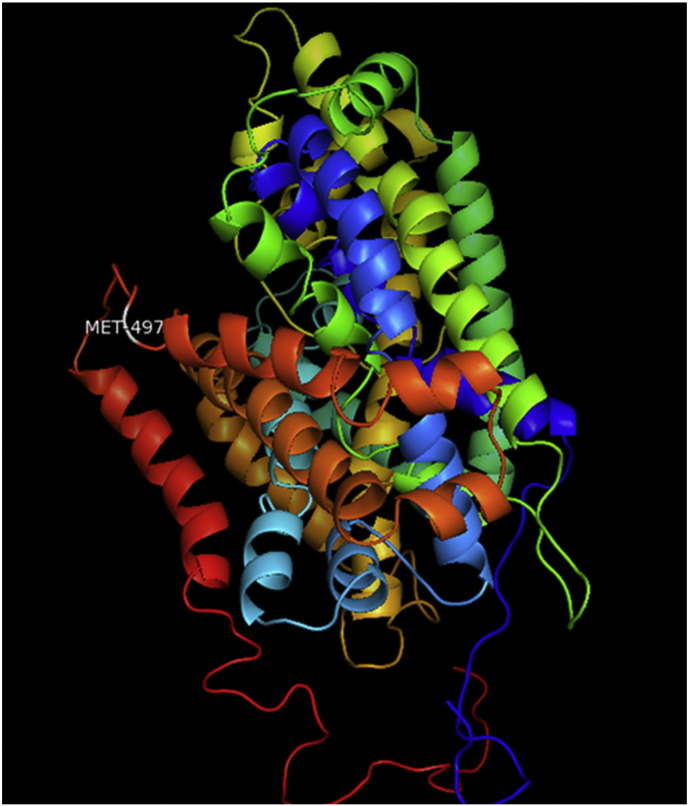
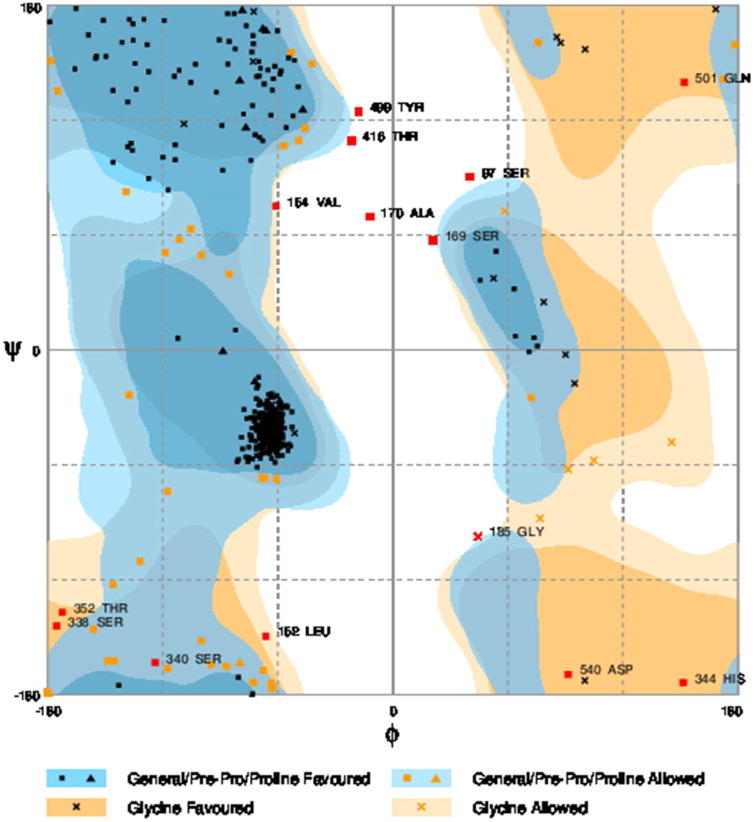
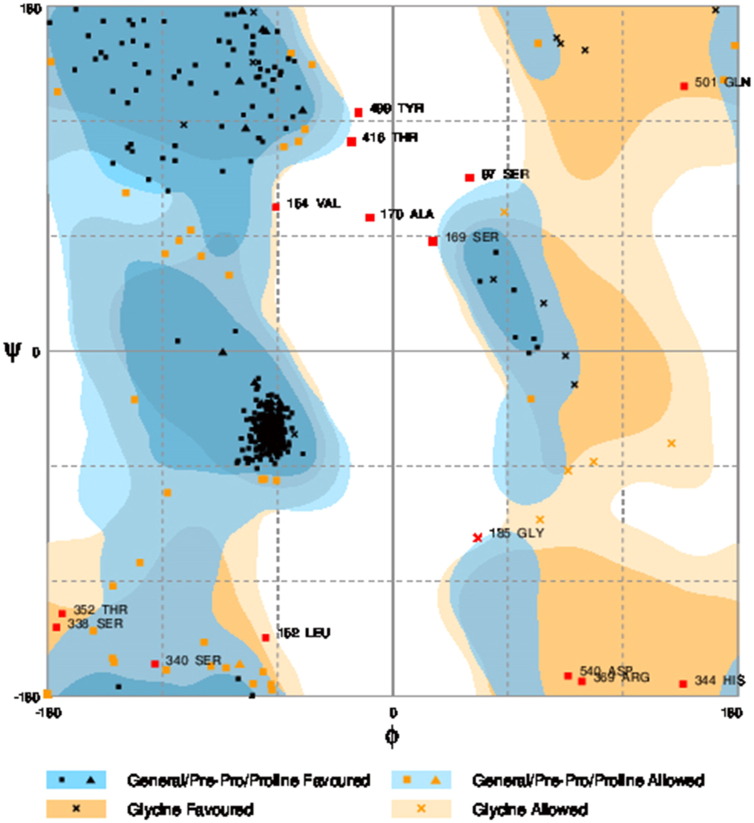
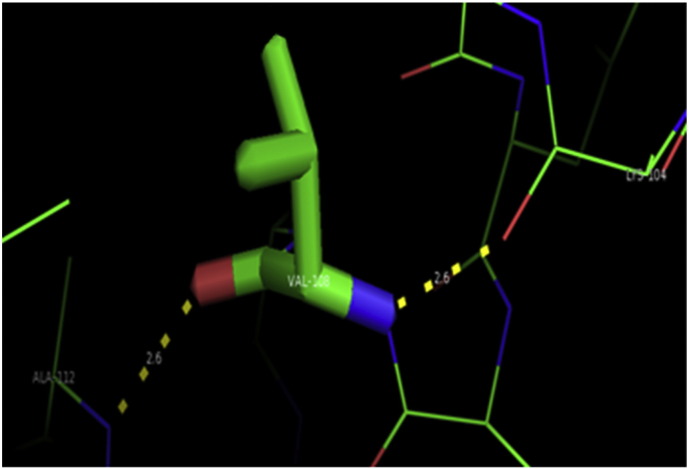
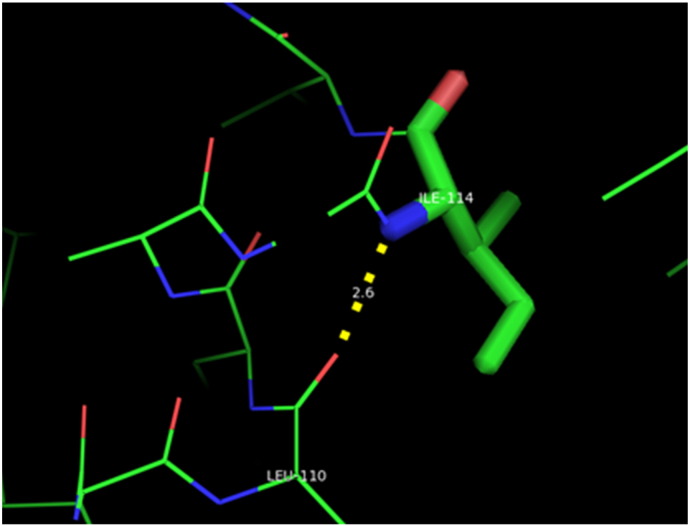
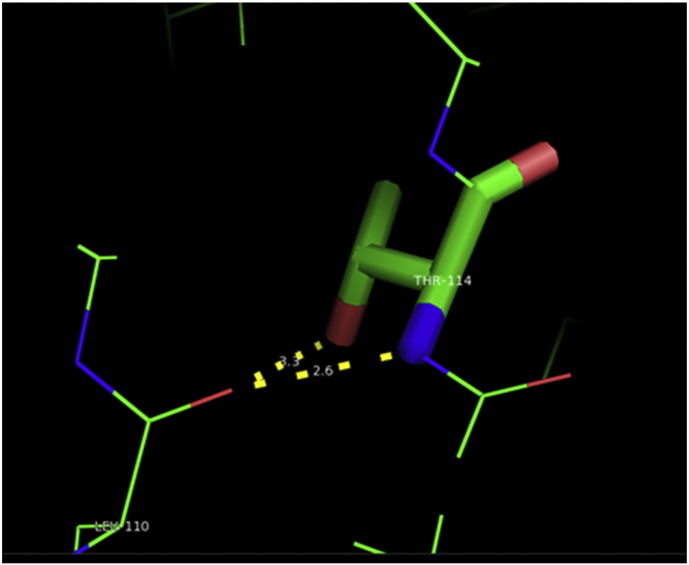
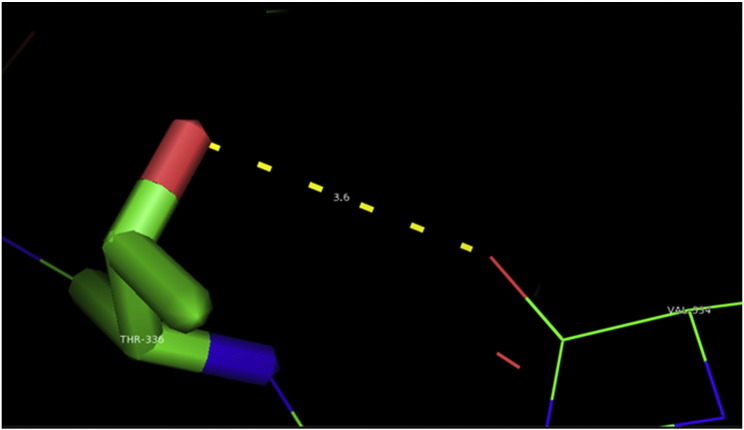
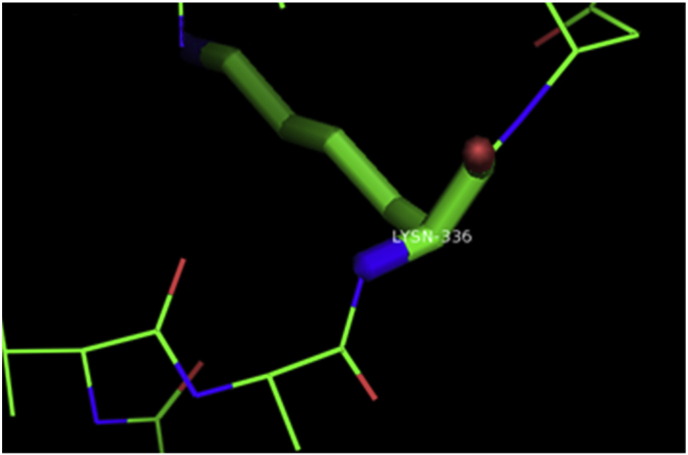
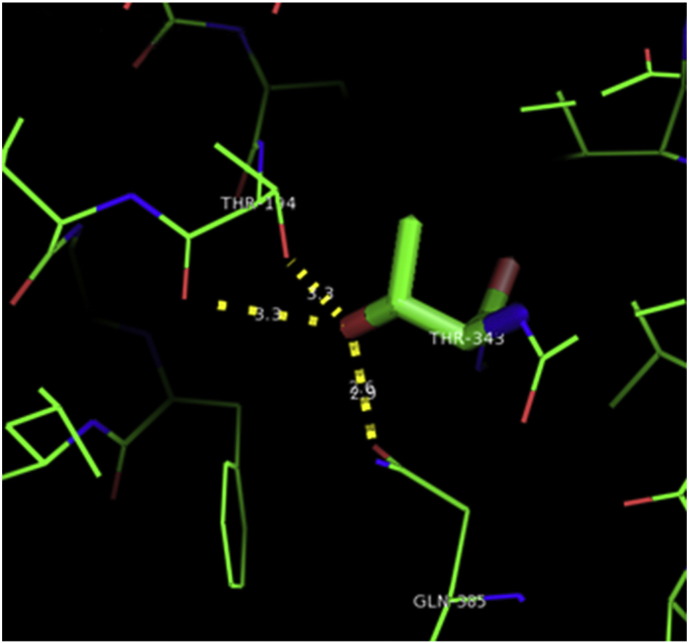
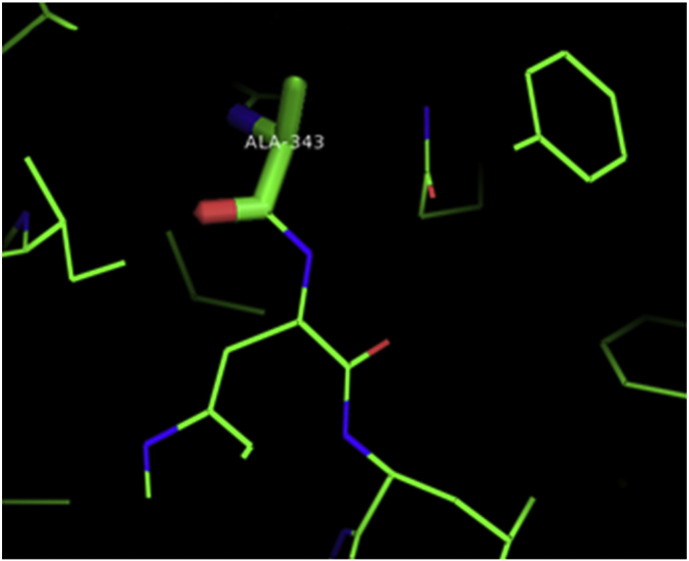
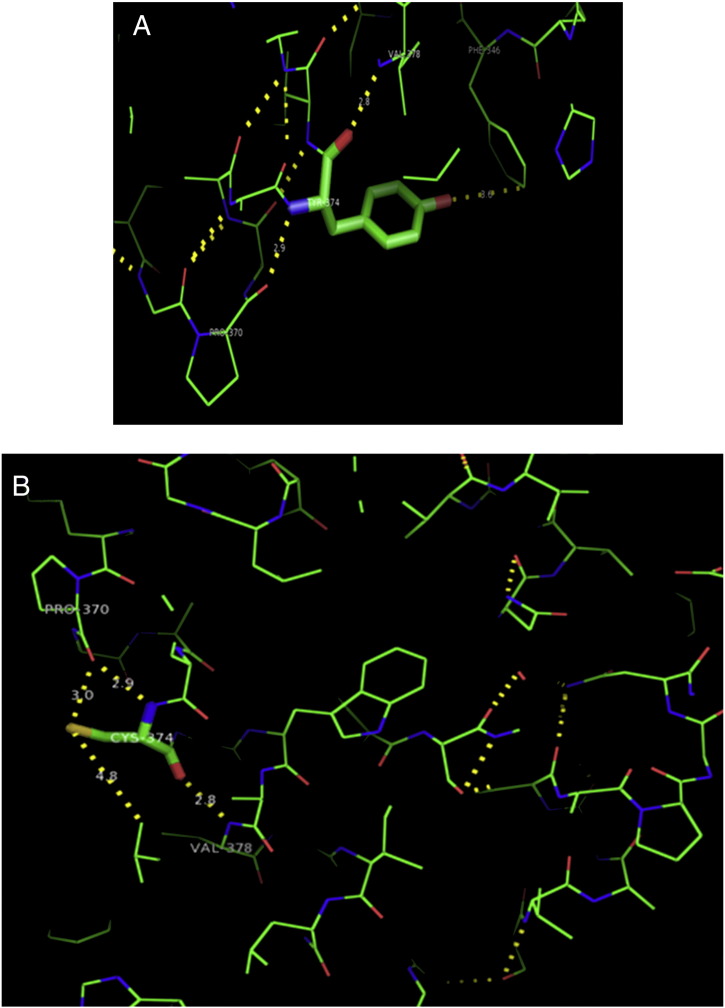
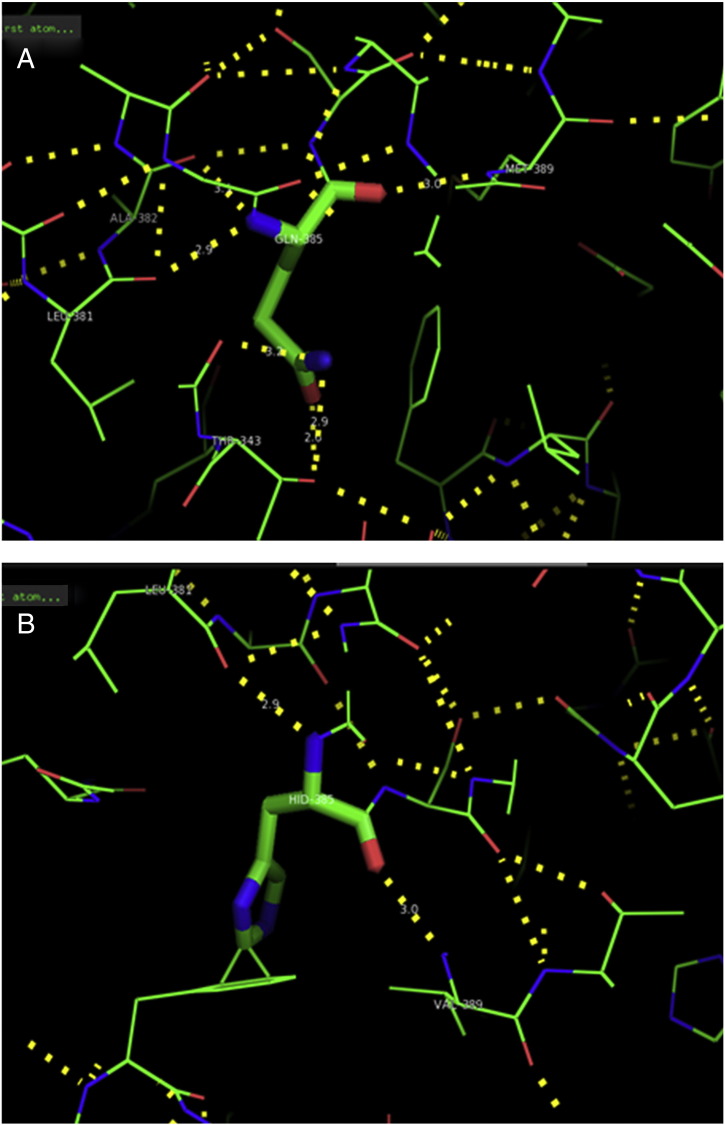
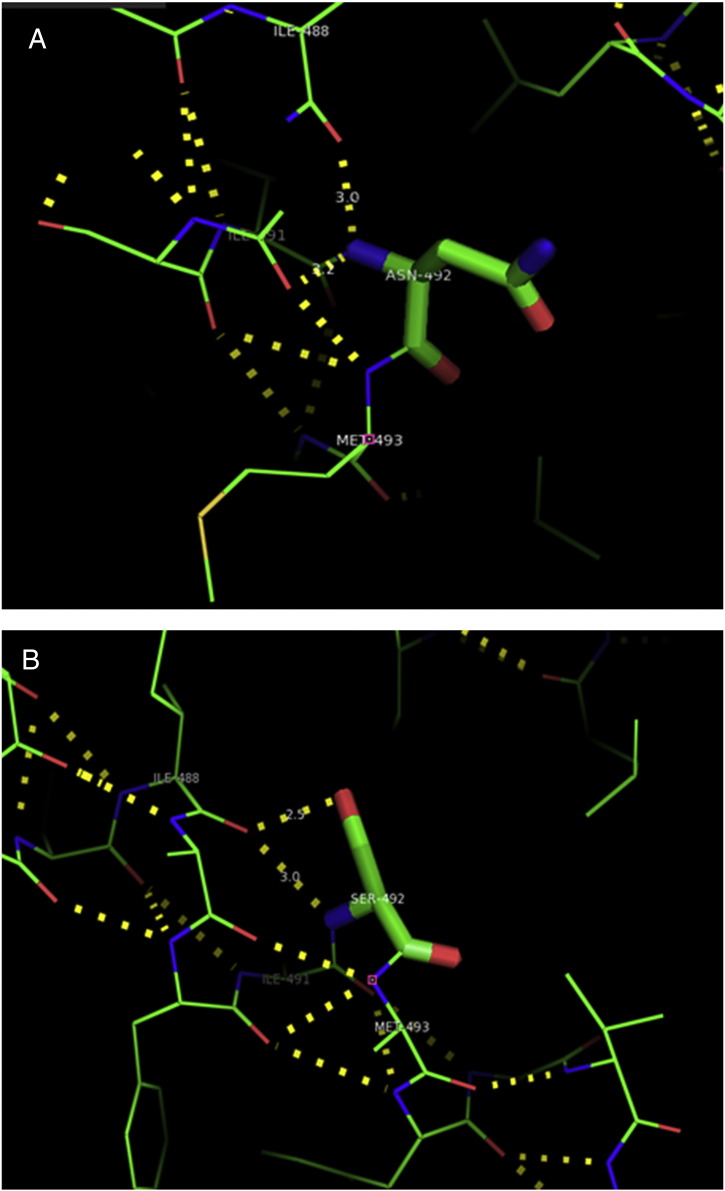
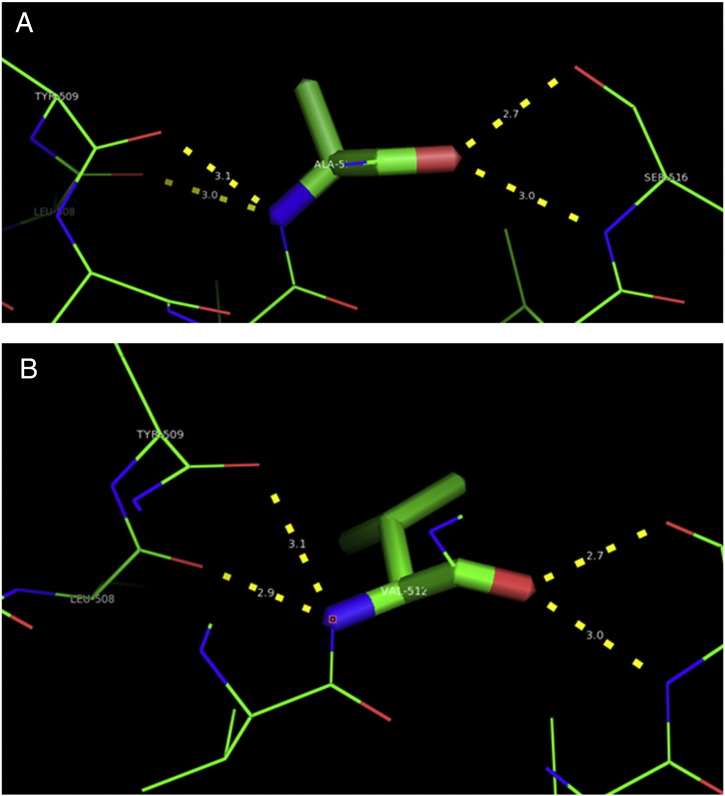
Ramachandran plot of native protein showed 442 residues (88.8%) in favored region, 42 residues (8.4%) in allowed region and 14 residues (2.8%) in outlier region (Supplementary Fig. 24). While in case of 22 altered proteins, in case of 21 nsSNPs, Ramachandran plot showed similar pattern as native protein but for nsSNP G369R, one residue from favored region was shifted to outlier region. So in this case, 441 residues (88.6%) were in favored region and 15 residues (3.0%) were in outlier region (Supplementary Fig. 25). While checking RMSD value, it was observed that there was not much deviation from native protein. The higher the RMSD value, the more the deviation between the two structures which in turn changes their functional activity [23]. V108I, R465Q, W477L, and V517I showed somewhat higher RMSD values of 0.053, 0.057, 0.033, and 0.036 respectively which are given in Table 3. While tested for polar interactions, in case of some nsSNPs, there is a change in interaction patterns compared to native protein. SNPs V108I, I114T, T336K, T343A, Y374C, Q385H, N492S and A512V showed different interaction patterns than native protein's amino acids. In V108I, V108 formed two polar interactions with K104 and A112 while I108 in altered protein formed three interactions. Two were with K104, I108 with altered bond lengths and third extra interaction with L105 (Supplementary Fig. 26 & B). I114 in case of I114T having interaction with L110 with a bond length of 2.6 Å, but T114 had additional interaction to same amino acid with 3.3 Å bond length due to change of R group from non-polar to polar (Supplementary Fig. 27A & B). K336 which is having positively charged R group in altered protein forming 3.6 Å long interaction with V334, while T334 proposed polar R group not forming any interaction (Supplementary Fig. 28. & B). SNP T343A showed alteration of R group from polar to non-polar, which changed interactions (Supplementary Fig. 29. & B). Y374 in native protein forming one polar interaction with P370 and one with V378 of 2.9 Å and 2.8 Å length respectively. While altered amino acid C374 forming two polar interactions with P370 of 2.9 Å, 3.0 Å lengths and with V378 of 2.8 Å, 4.8 Å lengths (Fig. 1 & B). In Q385C, Q385 formed three polar interactions with T343 of 2.8 Å, 2.9 Å, and 3.2 Å lengths, one with L381 of 2.9 Å length, one with A382 of 3.1 Å length and one with M389 of 3.0 Å length but altered residue H385 had interaction with L381 of 2.9 Å length and one with V389 of 3.0 Å length because of the change in R group (Fig. 2 & B). Native residue N492 showed one interaction with I488 of 3.0 Å length and one with I491 of 3.2 Å length and altered residue S492 forming two polar interactions with I488 of 2.5 Å and 3.0 Å lengths (Fig. 3 & B).Similarly, A512V showed the change in bond length (Fig. 4 & B). While verified further for energy change, T336K, Y374C, Q385H, M389V, R465Q, W477L, L484V, S490F, N492S, D502G, V510M, A512V and V517I showed higher total energy after energy minimization than native protein which are given in Table 3.
Supplementary Fig. 24.
Ramachandran plot of native protein.
Number of residues in favored region (~ 98.0% expected):442 (88.8%), number of residues in allowed region (~ 2.0% expected):42 (8.4%), number of residues in outlier region: 14 (2.8%).
Supplementary Fig. 25.
Ramachandran plot of altered protein (G369R).
Number of residues in favored region (~ 98.0% expected): 441 (88.6%), number of residues 489 in allowed region (~ 2.0% expected):42 (8.4%), number of residues in outlier region: 15(3.0%).
Table 3.
RMSD value and total energy after minimization of altered model.
| Amino acid change | RMSD value of altered protein | Total energy after energy minimization (kJ/mol) |
|---|---|---|
| Native protein | – | − 5449.571 |
| V108I | 0.053 | − 12,538.697 |
| I114T | 0.009 | − 5508.284 |
| V334A | 0.002 | − 5551.974 |
| T336K | 0.001 | − 5373.680 |
| T343A | 0.002 | − 5522.909 |
| G369R | 0.001 | − 5682.057 |
| A371S | 0.002 | − 5461.978 |
| Y374C | 0.002 | − 5419.823 |
| A377V | 0.000 | − 5492.021 |
| Q385H | 0.001 | − 5153.123 |
| M389V | 0.002 | − 5107.444 |
| R465Q | 0.057 | − 5345.201 |
| W477L | 0.033 | − 5294.021 |
| L484V | 0.001 | − 5488.854 |
| S490F | 0.001 | − 5314.866 |
| N492S | 0.001 | − 5291.899 |
| V497M | 0.002 | − 5469.048 |
| D502G | 0.001 | − 5381.310 |
| V507A | 0.002 | − 5449.402 |
| V510M | 0.001 | − 2574.750 |
| A512V | 0.001 | − 5448.742 |
Supplementary Fig. 26.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP V108I.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP V108I.
Supplementary Fig. 27.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP I114T.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP I114T.
Supplementary Fig. 28.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP K336T.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP K336T.
Supplementary Fig. 29.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP T343A.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP T343A.
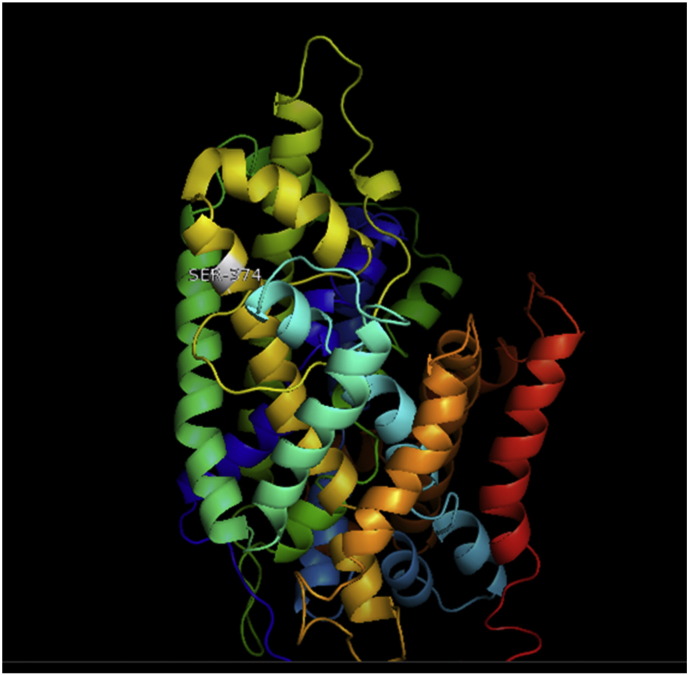
Fig. 1.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP Y374C.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP Y374C.
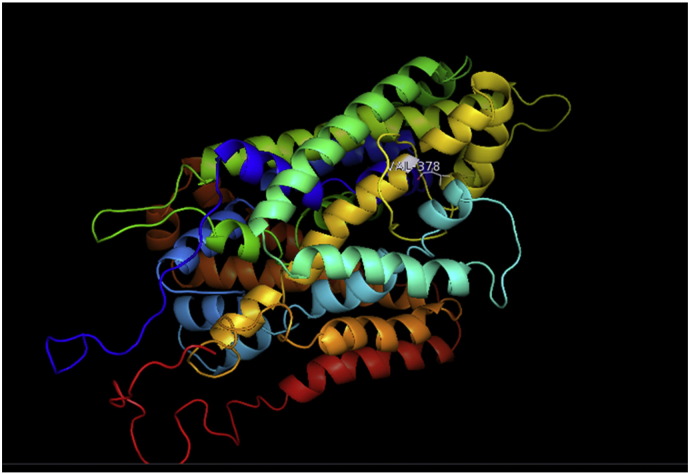
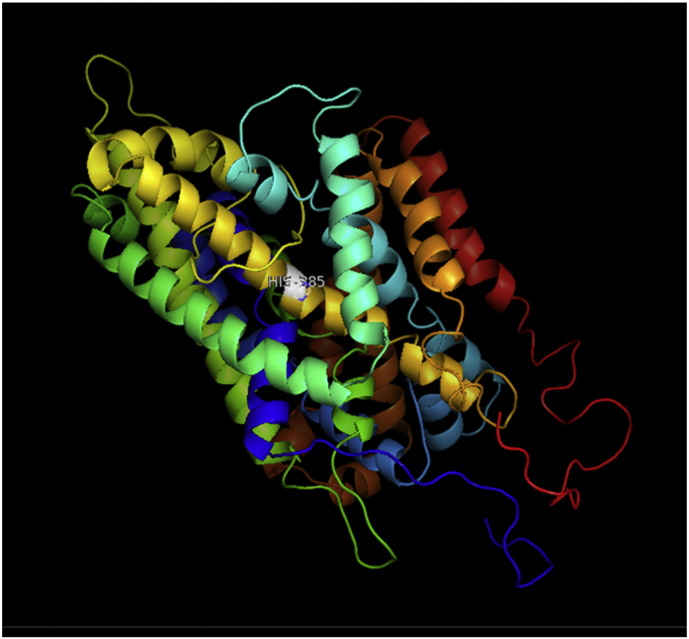
Fig. 2.
A. Interaction of native residue with vicinal residue (yellow dotted line) for SNP Q385H.
B. Interaction of altered residue with vicinal residue (yellow dotted line) for SNP Q385H.
Fig. 3.
A. Interaction of native residue with vicinal residues (yellow dotted line) for SNP N492S.
B. Interaction of altered residue with vicinal residues (yellow dotted line) for SNP N492S.
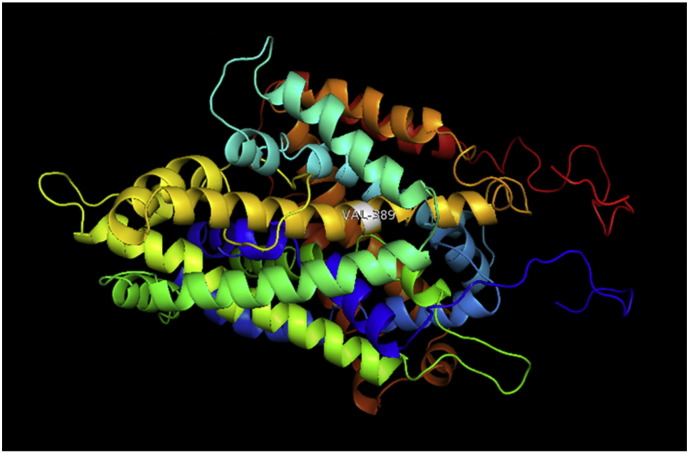
Fig. 4.
A. Interaction of native residue with vicinal residues (yellow dotted line) for SNP A512V.
B. Interaction of altered residue with vicinal residues (yellow dotted line) for SNP A512V.
3.7. Binding site and ligand prediction
Further when analyzed for binding site regions using several tools, results revealed ligands and ligand binding sites which are shown in Supplementary Tables 2 and 3. However, none of above nsSNPs resided in the above identified binding sites.
4. Discussion
In order to investigate structural and functional impact of nsSNPs present in coding region of Slc11a2, we performed extensive computational analysis. Non-synonymous SNPs in coding region can cause amino acid change further altering protein function which may lead to susceptibility to disease. Identification of deleterious nsSNPs from tolerant nsSNPs is ideal for analyzing individual susceptibility to disease. It is not necessary that all variants have a major deleterious functional impact and some may be well tolerated. However, nsSNPs which are linked to diseases or other phenotypes often have some molecular significance [4]. They may modify enzyme activity, destabilize protein structures or disrupt protein interactions.
Nowadays, major concern relating to nsSNP in molecular biology and population genetics is to identify and characterize the nsSNPs that are functionally related from those that are not [24]. To determine the functional effects of nsSNPs in Slc11a2 gene, we employed four widely used in silico tools specifically I-Mutant3, SIFT, PROVEAN and PANTHER. If a marker is found to be associated with the disease and the marker is a nsSNP, prediction tools can provide independent evidence as to whether the nsSNP itself contributes to disease. Because carrying out the appropriate assays may be time-consuming, these tools can filter out nsSNPs that are unlikely to affect protein function before experimentation. The difference in the results of these four prediction tools is due to the difference in features utilized by the methods therefore we would expect the outcomes to occur dissimilar at some point [6]. If the prediction results of all four tools for these identified nsSNPs in this ion transport innate immune gene Slc11a2 would be combined, it would provide high reliability. One of the nsSNP G185R was observed which abrogated iron transport in one of the phenotype of Belgrade rats [25]. This SNP was not observed in this analysis.
To test the effect of these nsSNPs on structural stability of protein, protein modeling proved to be an efficient in silico means using several bioinformatic tools. Change in amino acid can be further modeled and this altered modeled protein structure can be utilized during in silico approach to confirm the effect of particular nsSNP on stability of protein before validating in vitro. However here, nsSNPs are not falling in the epitope region according to the results of FT site and RaptorX Binding which identified potential binding sites in the protein structure. But change in amino acid affects polar–polar interactions within the protein molecule itself which further altered energy of stabilization and further destabilized the protein [26]. Here, as observed, in some amino acid changes, number of polar interactions changed which ultimately affected total energy of protein indicating decrease in protein stability. These imperative results indicate that identified nsSNPs in this protein might alter its stability and might affect the protein–protein interaction and metal binding sites.
By comparing the results of above 4 methods and total energy, we can conclude that nsSNPs viz. Y374C, Q385H and N492S should be further confirmed for their association with disordered Slc11a2 function in addition to existing nsSNPs of this gene. However, RMSD values were not that much higher and these nsSNPs were not residing in the metal binding site regions, suggesting that these nsSNPs might not be too strong candidate for disease association of this gene.
5. Conclusion
Nowadays, the next generation sequencing techniques are generating high throughput of data related to SNPs, but the evaluation of biologically functional SNPs using this in vitro studies is quite tedious, time consuming and economically less significant. On the other way, in silico approach can help us to predict the consequences of mutations and explain their affecting role in biological mechanisms. Out of 22, 8(36.36%) nsSNPs were revealed to be deleterious using SIFT. Similarly PROVEAN identified 8 nsSNPs deleterious. Additionally, I-Mutant3 predicted all 22 substitutions which affected the stability of protein. From the above 7 nsSNPs, PANTHER predicted 5 (22.72%) as damaging. G369R, Y374C, A377V, Q385H and N492S were predicted deleterious using abovementioned tools. Also, these nsSNPs were observed for altered interaction patterns and verified by calculating total energy change after energy minimization which confirmed Y374C, Q385H and N492S as damaging.
The following are the supplementary data related to this article.
Interaction of residues.
Supplementary tables
Conflict of interest
The authors have no conflict of interest.
Acknowledgment
The authors are thankful to the Department of Biotechnology, Government of India, New Delhi (grant no. BT/PR3111/AAQ/1/474/2011) for providing financial support.
References
- 1.Yates C.M., Sternberg M.J. The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J. Mol. Biol. 2013;425:3949–3963. doi: 10.1016/j.jmb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Rajasekaran R., Sudandiradoss C., Doss C.G., Sethumadhavan R. Identification and in silico analysis of functional SNPs of the BRCA1 gene. Genomics. 2007;90:447–452. doi: 10.1016/j.ygeno.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Gfeller D., Ernst A., Jarvik N., Sidhu S.S., Bader G.D. Prediction and experimental characterization of nsSNPs altering human PDZ-binding motifs. PloS ONE. 2014;9:e94507. doi: 10.1371/journal.pone.0094507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson M.M., Houck J., Chen C. Vol. 14. 2005. Environmental aspects selection for EEE using ANP method; pp. 1326–1329. (Screening for Deleterious Nonsynonymous Single-nucleotide Polymorphisms in Genes Involved in Steroid Hormone Metabolism and Response). [DOI] [PubMed] [Google Scholar]
- 5.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R. PANTHER: a library of protein families and subfamilies indexed by function. Genom. Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Alencar S.A., Lopes J.C. A comprehensive in silico analysis of the functional and structural impact of SNPs in the IGF1R genel. J. Biomed. Biotechnol. 2010;(2010):715139. doi: 10.1155/2010/715139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George Priya Doss C., Rajith B. Computational refinement of functional single nucleotide polymorphisms associated with ATM gene. PloS ONE. 2012;7:e34573. doi: 10.1371/journal.pone.0034573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanazi M., Abduljaleel Z., Khan W., Warsy A.S., Elrobh M., Khan Z. In silico analysis of single nucleotide polymorphism (SNPs) in human beta-globin gene. PloS one. 2011;6:e25876. doi: 10.1371/journal.pone.0025876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George Priya Doss C., Rajasekaran R., Sudandiradoss C., Ramanathan K., Purohit R., Sethumadhavan R. A novel computational and structural analysis of nsSNPs in CFTR gene. Genom. Med. 2008;2:23–32. doi: 10.1007/s11568-008-9019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain M.R., Shaik N.A., Al-Aama J.Y., Asfour H.Z., Khan F.S., Masoodi T.A. In silico analysis of Single Nucleotide Polymorphisms (SNPs) in human BRAF gene. Genetics. 2012;508:188–196. doi: 10.1016/j.gene.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Kamaraj B., Purohit R. In silico screening and molecular dynamics simulation of disease-associated nsSNP in TYRP1 gene and its structural consequences in OCA3. BioMed Res. Int. 2013;2013:697,051. doi: 10.1155/2013/697051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamoud H.S., Hussain M.R., El-Harouni A.A., Shaik N.A., Qasmi Z.U., Merican A.F. First comprehensive in silico analysis of the functional and structural consequences of SNPs in human GalNAc-T1 gene. Comput. Math. Methods Med. 2014;2014:904,052. doi: 10.1155/2014/904052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czachorowski M., Lam-Yuk-Tseung S., Cellier M., Gros P. Transmembrane topology of the mammalian Slc11a2 iron transporter. Biochemistry. 2009;48:8422–8434. doi: 10.1021/bi900606y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper C.A., Shayeghi M., Techau M.E., Capdevila D.M., MacKenzie S., Durrant C. Analysis of the rainbow trout solute carrier 11 family reveals iron import ≤ pH 7.4 and a functional isoform lacking transmembrane domains 11 and 12. FEBS lett. 2007;581:2599–2604. doi: 10.1016/j.febslet.2007.04.081. [DOI] [PubMed] [Google Scholar]
- 15.Pinner E., Gruenheid S., Raymond M., Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J. Biol. Chem. 1997;272:28,933–28,938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- 16.Mackenzie B., Ujwal M.L., Chang M.H., Romero M.F., Hediger M.A. Divalent metal-ion transporter DMT1mediates both H+ coupled Fe2 + and uncoupled fluxes. PflügersArchiv - Eur. J. Physiol. 2006;451:544–558. doi: 10.1007/s00424-005-1494-3. [DOI] [PubMed] [Google Scholar]
- 17.Gunshin H., Mackenzie B., Berger U.V., Gunshin Y., Romero M.F., Boron W.F. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 18.Lee P.L., Gelbart T., West C., Halloran C., Beutler E. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol. Dis. 1998;24:199–215. doi: 10.1006/bcmd.1998.0186. ss. [DOI] [PubMed] [Google Scholar]
- 19.Gruenheid S., Canonne-Hergaux F., Gauthier S., Hackam D.J., Grinstein S., Gros P. The iron transport protein NRAMP2 is an integral membrane glycoprotein that colocalizes with transferrin in recycling endosomes. J. Exp. Med. 1999;189 doi: 10.1084/jem.189.5.831. 831-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauline C.N., Steven H. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PloS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acid Res. 2005;33:306–310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reva B.A., Finkelstein A.V., Skolnick J. What is the probability of a chance prediction of a protein structure with an RMSD of 6 Å? Fold. Des. 1998;3:141–147. doi: 10.1016/s1359-0278(98)00019-4. [DOI] [PubMed] [Google Scholar]
- 24.George Priya Doss C., Nagasundaram N., Chakraborty C., Chen L., Zhu H. Extrapolating the effect of deleterious nsSNPs in the binding adaptability of flavopiridol with CDK7 protein: a molecular dynamics approach. Hum. Genom. 2013;7:10. doi: 10.1186/1479-7364-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming M., Romando M., Su M., Garrick L., Garrick M., Andrews N. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. PNAS. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Y., Zhaolong L., John M. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interaction of residues.
Supplementary tables